Abstract
Alginate biosynthesis by Pseudomonas aeruginosa was shown to be regulated by the intracellular second messenger bis-(3′-5′)-cyclic-dimeric-GMP (c-di-GMP), and binding of c-di-GMP to the membrane protein Alg44 was required for alginate production. In this study, PA1727, a c-di-GMP-synthesizing enzyme was functionally analyzed and identified to be involved in regulation of alginate production. Deletion of the PA1727 gene in the mucoid alginate-overproducing P. aeruginosa strain PDO300 resulted in a nonmucoid phenotype and an about 38-fold decrease in alginate production; thus, this gene is designated mucR. The mucoid alginate-overproducing phenotype was restored by introducing the mucR gene into the isogenic ΔmucR mutant. Moreover, transfer of the MucR-encoding plasmid into strain PDO300 led to an about sevenfold increase in alginate production, wrinkly colony morphology, increased pellicle formation, auto-aggregation, and the formation of highly structured biofilms as well as the inhibition of swarming motility. Outer membrane protein profile analysis showed that overproduction of MucR mediates a strong reduction in the copy number of FliC (flagellin), required for flagellum-mediated motility. Translational reporter enzyme fusions with LacZ and PhoA suggested that MucR is located in the cytoplasmic membrane with a cytosolic C terminus. Deletion of the proposed C-terminal GGDEF domain abolished MucR function. MucR was purified and identified using tryptic peptide fingerprinting and matrix-assisted laser desorption ionization-time of flight mass spectrometry. Overall, experimental evidence was provided suggesting that MucR specifically regulates alginate biosynthesis by activation of alginate production through generation of a localized c-di-GMP pool in the vicinity of Alg44.
Pseudomonas aeruginosa is a ubiquitous opportunistic pathogen which is responsible for systemic infection of immunocompromised patients and severe chronic infections of the lungs of cystic fibrosis (CF) patients (9, 10). Establishment of a chronic CF lung infection coincides with production of copious amounts of the exopolysaccharide alginate leading to the formation of persistent biofilms which prevent the diffusion of antibiotics and protect cells from the host immune response (9, 30). Early after the onset of infection, P. aeruginosa switches to a genetically stable alginate-overproducing mucoid variant capable of forming persistent biofilms (32, 33, 43). Alginates are linear unbranched exopolysaccharides consisting of β-1,4-linked monomers of β-d-mannuronic acid and its C5-epimer α-l-guluronic acid (44, 46). The switch to mucoidity and the complex transcriptional regulation of alginate biosynthesis have extensively been investigated (4).
Recently it has become apparent that an additional posttranscriptional level of regulation is playing a role in alginate biosynthesis in P. aeruginosa. The membrane-anchored alginate biosynthesis protein Alg44, which is essential for alginate production, contains a bis-(3′-5′)-cyclic-dimeric-GMP (c-di-GMP) binding/sensing PilZ domain in its C terminus (45). This PilZ domain was demonstrated to bind c-di-GMP and was essential for alginate biosynthesis (1, 34).
The secondary messenger c-di-GMP is a central regulator of bacterial physiology and has been linked to diverse physiological responses, such as exopolysaccharide production, motility, biofilm formation, and the production of adhesive surface organelles (48). P. aeruginosa PAO1 contains at least 38 genes encoding proteins shown to be involved in the production (diguanylate cyclases [DGC]) and/or the breakdown (phosphodiesterases [PDE]) of c-di-GMP (26). Sequence analysis of these enzymes indicated the presence of domains involved in signal transduction, which suggested that these proteins function in response to diverse signals. It was also suggested that localized c-di-GMP pools might exist inside the cell, enabling spatially resolved regulation of physiological responses (26). At present it is unclear how or if any of these different proteins influence alginate biosynthesis. Since the c-di-GMP-binding Alg44 protein is localized in the cytoplasmic membrane, presumably as a subunit of the alginate polymerization/secretion multiprotein complex, membrane-anchored DGCs could provide localized pools of c-di-GMP for activation of alginate production. Putative membrane-anchored PA1727 (MucR) comprises a DGC (GGDEF) as well as a PDE (EAL) and a conserved integral membrane-sensing MHYT domain, which was proposed to sense oxygen, CO, or NO (16). DGC activity but not PDE activity has previously been demonstrated (26). Oxygen and NO have previously been shown to influence alginate production (6, 15) In this study, MucR was assessed with respect to its involvement in regulation of alginate production.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. All Escherichia coli strains were grown in LB medium at 37°C, and E. coli S17-1 (54) was used for conjugative transfer of mob site-containing pBBR1MCS-5 (23) derivatives. When required, antibiotics were added to the media at the following concentrations: ampicillin, 100 μg/ml; and gentamicin, 10 μg/ml. P. aeruginosa was cultivated in LB or Pseudomonas isolation agar (PIA) medium at 37°C, and, if required, gentamicin was added at a concentration of 100 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description or sequence | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Prototrophic wild-type strain; Alg− | 21 |
| PDO300 | mucA22 isogenic mutant derived from PAO1 | 33 |
| PAO1ΔPA1727 | Isogenic mucR (PA1727) deletion mutant derived from PAO1 | This study |
| PDO300ΔPA1727 | Isogenic mucR (PA1727) deletion mutant derived from PDO300 | This study |
| PDO300Δalg8 | Isogenic alg8 deletion mutant derived from PDO300; cannot produce alginate | 47 |
| PAO1ΔpslA | Isogenic pslA deletion mutant derived from PDO300; does not produce the Psl polysaccharide | 40 |
| E. coli | ||
| TOP10 | E. coli cloning strain | Invitrogen |
| S17-1 | thi-1 proA hsdR17 (rK− mK+) recA1; tra gene of plasmid RP4 integrated in chromosome | 54 |
| Plasmids | ||
| pBBR1MCS-5 | Gmr; broad-host-range vector; Plac | 23 |
| pBBR1MCS-5:mucR | KpnI-ClaI fragment comprising mucR inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:mucRhis | Translational MucR-hexahistidine tag fusion, inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:mucRlacZ | Translational MucR-LacZ fusion, inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:mucRphoA | Translational MucR-PhoA fusion, inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:mucR(277)lacZ | Truncated translational MucR-LacZ fusion, inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:mucR(277)phoA | Truncated translational MucR-PhoA fusion, inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:wspR | XbaI-SacI fragment comprising wspR inserted into vector pBBR1MCS-5 | This study |
| pBBR1MCS-5:rocR | XbaI-SacI fragment comprising rocR inserted into vector pBBR1MCS-5 | This study |
| pEX100T | Apr Cbr, gene replacement vector containing sacB gene for counterselection | 20 |
| pEX100TΔmucRGm | Apr Cbr Gmr; vector pEX100T with SmaI-inserted mucR deletion construct | This study |
| pPS865 | Apr Gmr; source of 1,100-bp BamHI fragment comprising aacC1 gene flanked by Flp recombinase target site signal sequences | 20 |
| pPFLP2 | Apr Cbr; broad-host-range vector encoding Flp recombinase | 20 |
| pPHO7 | Apr; phoA without signal sequence | 17 |
| pJE608 | LacZ lacking the first eight amino acids with promoter Ptac in pMMB67EH | 12 |
| Primers | ||
| PA1727C1-Bbr | TATACCACGTGCTGGCGAGCAACTGCTCGGGCATCGGCCTG | |
| PA1727C2-Ba | TCAATGGATCCGCAGATCGGCGAGAGGGTGCTCGACGAAGC | |
| PA1727N1-Ba | AAGTGGGATCCGATCACCGCGATCAGGATGGAGAGG | |
| PA1727N2-Bbr | CTGCTCACGTGCAGGTTCTTGTCGCCTTTTCCCTGATTGTGG | |
| PA1727C(Cla) | GAGTAATCGATAAATCAGGCGACGATGGCGAGCAACTGCTCG | |
| PA1727N (KpSDNd) | AGCAAGGTACCAGGAGACGCTCATATGCTTATCAGCAGCTACACCCAGGTTATT | |
| PA1727C-tga(BamHI-ClaI) | GAGTAATCGATAAATCAAGGATCCGCGGCGACGCTGGCGAGCAACTGCTGCGCCGGCATCGG | |
| PA1727(277) Fus(BamHI-ClaI) | GAGTAATCGATAAATCAAGGATCCGCGAGGAGCATGCGGTTGGGCAGCTTGGTCAGGTTGTCGTGCAGG | |
| PA3702N(XbSDNd) | GCGTCGTCTAGAAGGAGAGAGACATATGCACAACCCTCATGAGAGCAAGACCGACC | |
| PA3702C(SacI) | GGCTGGAGCTCAAATCAGCCCGCCGGGGCCGGCGGCACC | |
| PA3947N(XbSDNd) | GCGTCGTCTAGAAGGAGGGACCCATATGAATGATTTGAATGTTCTGGTGTTGGAGG | |
| PA3947C(SacI) | GGCTGGAGCTCAAATCAGGATCCGGAGCAATAGTCGAGAAAGTGC | |
| LumioHis (BaXbdirect) | GATCCATGTTGTCCTGGCTGTTGCGGTGGCGGCACCGGTCATCATCACCATCACCATTGAT | |
| LumioHis (BaXbcomplement) | CTAGATCAATGGTGATGGTGATGATGACCGGTGCCGCCACCGCAACAGCCAGGACAACATG |
Isolation, analysis, and manipulation of DNA.
General cloning procedures were performed as described previously (51). Deoxynucleoside triphosphate, Taq, and Platinum Pfx polymerases were purchased from Invitrogen. DNA sequences of new plasmid constructs were confirmed by DNA sequencing according to the chain termination method using the model ABI310 automatic sequencer.
Construction of mucR deletion mutant.
Two regions of the mucR gene were amplified by using Taq polymerase with primers PA17271N2-Bbr, PA17271N1-Ba, PA17272C2-Ba, and PA17272C1-Bbr. Region PA1727N (432 bp) comprised bases 21 to 452, and region PA1727C (555 bp) comprised bases 1493 to 2047, relative to the designated mucR coding region (55). Both PCR products were hydrolyzed with BamHI, ligated together, and inserted into vector pGEM-TEasy (Promega). Vector pPS856 (20) was hydrolyzed with BamHI, releasing an about 1,100-bp fragment containing the aacC1 gene (encoding gentamicin acetyltransferase) flanked by two Flp recombinase target sites. The 1,100-bp BamHI fragment (aacC1 gene) was inserted into the BamHI site of plasmid pGEM-TEasy:ΔmucRNC, resulting in plasmid pGEM-TEasy:ΔmucRGm. The ΔmucRGm-comprising DNA fragment was excised using restriction endonuclease BbrP1, and the corresponding 2,087-bp fragment was inserted into the SmaI site of vector pEX100T (20, 53), resulting in plasmid pEX100TΔmucRGm. E. coli S17-1 was used as a donor for the transfer of plasmid pEX100TΔmucRGm into P. aeruginosa strains, and transconjugants were selected on mineral salt medium (52) containing 300 μg of gentamicin/ml and 5% (wt/vol) sucrose. Cells growing on this selective medium should have emerged from double-crossover events. Gene replacement was confirmed after the subculture of cells on PIA medium containing 300 μg of gentamicin/ml and using PCR with primers PA1727up and PA1727down. E. coli S17-1 was used to transfer the Flp recombinase-encoding vector pFLP2 (20) into P. aeruginosa ΔmucRGm strains, and after 24 h of cultivation on PIA medium containing 5% (wt/vol) sucrose, gentamicin- and carbenicillin-sensitive cells were analyzed by PCR for loss of the gentamicin resistance cassette.
Complementation of the ΔmucR mutant.
The mucR gene of P. aeruginosa PAO1 was amplified by PCR using the primers PA1727N(KpSDNd) and PA1727C(Cla). The PCR product was hydrolyzed with KpnI and ClaI and was inserted into the KpnI and ClaI sites of the broad-host-range vector pBBR1MCS-5, resulting in plasmid pBBR1MCS-5:mucR. In addition a C-terminally hexahistidine-tagged MucR-encoding plasmid was constructed by using primers PA1727N(KpSDNd) and PA1727C-tga(BamHI-ClaI), and the resulting PCR product was inserted into pBBR1MCS-5 as described above. The resulting plasmid was then hydrolyzed with BamHI and XbaI; a hexahistidine-encoding sequence was ligated into this using the complementary primer dimer pair LumioHis(BaXbdirect) and LumioHis(BaXbcomplement), resulting in plasmid pBBR1MCS-5:mucRHis. In addition two plasmids carrying two other genes known to influence c-di-GMP levels were constructed. The genes wspR (PA3702) (encoding a highly active DGC) and PA3947 (encoding a highly active PDE) were amplified from the P. aeruginosa PAO1 genome using the 5′-end primers PA3702N(XbSDNd) and PA3947N(XbSDNd) and the 3′-end primers PA3702C(SacI) and PA3947C(SacI), respectively. These PCR products were then hydrolyzed with XbaI and SacI and inserted into the respective sites of pBBR1MCS-5, resulting in plasmids pBBR1MCS-5:PA3702 and pBBR1MCS-5:PA3947, respectively.
Construction of plasmids encoding reporter enzyme fusion proteins.
For the generation of a plasmid encoding full-length MucR fused either to β-galactosidase (LacZ) or alkaline phosphatase (PhoA), the 3′ end of mucR (mucR minus the stop codon) was amplified by PCR using the 5′-end primer PA1727N(KpSDNd) and the 3′-end primer PA1727C-tga(BamHI-ClaI). The PCR product was hydrolyzed with KpnI and BamHI and was inserted into the respective sites of broad-host-range vector pBBR1MCS-5, resulting in plasmid pBBR1MCS-5:mucR(Δstop). XbaI-BamHI fragments of vectors pPHO7 (12) and pJE608 (17) were inserted into XbaI/BamHI-hydrolyzed pBBR1MCS-5:mucR(Δstop) to construct translational LacZ (pBBR1MCS-5:mucRLacZ) and PhoA (pBBR1MCS-5:mucRPhoA) fusions, respectively. In addition, a translational fusion was constructed that encodes the first 277 amino acids of MucR fused to either LacZ or PhoA. The 3′ region was amplified by using the 5′-end primer PA1727N(KpSDNd) and the 3′-end primer PA1727277(BamHI-ClaI). Fusion protein-encoding plasmids pBBR1MCS-5:mucR277LacZ and pBBR1MCS-5:mucR277PhoA were constructed as described above.
Subcellular localization of MucR.
Strains of P. aeruginosa were grown overnight in LB medium containing the appropriate antibiotics. The cells were harvested by centrifugation (1 h at 5,000 × g) and washed with 1 volume of 10 mM HEPES (pH 7.4). Cells were placed in 15 ml 10 mM HEPES with Roche Complete Mini EDTA-free protease inhibitor, sonicated on ice for 12 cycles of 15-s sonication, and then cooled for 15 s. Cellular debris and the remaining intact cells were sedimented by centrifugation (1 h at 5,000 × g). The supernatant was centrifuged at 100,000 × g for 2 h. The supernatant (soluble fraction) was removed, and the sediment was washed in 1 volume of 10 mM HEPES and centrifuged under the same conditions. The resulting sediment represents the envelope fraction, which was dissolved in 10 mM HEPES. The total protein concentration of the respective fractions was determined using the Bradford method (5).
Outer membrane protein analysis.
Cells were fractionated as described above. The cytosolic membrane fraction was then selectively solubilized from the total envelope fraction using 0.7% (wt/vol) n-lauroylsarcosine. The insoluble (outer membrane) fraction of the envelope fraction was then obtained by centrifugation at 100,000 × g for 1 h. Resulting outer membrane proteins were then separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and identified by tryptic peptide fingerprinting using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry.
β-Galactosidase and alkaline phosphatase activity assays.
β-Galactosidase and alkaline phosphatase enzymatic assays were performed according to the methods by Miller (37) and Manoil (31), respectively. Between 2 and 20 μl of the various subcellular fractions was added to the reaction mixtures, and the specific activity was determined in U/mg total protein, with 1 U corresponding to the hydrolysis of 1 μmol of substrate (o-nitrophenyl-β-galactoside and p-nitrophenylphosphate for alkaline phosphatase and β-galactosidase, respectively) per 1 min at 37°C. The results are given as average values of at least four independent experiments.
Uronic acid assays.
Alginate concentrations were assayed by a modification of the Blumenkrantz and Asboe-Hansen protocol (3), using purified P. aeruginosa PDO300 alginate (100% [wt/wt] uronic acid content) as a standard, as previously described (45). The uronic acid concentrations were determined spectrophotometrically at a wavelength of 520 nm.
Swarming motility.
Swarming motility was assessed by the method of Tremblay et al. (57). Briefly, plates consisted of modified M9 medium (20 mM NH4Cl; 12 mM Na2HPO4; 8.6 mM NaCl; 1 mM MgSO4; 1 mM CaCl2·2H2O; and 10 mM dextrose, supplemented with 0.5% [wt/vol] Casamino Acids [Difco]) solidified with 0.5% (wt/vol) Bacto agar (Difco). Autoclaved medium was poured in petri dishes and dried under laminar flow for 60 min. Then swarm plates were immediately inoculated with 5 μl of stationary-phase bacterial culture and incubated at 30°C for 16 h.
Solid surface attachment assay.
Attachment to a solid surface was assessed by a modified method described by Merritt et al. (36). Briefly, relevant strains were grown to saturation in LB medium. A total of 100 μl of these cultures was transferred to 4 wells of a sterile 96-well microtiter plate and incubated at 37°C for 1 h. Planktonic/nonadherent bacteria were removed by inverting the plate, and the plates were washed with water. A total of 125 μl of 0.1% crystal violet was added to each well, incubated at room temperature for 10 min, subsequently washed twice as described above, and allowed to air dry. Bound crystal violet was solubilized with the addition of 200 μl of 100% dimethyl sulfoxide. Absorbance was measured at 595 nm.
Continuous-culture flow cell biofilms.
For biofilm analysis, P. aeruginosa strains were grown in continuous-culture flow cells (channel dimensions of 4 mm by 40 mm by 1.5 mm) at 37°C as previously described (7). Channels were inoculated with 0.5 ml of early-stationary-phase cultures containing approximately 2 × 109 cells ml−1 and incubated without flow for 4 h at room temperature. Flow was then started with a mean flow of 0.3 ml min−1, corresponding to a laminar flow with a Reynolds number of 5. The flow cells were then incubated at 37°C for 20 h. Biofilms were stained using the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Inc., Eugene, OR) and visualized first using phase-contrast microscopy and then confocal laser scanning microscopy (Leica SP5 DM6000B).
Detection and purification of MucR.
The envelope fraction of the PDO300ΔmucR strain harboring plasmid pBBR1MCS-5:mucRhis was solubilized by the addition of 1% (vol/vol) Triton X-100 and incubated at room temperature for 2 h. The solubilized envelope fraction was subjected to affinity purification using Ni-NTA agarose (Qiagen). Briefly, the solubilized envelope fraction from 1 liter of a stationary-phase culture was resuspended in 20 ml of buffer A (50 mM NaH2PO4, 300 mM NaCl, 1% Triton X-100, pH 8.0) containing 10 mM imidazole, and 1 ml of a 50% (wt/vol) Ni-NTA agarose (Qiagen) was added and incubated with shaking at 4°C for 1 h. The Ni-NTA agarose was then loaded onto a gravity flow column, and the agarose was washed four times with 5 ml of buffer A containing 20 mM imidazole and four times with 5 ml of buffer A containing 50 mM imidazole. The bound proteins were eluted four times with 500 μl of buffer A containing 250 mM imidazole. Fractions were then subjected to SDS-PAGE analysis and immunoblotting analysis. In order to identify proteins, protein bands of interest were cut off the gel. Proteins were subjected to tryptic peptide fingerprinting. Identification of tryptic peptides was performed by collision-induced dissociation tandem mass spectrometry and enabled identification of proteins. For immunoblotting, proteins were transferred to a nitrocellulose membrane and hexahistidine-tagged proteins were detected using a SuperSignal HisProbe-HRP kit (Pierce).
RESULTS
Primary structure analysis of MucR.
The MucR (PA1727) sequence was attained from the complete P. aeruginosa PAO1 genome sequence (55). TMHMM (24) predicts that MucR has seven transmembrane regions, all located in the N-terminal 240 residues of the protein, with the N terminus residing in the periplasm and the C terminus residing in the cytosol. SMART (28) and Pfam (13) predicted an MHYT domain (16) in the second to seventh transmembrane regions. This domain consists of six transmembrane segments connected by short arginine-rich cytosolic loops. The second, fourth, and sixth transmembrane segments in the domain have a highly conserved amino acid motif, MHYTXM, located near the outer side of the inner membrane. It has been suggested that the MHYT domain serves as a sensing domain. The C terminus of MucR is predicted to contain a GGDEF domain, common to DGCs, at amino acids 252 to 423 and an EAL domain, conserved among PDE, at amino acids 433 to 679. A sequence database search (BLASTP) revealed the strongest similarity of 73% identity with the hypothetical protein ZP_00417207 from Azotobacter vinelandii belonging to one of the two bacterial genera (Pseudomonas and Azotobacter) comprising species capable of alginate production.
The ΔmucR mutant is defective in alginate biosynthesis.
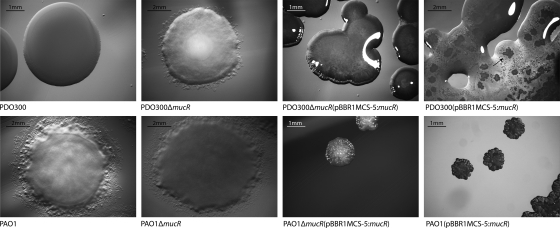
Deletion of 1,041 bp of the mucR gene in the alginate-overproducing P. aeruginosa strain PDO300 resulted in a nonmucoid colony phenotype on solid medium (Fig. 1). Interestingly, strains PDO300 and PAO1, each harboring plasmid pBBR1MCS-5:mucR and thus additional mucR gene copies, showed small wrinkly colonies, while PDO300 was additionally surrounded by copious amounts of transparent extracellular alginate (Fig. 1). Alginate is the primary exopolymeric substance responsible for the mucoid phenotype, which suggested that MucR plays a role in biosynthesis or regulation of alginate biosynthesis in P. aeruginosa. To assess whether the ΔmucR mutant of strain PDO300 is still able to produce alginate, the extracellular polysaccharide was purified and the uronic acid (alginate) content was determined. The ΔmucR mutation caused a reduction in alginate production to nearly undetectable levels (Table 2). To confirm this was not due to polar effects, the mucR gene (pBBR1MCS-5:mucR) was introduced into the PDO300ΔmucR strain, and alginate production was restored and increased by about 4.4-fold compared with that of strain PDO300. The ΔmucR mutant harboring only the vector pBBR1MCS-5 showed an about 1.6-fold increase in alginate production compared with strain PDO300 (Table 2).
FIG. 1.
MucR is essential for the mucoid colony morphology. Colonies of various P. aeruginosa strains were grown on PIA medium for 24 h. The top row shows the mucoid strain PDO300 and the strains derived from it; the bottom row shows the nonmucoid strain PAO1 and the plasmids derived from it. Plasmids harbored by certain strains are indicated in parentheses.
TABLE 2.
Alginate quantification of various P. aeruginosa strains
| Strain | Alginate production (g/g CDM)a | SDb |
|---|---|---|
| PDO300 | 0.225 | ± 0.047 |
| PDO300ΔmucR | 0.006 | ± 0.001 |
| PDO300(pBBR1MCS-5) | 0.584 | ± 0.023 |
| PDO300ΔmucR(pBBR1MCS-5) | 0.386 | ± 0.021 |
| PDO300(pBBR1MCS-5:mucR) | 1.54 | ± 0.130 |
| PDO300ΔmucR(pBBR1MCS-5:mucR) | 0.958 | ± 0.170 |
| PAO1 | 0 | ± 0 |
| PAO1ΔmucR | 0 | ± 0 |
| PAO1(pBBR1MCS-5) | 0 | ± 0 |
| PAO1ΔmucR(pBBRMCS-5) | 0 | ± 0 |
| PAO1(pBBR1MCS-5:mucR) | 0 | ± 0 |
| PAO1ΔmucR(pBBRMCS-5:mucR) | 0 | ± 0 |
| PDO300(pBBR1MCS-5:wspR) | 0.295 | ± 0.006 |
| PDO300(pBBR1MCS-5:rocR) | 0 | ± 0 |
CDM, cell dry mass.
SD, standard deviation. Results represent the data of four independent experiments.
To assess whether the increase in alginate production was due to a general increase in intracellular c-di-GMP levels based on the presence of multiple mucR gene copies or was the result of a more specific function of MucR, additional c-di-GMP-influencing genes were introduced. The DGC gene (wspR/PA3702) shown to encode a highly active DGC was expressed in PDO300 (18, 26). This resulted not in an increase but in a twofold reduction in alginate production (Table 2). Overexpression of rocR (PA3947), which encodes a highly active PDE, abolished alginate production (26, 27) (Table 2).
MucR impacts colony morphology, auto-aggregation, pellicle formation, attachment to surfaces, and swarming motility.
To further assess the function of MucR, the ΔmucR mutants were characterized with respect to several phenotypes known to be influenced by c-di-GMP levels.
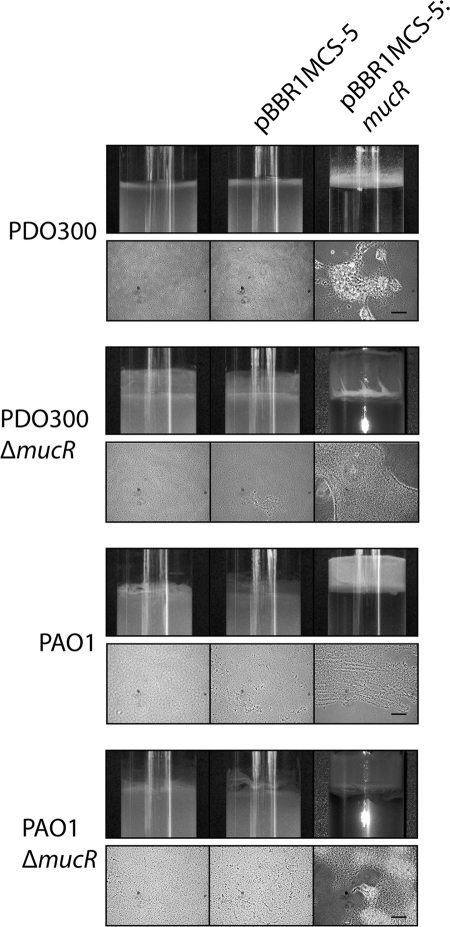
Strains harboring additional copies of the mucR gene showed an altered phenotype. On solid medium these strains typically formed compact wrinkly colonies, which were surrounded by alginate when PDO300 was considered (Fig. 1). In liquid culture, cells harboring plasmids encoding MucR formed large aggregates (pellicles) at the air-liquid interface (Fig. 2). In order to assess if any of these phenotypic changes were due to increased alginate production mediated by MucR, the Δalg8 mutant of PDO300, which is defective in alginate biosynthesis, was employed (47). Multiple copies of the mucR gene in the alginate-negative Δalg8 mutant resulted in small wrinkly colonies and the same auto-aggregation phenotype that was observed for the nonmucoid strain PAO1 (data not shown and Fig. 2).
FIG. 2.
Multiple copies of mucR induce auto-aggregation and the formation of pellicles. Various strains were grown in LB for 16 h. The top row for each strain indicated on the left contains photographs of overnight cultures grown in LB overnight showing pellicle formation. The bottom row for each strain contains phase-contrast microscopic images showing the cell aggregation; the black bars represent 50 μm. Plasmids (where present) are indicated along the top.
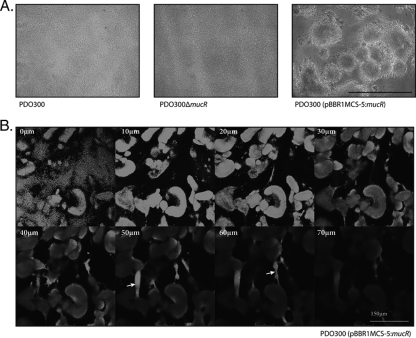
The solid surface assay was applied to assess attachment and biofilm formation. In general, MucR mediated increases in initial attachment of about 8.4-fold and 8.9-fold when produced in PDO300 and PAO1, respectively (Table 3). These results were mimicked by continuous-culture flow cell biofilm analysis. Cellular layers, 1or 2 cells thick, were observed after 20 h on the glass surface for the PDO300 and PDO300ΔmucR strains (Fig. 3). Multiple copies of the mucR gene in PDO300 resulted in an initial increased attachment which developed into highly structured biofilms after 20 h (Fig. 3).
TABLE 3.
Rapid solid surface attachment assay
| Strain | OD550a | SDb |
|---|---|---|
| PDO300(pBBR1MCS-5) | 0.088 | ± 0.046 |
| PDO300ΔmucR(pBBR1MCS-5) | 0.198 | ± 0.179 |
| PDO300ΔmucR(pBBR1MCS-5:mucR) | 0.742 | ± 0.141 |
| PAO1(pBBR1MCS-5) | 0.114 | ± 0.078 |
| PAO1ΔmucR(pBBR1MCS-5) | 0.095 | ± 0.028 |
| PAO1ΔmucR(pBBR1MCS-5:mucR) | 0.983 | ± 0.250 |
OD550, optical density at 550 nm.
SD, standard deviation. Results represent the data of three independent experiments.
FIG. 3.
Multiple copies of mucR lead to the formation of highly structured biofilms. (A) Phase-contrast images of 24-h-old biofilms grown in a continuous-culture flow cell. Black bar represents 200 μm. (B) Confocal laser scanning microscopy images of a 24-h-old biofilm of the strain PDO300(pBBR1MCS-5:mucR). Each frame represents a picture 10 μm away from the last. The white bar represents 150 μm; the white arrow shows the formation of bridge-like structures connecting adjacent microcolonies.
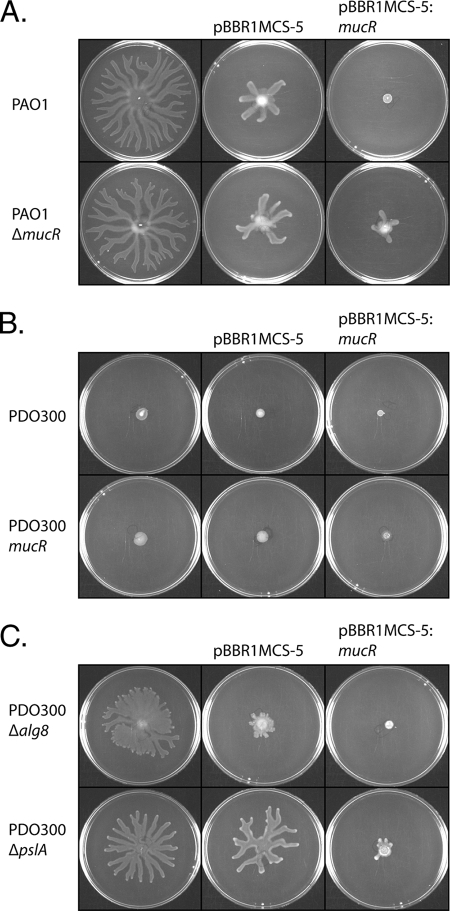
Recent studies have described a link between c-di-GMP levels and swarming motility in P. aeruginosa (25, 35). The alginate-overproducing strain PDO300 did not show a swarming phenotype, whereas PAO1 did. The lack of swarming shown in the PDO300 strains could be due to the presence of alginate. Thus, when the Δalg8 mutant of PDO300 was analyzed, this mutant showed swarming motility (Fig. 4). Deletion of the mucR gene did not significantly change the level of swarming motility in any of the strains (Fig. 4). In strains harboring only the vector and where gentamicin was added to the media, the swarming phenotype was reduced but detectable (Fig. 4). The presence of plasmids containing the mucR gene in the PAO1, PAO1ΔmucR, and PDO300Δalg8 strains resulted in a strong inhibition of swarming motility (Fig. 4).
FIG. 4.
Loss of mucR does not influence swarming motility, but multiple copies of mucR inhibit swarming motility. (A) Swarming motility of the nonmucoid P. aeruginosa strain PAO1 and its isogenic mucR knockout mutant. (B) Swarming motility of the mucoid P. aeruginosa strain PDO300 and its isogenic mucR knockout mutant. (C) Swarming phenotype of a mutant derived from the mucoid PDO300 strain incapable of producing alginate due to the loss of the alg8 gene, which is essential for alginate biosynthesis.
MucR affects the outer membrane protein profile.
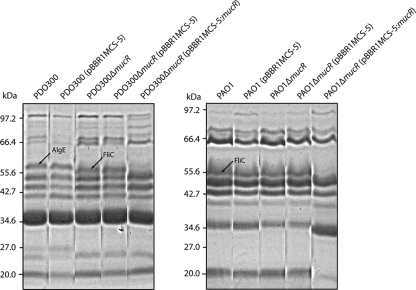
The altered phenotypes described above suggested that MucR influences cell surface properties. Thus, the outer membrane protein profiles were analyzed. The protein profiles differed between the mucoid strain PDO300 and the nonmucoid strain PAO1 with respect to two proteins (Fig. 5). A protein with an apparent molecular mass of 56 kDa was present only in PDO300 and was identified as AlgE by using tryptic peptide fingerprinting employing MALDI-TOF mass spectrometry. AlgE is essential for alginate production, presumably enabling the export of alginate (42, 43). Another protein with an apparent molecular mass of 53 kDa was found in PAO1 and absent in PDO300 (Fig. 5). This protein was identified as FliC, a flagellin type B protein.
FIG. 5.
Outer membrane protein profiles of various P. aeruginosa strains. (Left) Outer membrane protein profiles of the ΔmucR mutant derived from PDO300; (right) outer membrane protein profiles of the ΔmucR mutant derived from PAO1. The arrows indicate the identity of proteins whose levels differed between mutants. Plasmids harbored by certain strains are indicated in parentheses.
In the ΔmucR mutant of PDO300, AlgE becomes less abundant while FliC becomes apparent. The PAO1ΔmucR showed no significant change in the outer membrane protein profile. Introduction of a plasmid encoding MucR significantly reduced the levels of FliC in the outer membrane of both PAO1 and PDO300 but did not affect the levels of AlgE in PDO300 (Fig. 5).
The GGDEF/EAL domain is required for mucR function.
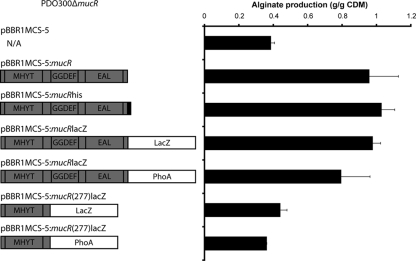
To establish whether MucR function in the regulation of alginate production was dependent on the GGDEF/EAL domain containing the C terminus of MucR, a hybrid gene encoding only the first 277 amino acids of MucR was constructed. This region corresponded to the seven-transmembrane putative MHYT-sensing domain but lacked the GGDEF and EAL domains. This N-terminal region and full-length MucR were each translationally fused to the reporter enzymes LacZ and PhoA. Full-length MucR but not C-terminally truncated MucR restored alginate production to levels exceeding wild-type levels as well as mediated the formation of wrinkly colonies and an auto-aggregation phenotype when produced in the ΔmucR mutant (Fig. 6; data not shown).
FIG. 6.
The alginate stimulating activity of MucR is dependent on the GGDEF and EAL domains containing the C terminus. (Left) Schematic representations of the various MucR fusion proteins. Plasmids harbored by the PDO300ΔmucR strain are indicated in parentheses. (Right) Alginate production levels of the PDO300ΔmucR strain harboring the genes encoding the various MucR fusion proteins. Right, CDM, cell dry mass. Alginate quantification data represent the results of three independent experiments.
Subcellular localization, membrane topology, and purification of MucR.
The fusion proteins described above were employed to test the predicted membrane topology. P. aeruginosa cells expressing the various fusion proteins were fractionated into the cell lysate, soluble cytosol, and insoluble envelope fraction. No significant alkaline phosphatase activity could be detected for either the full-length or truncated MucR protein fused to PhoA in any of the fractions (Table 4). β-Galactosidase activity could be detected for both the full-length and truncated MucR-LacZ fusions, with the highest specific activity in the envelope fraction (Table 4).
TABLE 4.
β-Galactosidase and alkaline phosphatase activities of MucR fusion proteins in various subcellular fractions
| Fusion protein | Subcellular fractiona | LacZ U (μM/min)/mg ± SDb | PhoA U (μM/min)/mg ± SD |
|---|---|---|---|
| MucR | WCL | 11.06 ± 0.77 | 0.01 ± 0.00 |
| SOL | 11.54 ± 1.42 | 0.01 ± 0.00 | |
| MEM | 48.68 ± 0.22 | 0.03 ± 0.00 | |
| MucR(277) | WCL | 135.45 ± 2.41 | 0.11 ± 0.01 |
| SOL | 34.10 ± 3.90 | 0.12 ± 0.01 | |
| MEM | 559.40 ± 20.36 | 0.07 ± 0.00 |
WCL, whole-cell lysate; SOL, soluble; MEM, membrane.
SD, standard deviation. Results represent data of three independent experiments.
In addition, plasmid pBBR1MCS-5:mucRhis was constructed, encoding C-terminally hexahistidine-tagged MucR. The PDO300ΔmucR strain harboring pBBR1MCS-5:mucRhis showed restored alginate production and was used to detect hexahistidine-tagged MucR via immunoblotting using anti-hexahistidine antibodies (Fig. 7). Immunoblot analysis of the cell lysate, cytosol, and envelope enabled detection of two protein bands in both the cell lysate and the envelope but not in the soluble cytosol fraction. These proteins showed an apparent molecular mass of 65 kDa and 50 kDa, respectively (Fig. 7). The theoretical molecular mass of full-length MucR is 74.4 kDa. The detected proteins could not be separated from the complex mixture of proteins in the gel. To confirm that these proteins corresponded to MucR, the solubilized total membrane fraction was subjected to Ni-NTA affinity purification, resulting in partial purification of the two detected proteins, enabling identification via tryptic peptide fingerprinting (Fig. 7). Both proteins were identified as MucR; however, only peptides corresponding to the C-terminal half of MucR and none from the transmembrane regions were detected. In addition, a protein with an apparent molecular mass of 330 kDa could be detected by immunoblotting, and peptide fingerprinting analysis confirmed that it belongs to MucR; this is presumably aggregate formation due to the hydrophobicity in the N terminus (Fig. 7).
FIG. 7.
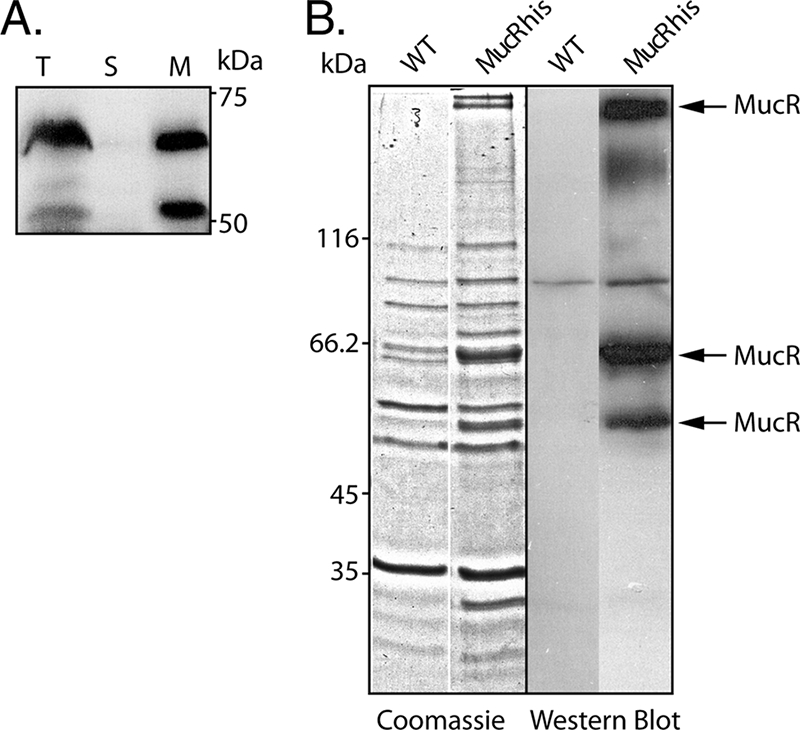
Subcellular localization and purification of MucR. (A) Immunoblot analysis of whole-cell lysate (T), soluble cytosol (S), and envelope (M) fractions. (B) SDS-PAGE and immunoblot analysis of hexahistidine-tagged MucR (MucRhis) purified from the solubilized membrane fraction of the PDO300ΔmucR(pBBR1MCS-5:mucRhis) strain using Ni-NTA affinity purification. MucR was identified by tryptic peptide fingerprinting and MALDI-TOF mass spectrometry. WT, wild type.
DISCUSSION
Recently it has been shown that c-di-GMP binding to membrane-anchored Alg44, an essential alginate biosynthesis protein, is required for alginate production in P. aeruginosa (1, 34, 45). It has been suggested that some of the DGC and PDE proteins respond to different signals by controlling intracellular c-di-GMP levels in localized pools. Thus, c-di-GMP sensing/binding proteins may be colocalized in the membrane with proteins producing and/or degrading c-di-GMP.
Experimental evidence to support this hypothesis had previously been obtained for Gluconacetobacter xylinus, where the DGC DgcA and the PDE PdeA were shown to copurify with the c-di-GMP binding cellulose synthase (50), and for Caulobacter crescentus, where the GGDEF-containing protein PleD localized to the base of the flagellum where its DGC activity is required for flagellum ejection (41).
Twenty-two of the 38 DGCs and PDEs contain predicted transmembrane regions, and 14 of these contained domains recognized by Pfam (14). These domains were the PAS and PAC domains, involved in numerous signaling processes sensing diverse signals; the CHASE domains, sensing domains predicted to bind diverse low-molecular-weight ligands, such as the cytokinin-like adenine derivatives or peptides (38); the 7TM-DISM2 domains, thought to act as a receptor for carbohydrates (2); the MASE1 domain, with unknown function; and the MHYT domains, thought to sense oxygen, CO, or NO through the coordinated binding of one or two copper atoms (16). Both oxygen and NO have previously been shown to influence alginate production in P. aeruginosa (6, 15, 58, 59). Therefore, the two MHYT domain-containing proteins (MucR and PA3311) were considered. However, in a recent study, PA3311 showed no detectable DGC or PDE activity, whereas MucR showed DGC activity (26). Consequently, in this study MucR was functionally characterized.
The isogenic ΔmucR mutant of alginate-overproducing P. aeruginosa PDO300 showed a nonmucoid colony morphology and was strongly impaired in alginate production, and restoration of alginate production by the mucR gene suggested that MucR is involved in regulation of alginate biosynthesis (Fig. 1 and 2). The observed induction of alginate production by only the vector pBBR1MCS-5 had previously been described to be mediated by gentamicin (45, 47).
Introduction of the mucR gene-containing plasmid into PDO300 mediated a wrinkly colony morphology and strongly increased alginate production (Fig. 1 and 2). The small wrinkly colony morphology is very similar to the “small-colony variant” observed with the overexpression of the DGC WspR protein and commonly found among isolates from CF patients (11, 19).
The finding that the introduction of the wspR gene, encoding a cytosolic DGC showing 18.5-fold higher activity than MucR, did not cause an increased alginate production (26; Table 2) supports the premise of a specific role of MucR in the regulation of alginate biosynthesis and the hypothesis of localized pools of c-di-GMP.
Previous studies showed that various c-di-GMP concentration-controlling enzymes, e.g., SadC and BifA, inversely regulate attachment/biofilm formation and swarming motility in P. aeruginosa (8, 22, 25, 35). In general, DGCs increase intracellular c-di-GMP levels and correlate with enhanced aggregation, attachment, and biofilm formation, but with repressed swarming motility, whereas PDEs lower levels of c-di-GMP and correlate with the contrary. Overproduction of MucR followed this trend, but loss of the mucR gene did not result in any change in these phenotypes, as would be expected of a DGC directly involved in regulation of these phenotypic properties.
Although transfer of pBBR1MCS-5:mucR, i.e., multiple mucR gene copies, into various P. aeruginosa strains led to multiple phenotypic changes, many of these phenotypic changes resemble the overexpression of many DGCs and the corresponding increase in c-di-GMP levels (11, 19, 35, 48, 49, 56).
Since MucR impacted phenotypic properties which often depend on proteinaceous cell surface structures, the outer membrane protein profiles of the various strains were analyzed (Fig. 5). The strong decrease of the copy numbers of the flagellar filament protein FliC in strains PAO1 and PDO300 harboring plasmid pBBR1MCS-5:mucR correlated with the loss of swarming motility (Fig. 4). FliC has been described to be required for flagellum-based swarming motility (39). However, plasmid pBBR1MCS-5:mucR caused not an increase but a slight decrease of the copy numbers of the alginate export protein AlgE, while mediating alginate overproduction (Fig. 5).
The loss of MucR function after removal of the C-terminal region comprising the GGDEF domain suggested that DGC activity is required for MucR function (Fig. 6). As LacZ can fold correctly and become active only when exposed to the cytosol and as PhoA can correctly fold as an active enzyme only in the periplasm (29), the reporter enzyme activities support the prediction that MucR is localized to the cytoplasmic membrane with its GGDEF/EAL C terminus exposed to the cytosol (Table 3).
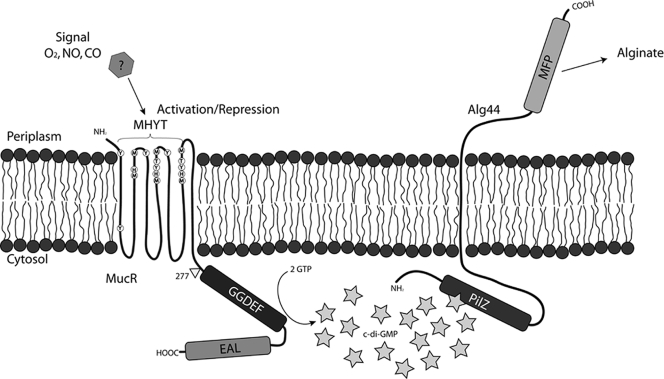
Overall this study suggested that MucR is a membrane-anchored DGC which is a positive regulator of alginate biosynthesis. In accordance with the recent findings by Merighi et al. (34) that the c-di-GMP-binding PilZ domain of the Alg44 membrane protein is essential for alginate biosynthesis, a model for c-di-GMP-dependent regulation of alginate biosynthesis was developed (Fig. 8). The presence of the proposed localized c-di-GMP pool was supported by the finding that a single gene copy of mucR is required for normal levels of alginate biosynthesis in mucoid P. aeruginosa, whereas multiple copies of mucR led to phenotypes common to overexpression of DGCs (25). Further support that MucR regulated alginate biosynthesis via localized c-di-GMP pools in the vicinity of Alg44 was obtained by the finding that MucR but not the highly active cytosolic DGC WspR protein significantly increased alginate production (Fig. 8). The model proposed that an as yet unidentified signal (possibly CO, NO, or O2 [16]) is detected via the MHYT domain of MucR. Reception of this signal might influence the activity of MucR by modulating the activity of the DGC, and possibly the PDE, domain of MucR.
FIG. 8.
Proposed model for the MucR-mediated regulation of alginate biosynthesis in P. aeruginosa. The membrane topology and domain structure of MucR, as predicted by SMART and Pfam, are shown. The circles represent the conserved MHYT residues suggested to be involved in binding a copper atom and sensing oxygen, NO, or CO. The hexagon represents the putative signal. The stars represent c-di-GMP. Various domains are shown. MFP, membrane fusion protein domain, similar to multidrug efflux systems.
Acknowledgments
This study was supported by research grants to B.H.A.R. from the Institute of Molecular BioSciences at Massey University and the Deutsche Forschungsgemeinschaft (Re 1097/6-1). I.D.H. is funded by a Massey University doctoral scholarship.
Footnotes
Published ahead of print on 16 December 2008.
REFERENCES
- 1.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 2.Anantharaman, V., and L. Aravind. 2003. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumenkrantz, N., and G. Asboe-Hansen. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484-489. [DOI] [PubMed] [Google Scholar]
- 4.Boucher, J. C., H. Yu, M. H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bragonzi, A., D. Worlitzsch, G. B. Pier, P. Timpert, M. Ulrich, M. Hentzer, J. B. Andersen, M. Givskov, M. Conese, and G. Doring. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J. Infect. Dis. 192:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campisano, A., C. Schroeder, M. Schemionek, J. Overhage, and B. H. Rehm. 2006. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:3066-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choy, W. K., L. Zhou, C. K. Syn, L. H. Zhang, and S. Swarup. 2004. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol. 186:7221-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 9:50-52. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 11.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ethier, J., and J. M. Boyd. 2000. Topological analysis and role of the transmembrane domain in polar targeting of PilS, a Pseudomonas aeruginosa sensor kinase. Mol. Microbiol. 38:891-903. [DOI] [PubMed] [Google Scholar]
- 13.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H.-R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firoved, A. M., S. R. Wood, W. Ornatowski, V. Deretic, and G. S. Timmins. 2004. Microarray analysis and functional characterization of the nitrosative stress response in nonmucoid and mucoid Pseudomonas aeruginosa. J. Bacteriol. 186:4046-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galperin, M. Y., T. A. Gaidenko, A. Y. Mulkidjanian, M. Nakano, and C. W. Price. 2001. MHYT, a new integral membrane sensor domain. FEMS Microbiol. Lett. 205:17-23. [DOI] [PubMed] [Google Scholar]
- 17.Gutierrez, C., and J. C. Devedjian. 1989. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 17:3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guvener, Z. T., and C. S. Harwood. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66:1459-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haussler, S. 2004. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ. Microbiol. 6:546-551. [DOI] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Holloway, B. W., H. Matsumoto, and P. V. Phibbs, Jr. 1986. The chromosome map of Pseudomonas aeruginosa PAO1. Acta Microbiol. Pol. 35:161-164. [PubMed] [Google Scholar]
- 22.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60:1026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 24.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 25.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulasekara, H. D., I. Ventre, B. R. Kulasekara, A. Lazdunski, A. Filloux, and S. Lory. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368-380. [DOI] [PubMed] [Google Scholar]
- 28.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 32.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathee, K., C. Sternberg, O. Ciofu, P. Jensen, J. Campbell, M. Givskov, D. E. Ohman, N. Hoiby, S. Molin, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 34.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876-895. [DOI] [PubMed] [Google Scholar]
- 35.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merritt, J. H., D. E. Kadouri., and G. A. O'Toole. 2005. Growing and analyzing static biofilms, p. 1B.1.1-1B.1.7. In R. Coico, T. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology, vol. 1. J. Wiley & Sons, Hoboken, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Mougel, C., and I. B. Zhulin. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 26:582-584. [DOI] [PubMed] [Google Scholar]
- 39.Murray, T. S., and B. I. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overhage, J., M. Schemionek, J. S. Webb, and B. H. Rehm. 2005. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl. Environ. Microbiol. 71:4407-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rehm, B. H., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rehm, B. H., E. Grabert, J. Hein, and U. K. Winkler. 1994. Antibody response of rabbits and cystic fibrosis patients to an alginate-specific outer membrane protein of a mucoid strain of Pseudomonas aeruginosa. Microb. Pathog. 16:43-51. [DOI] [PubMed] [Google Scholar]
- 44.Rehm, B. H., and S. Valla. 1997. Bacterial alginates: biosynthesis and applications. Appl. Microbiol. Biotechnol. 48:281-288. [DOI] [PubMed] [Google Scholar]
- 45.Remminghorst, U., and B. H. Rehm. 2006. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 580:3883-3888. [DOI] [PubMed] [Google Scholar]
- 46.Remminghorst, U., and B. H. Rehm. 2006. Bacterial alginates: from biosynthesis to applications. Biotechnol. Lett. 28:1701-1712. [DOI] [PubMed] [Google Scholar]
- 47.Remminghorst, U., and B. H. A. Rehm. 2006. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:298-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218-228. [DOI] [PubMed] [Google Scholar]
- 49.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 50.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch. Mikrobiol. 38:209-222. (In Russian.) [PubMed] [Google Scholar]
- 53.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 54.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 55.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 56.Tamayo, R., J. T. Pratt, and A. Camilli. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tremblay, J., A. P. Richardson, F. Lepine, and E. Deziel. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9:2622-2630. [DOI] [PubMed] [Google Scholar]
- 58.Wood, S. R., A. M. Firoved, W. Ornatowski, T. Mai, V. Deretic, and G. S. Timmins. 2007. Nitrosative stress inhibits production of the virulence factor alginate in mucoid Pseudomonas aeruginosa. Free Radic. Res. 41:208-215. [DOI] [PubMed] [Google Scholar]
- 59.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]