Abstract
Genetic fingerprinting methods, such as denaturing gradient gel electrophoresis (DGGE), are used in microbial ecology for the analysis of mixed microbial communities but are associated with various problems. In the present study we used a new alternative method: denaturing high-performance liquid chromatography (dHPLC). This method was previously shown to work with samples from water and gut flora but had not yet been applied to complex environmental samples. In contrast to other publications dealing with dHPLC, we used a commonly available HPLC system. Samples from different origins (fermentor sludge, compost, and soil), all ecologically significant, were tested, and the 16S rRNA gene was amplified via PCR. After optimization of the HPLC elution conditions, amplicons of pure cultures and mixed microbial populations could be separated successfully. Systematic differentiation was carried out by a cloning approach, since fraction collection of the peaks did not result in satisfactory fragment separation. dHPLC was evaluated as a tool for microbial community analysis on a genetic level and demonstrated major improvements compared to gel-based fingerprinting methods, such as DGGE, that are commonly used in microbial ecology.
The introduction of molecular biological methods in microbiology has changed research in microbial ecology fundamentally. The approach to characterize and classify microbial communities by cultivation methods switched to the genetic level, and the analysis of community structure became possible without any further need of cultivation for systematic analysis. This is especially important since only an estimated 1% of the naturally occurring bacteria have been isolated and characterized thus far (12, 15).
Genetic fingerprinting techniques, often in combination with a cloning approach, are often used. They provide a pattern or profile of the community diversity based upon the physical separation of unique nucleic acid species, while systematic assignment is often achieved by results from clone libraries. The fingerprinting methods are quite rapid, relatively easy to perform, and permit the simultaneous analysis of multiple samples. This allows comparison of the genetic diversity of microbial communities from different habitats, and in individual communities over time. The most commonly applied methods include denaturing gradient gel electrophoresis (DGGE), temperature gradient gel electrophoresis, single-strand conformation polymorphism, restriction fragment length polymorphism, and terminal restriction fragment length polymorphism. All of these methods provide information on community structure, and DGGE is probably the most frequently used method for this purpose. However, in daily lab use, many problems arise with any of these methods (e.g., the lack of reliable and fast quantification, difficulties in reproducibility, labor-intensive steps, etc.), so alternatives are necessary.
In the present study we describe an alternative fingerprinting method, denaturing high-performance liquid chromatography (dHPLC). dHPLC was applied at first—like DGGE—in medical research (4, 22). It works via ion-pair reversed-phase HPLC under partly denaturing conditions achieved by temperature and an increase in denaturing conditions over time via an organic modifier. For the amplification of DNA fragments by PCR, various primers can be used. Analysis of fragments of up to 1.4 kb is possible (13), and generally primers including GC clamps are preferable or necessary (21).
The use and applicability of dHPLC for the analysis of complex microbial populations in water samples (1) and the intestinal gut flora (6) was introduced previously. We wanted to go a step further and show the advantages of dHPLC compared to other profiling methods (especially DGGE) in difficult matrices such as soil, compost, and anaerobic digester residue, thus introducing dHPLC generally into microbial ecology. This was done by using an ordinary HPLC system instead of the special and very expensive dHPLC system used thus far.
MATERIALS AND METHODS
Samples and sample preparation.
Fermentor sludge (FS), compost (CT), and soil (SL) samples were used for community profiling. The FS sample originated from an anaerobic digester treating biowaste (Roppen, Austria) (some basic parameters of the sludge and the biogas plant can be found in references 10 and 19); the CT was from a composting plant in Roppen, Austria; and the SL samples were from a sampling site in the Tyrolean Inntal, Austria (sampling point 11 in reference 11).
Pure cultures were purchased from DSMZ (Germany) and cultivated according to the manufacturer's protocols in the recommended media.
Experimental procedures.
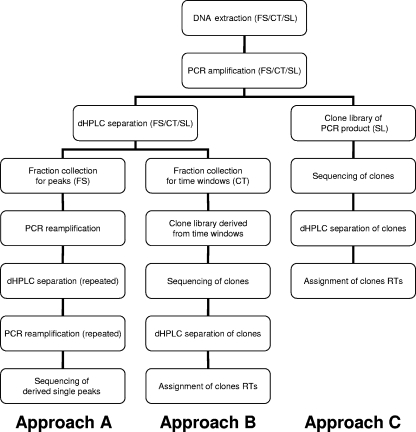
To demonstrate the applicability of dHPLC for community profiling using universal bacterial and/or archaeal primers, three ecologically interesting habitats—FS, CT, and SL—were chosen. After sample collection, DNA extraction and subsequent PCR amplification were performed as described above. PCR products were loaded into the dHPLC system to obtain a chromatogram indicative of the mixed microbial population of the respective habitats. Different fractions representing the tops of peaks were collected to systematically characterize the obtained profiles as described by Barlaan et al. (1). Since this turned out to be a problematic step, three different methods of assigning peaks to microbial species were examined (Fig. 1). The first method was fraction collection of the very top of a peak. These peaks were reamplified, assigned to peaks in the chromatogram of the mixed population, and sequenced. An attempt to assign chromatographic peaks to pure-culture amplicons was also made. The second method consisted of fraction collection by a time window and the construction of clone libraries for each fraction. Cloned fragments were examined by dHPLC, assigned to peaks in the chromatogram, and sequenced. The third method was the construction of clone libraries directly from the initial PCR product from the mixed population: cloned fragments were examined by dHPLC, assigned to peaks in the chromatogram, and sequenced.
FIG. 1.
Experimental procedure scheme.
DNA isolation and PCR amplification of 16S rRNA genes.
DNA was extracted by using a Power Soil DNA kit (MO BIO Laboratories, Inc.) according to the manufacturer's protocol (elution volume, 100 μl), and the residual DNA samples were frozen at −20°C and kept as DNA stock solutions. PCR was performed using primers listed in Table 1. All of the primer pairs used contained a GC clamp to prevent DNA fragments from melting completely (Table 1). PCR mixtures (total volume, 25 μl) were prepared with the following final concentrations: primer (0.2 μM each), deoxynucleoside triphosphates (200 μM each), MgCl2 (2 mM), 1× PCR buffer, 0.625 U of Taq DNA polymerase (BioTherm; GeneCraft), and 1 μl of template for environmental samples and up to 3 μl for the reamplification procedure. For all environmental samples, the addition of bovine serum albumin (BSA) at a final concentration of 0.08% was necessary to obtain a PCR product. As a template for a positive control, a sludge sample previously proven to contain different amplicons on DGGE by Malin and Illmer (14) was used. A PCR mixture without additional template DNA was used as a negative control.
TABLE 1.
Primers used for DNA amplification of environmental samples
| Organism | Primer pair | Orientationa | Sequence (5′-3′)b | Expected fragment length (bp) | Reference |
|---|---|---|---|---|---|
| Bacteria | FP6/MUI | F | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG* | 194 | 16 |
| R | ATTACCGCGGCTGCTGG | ||||
| 984GCf/1378r | F | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGAACGCGAAGAACCTTAC* | 394 | 8 | |
| R | CGGTGTGTACAAGGCCCGGGAACG | ||||
| Archaea | 934f/1390GCr | F | AGGAATTGGCGGGGGAGCA | 456 | 20 |
| R | ACGGGCGGTGTGTGCAACGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGC‡ | ||||
| 109GCf/934r | F | CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACKGCTCAGTAACACGT†‡ | 825 | 7 | |
| R | GTGCTCCCCCGCCAATTCCT |
F, forward; R, reverse.
*, 40-bp GC clamp; †, K = G+T; ‡, 32-bp GC clamp. Underscoring indicates the GC clamp.
The PCR conditions for the primers 934f/1390GCr (same as for the primer 109GCf/934r) and 984GCf/1378r are described elsewhere (14, as are the PCR conditions for FP6/MUI (1). Undesired heteroduplex formation was eliminated or reduced by reconditioning PCR as described by Thompson et al. (18) for all environmental samples by adding 5 μl of PCR product to 20 μl of fresh PCR mixture (1:5 dilution), followed by reamplification for five cycles. Environmental samples were purified prior to dHPLC analysis by using a NucleoSpin Extract II kit (Macherey-Nagel, Germany) to remove BSA from the reaction mixture.
For the reamplification of collected fractions (see below), 22 cycles instead of 30 cycles were performed. For testing the effect of acetamide (Sigma-Aldrich) on its ability to promote PCR amplification of GC-rich sequences as described earlier (9, 17), concentrations of from 1 to 5% (vol/vol) (in the PCR mixture) were evaluated.
dHPLC system and running conditions.
The dHPLC system used consisted of a conventional high-performance liquid chromatograph (Shimadzu Prominence) made up of a quaternary pump with a degassing unit, autosampler, column oven (accuracy, 0.1°C), and conventional UV detector. Samples were injected into the dHPLC system using a two-eluant buffer system (see below), subjected to partially denaturing conditions by temperature, retained in a special column according to the fragment's melting point defined by its GC content (1), and eluted by an increase in the concentration of an organic modifier (acetonitrile, see below). The detection of the fragments was carried out without any need of fluorescent dyes at a wavelength of 254 nm.
All lanes and fittings were made of PEEK (polyetheretherketon). In addition to the conventional system, an ∼1.8-m PEEK line was installed in the column oven ahead of the column to ensure partially denaturing conditions. DNA samples were analyzed by ion-pair reversed-phase HPLC using a DNA-Sep cartridge (Transgenomic, Berlin, Germany).
A two-eluant buffer system was used. Buffer A consisted of 100 mM triethylammonium acetate (pH 7; Applichem, Germany) in aqueous solution, and buffer B consisted of 100 mM triethylammonium acetate (pH 7) with 25% (vol/vol) acetonitrile (HPLC gradient grade; Merck, Germany). For the column wash, an aqueous solution with 75% (vol/vol) acetonitrile was used. For the preparation of aqueous solutions only ultrapure water with >18-MΩ quality was used.
Since retention and resolution are influenced mostly by column temperature, flow rate, denaturing gradient rate, and buffer B (1), within the experiment different oven temperatures (from 55 to 80°C), flow rates (from 0.5 to 0.9 ml min−1), and gradient rates were investigated for use with PCR amplicons of different primers to extract optimum running conditions (Table 2).
TABLE 2.
dHPLC running conditions for different primersa
| Time (min) | Concn of buffer B (%) with primer pair:
|
|||
|---|---|---|---|---|
| FP6/MUI | 984GCf/1378r | 934f/1390GCr | 109GCf/934r | |
| 0 | 46 | 42 | 42 | 40 |
| 31.1 | 60 | |||
| 31.2 | 100 | |||
| 31.8 | 100 | |||
| 31.9 | 46 | |||
| 36.0 | Stop | |||
| 27.0 | 53.25 | 53.25 | 56 | |
| 27.1 | 100 | 100 | 100 | |
| 27.2 | 100 | 100 | 100 | |
| 27.8 | 42 | 42 | 40 | |
| 32 | Stop | Stop | Stop | |
The concentration of buffer A (%) = 100% − the concentration of buffer B (%). For primer pairs FP6/MUI, 984GCf/1378r, 934f/1390GC, and 109GCf/934r, the oven temperatures were 64, 67, 68, and 72°C, respectively. The corresponding gradient rates were 0.450, 0.417, 0.417, and 0.592% buffer B min−1, respectively.
Fraction collection and sequencing.
Fractions were obtained by manually collecting 3 drops representing the top of the peak. To avoid or minimize the loss of DNA, tubes with a low DNA-binding capacity (Biozym, Germany) were used. Collected peaks were reamplified by PCR (22 cycles) after the removal gradient residues via a speed vacuum treatment (50°C). The resulting PCR product was then reloaded into dHPLC to check for the elimination of DNA other than the desired peak. Again, fraction collection and purification (see above) was performed until the result was satisfactory. For single peaks (pure cultures and pure culture mixtures) this procedure had to be repeated one to three times, and for non-baseline-separated peaks (environmental PCR products) it had to be repeated up to ten times (but even then this did not necessarily lead to a single peak). DNA prepared in this way was subsequently sequenced by MWG (Germany) after purification of the PCR products (NucleoSpin Extract II; Macherey-Nagel, Germany). Sequence comparison and alignment was done by using the NCBI databases and the CLC Workbench 3.6 program (CLCbio, Denmark).
Creation of clone libraries.
According to Fig. 1, clone libraries were built up from fractions collected within a time window for the CT sample and from the initial PCR product of the SL sample by using Qiagen PCR Cloning Plus kit (Hilden, Germany) according to the manufacturer's protocol, including blue or white screening. The transformation mixture from the SL sample was incubated at 37°C for 30 min prior to plating on LB agar. After overnight cultivation, white colonies were picked from the agar plates and transferred into both a PCR mixture containing standard vector primers T7/SP6 and a liquid cultivation medium (LB medium) to allow storage of clones containing different fragments. Clones were cultivated overnight at 37°C and 200 rpm. Subsequently, an equal amount of sterile 50% glycerol solution was added, and clones were frozen at −80°C.
Clone PCR was carried out according to standard protocols. The reaction mixture was the same as that described for amplification of 16S rRNA gene sequences, but the primer pair T7/SP6 was used. PCR conditions were as follows: initial denaturation at 94°C for 5 min; followed by 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; with a final cycle of 10 min at 72°C. Obtained amplicons were checked for specific amplification and correct size by agarose gel electrophoresis (1% agarose gel). Amplicons were screened for different retention times (RTs) on dHPLC, and chosen clones were sequenced using primer T7 (MWG-Biotech, Germany).
DGGE analysis was performed as described previously (14).
RESULTS AND DISCUSSION
PCR optimization.
Four primer pairs for the amplification of archaeal and bacterial DNA were tested for their suitability for dHPLC analysis (Table 1). Since the use of GC clamps was described as a basic necessity for dHPLC separation of amplicons derived from different bacterial species (1), all primer sequences included a 32- or 40-bp GC clamp to prevent amplicons from being denatured completely during dHPLC analysis. The primer pairs 984GCf/1378r for Bacteria and 934f/1390GCr for Archaea were tested (among others) and considered best suited, since fragments derived from other primers were either too short for a reliable systematic qualification after sequencing (FP6/MUI) or too long (109GCf/934r), resulting in poor chromatographic resolution. Chromatograms tend to become more diffuse as fragment length increases because the proportional effect of single-base-pair differences is smaller (data not shown). To keep the scope of the project realistic, the archaeal primer pair was chosen. This has been found in any case to amplify not only archaeal DNA but also some bacterial DNA as well (14).
The addition of different acetamide concentrations was evaluated for its potential to improve PCR fragment quality since it has been reported to promote amplification of GC-rich DNA (9, 17). Barlaan et al. (1) used acetamide to obtain generally better PCR products from environmental samples. On the one hand, for pure cultures up to 5% (vol/vol) acetamide partly improved the quality of the resulting PCR fragments, as seen by a better resolution of the peaks. However, it also significantly reduced the amount of PCR products obtained, resulting in a decrease in peak height as well (data not shown). On the other hand, the fragment quality of dominant species in environmental samples was improved using up to 2% acetamide, but small peaks disappeared completely. Therefore, the operator is able decide whether to improve fragment quality by concomitant loss of peak number and size (e.g., for peak collection for subsequent sequencing) or to improve detection sensitivity (e.g., for applications in microbial ecology) when it is desired to obtain the highest possible diversity.
Generally, optimization of PCR used for the amplification of DNA originating from environmental samples has a big potential for improving the quality of the subsequently obtained results. In our investigation, the addition of BSA to PCR mixtures turned out to be essential for obtaining high-quality PCR products from environmental samples. However, many problems remain. For example, replicate PCR amplifications of the same template DNA can result in peaks with different RTs, as previously observed by Goldenberg et al. (6).
dHPLC and dHPLC conditions.
In order to demonstrate the general applicability of dHPLC, we did not use the WAVE DNA fragment analysis system (Transgenomic) but instead used a conventional HPLC system that is less expensive and available in many laboratories. The detection of the fragments was carried out with a conventional UV detector at a wavelength of 254 nm without any fluorescent dyes or a fluorescence detection system. This saves major costs since conventional, nonfluorescent PCR reagents can be used, and UV detectors are common in laboratories and have the advantage of stable and robust performance.
dHPLC conditions had to be optimized for each primer pair prior to fragment analysis of complex microbial populations. The elution of the DNA fragments was influenced by several known factors, such as temperature, pump flow rate, and/or gradient rate of buffer B (1, 5), but also by the temperature of the sample at the time of injection (sample temperature) and the duration of the washing step with 100% of buffer B at the end of each run (data not shown). For each primer, various oven temperatures, flow rates, and gradients were tested. Eventually, it was found that oven temperatures between 64 and 72°C, depending on the fragment length generated by the various primers used, worked best. Generally, the lowest temperature possible should be preferred since then the gradient can be run more steeply, and the peaks are sharper. A flow rate of 0.90 ml min−1 gave the best results and the sharpest peaks. The optimum running conditions for the primers used are presented in Table 2.
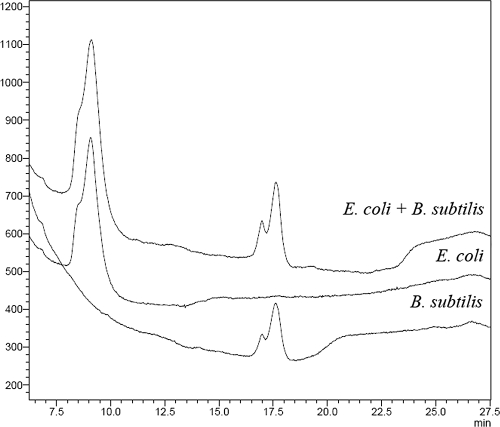
Amplicons of the 16S rRNA gene of pure cultures of several archaeal (Methanobrevibacter smithii [Deutsche Sammlung für Mikroorganismen und Zellkulturen DSM 861], Methanobacterium formicicium [DSM 1535], Methanothermobacter wolfeii [DSM 2970], Methanothermobacter thermoautotrophicus [DSM 1053], Methanomicrobium mobile [DSM 1539], Methanosaeta concilii [DSM 2139], Methanosarcina siciliae [DSM 3028], and Methanoculleus thermophilus [DSM 2640]) and bacterial (Escherichia coli [DSM 2670], Bacillus subtilis [DSM 10], Bacillus psychrophilus [DSM 3], Azotobacter chroococcum [DSM 368], Micrococcus luteus [ATCC 9341], Micrococcus lysodeicticus [DSM 20030], Listeria monocytogenes [ATCC 1918], and Clostridium perfringens [DSM 756]) species were generated via PCR and analyzed on the dHPLC system. In Fig. 2, the different RTs of pure culture amplicons of E. coli and B. subtilis are demonstrated when injected separately and in combination. The separation of amplicons derived from the two different bacteria can be seen clearly, as well as the reproducibility when injected together.
FIG. 2.
Separation of two pure culture amplicons (E. coli (RT = 9.07 min) and B. subtilis (RT = 16.99 min and 17.61 min) in separated dHPLC runs and injected together. The PCR product of B. subtilis shows a double peak (for an explanation, see the text).
Most pure cultures gave a single peak with defined RT, but a few resulted in more than one peak, an effect that was also found previously for analysis on DGGE gels (3) and on dHPLC as well (6). An explanation might be the inaccuracy of polymerase with PCR amplification, but more likely there are sequence variations within the template DNA (2).
Application of dHPLC for analysis of microbial communities in FS, CT, and SL.
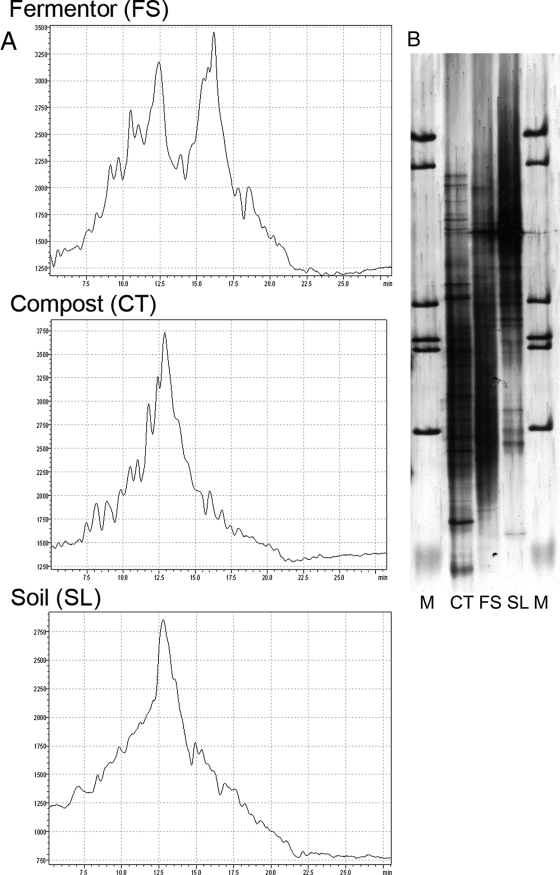
DNA extracts of samples derived from three diverse, ecologically interesting habitats were amplified via PCR with the primer pair 934f/1390GCr and subsequently applied to dHPLC using optimized running conditions as described above. In order to be able to compare dHPLC chromatograms (Fig. 3A) to a standard gel-based method, a DGGE analysis was performed additionally (Fig. 3B) with an aliquot of samples used for dHPLC. DGGE and other gel-based methods that have commonly been used as standard methods in our laboratory have demonstrated various problems in analyzing complex environmental samples such as those investigated here. This was one of the reasons we sought to establish a dHPLC system as an alternative to DGGE. As can be seen in Fig. 3B, a clear and distinct band pattern could not be obtained with the gel-based method. However, although dHPLC chromatograms of the investigated samples did not result in a baseline separation of amplicons, dHPLC was effective with such complex samples and therefore represents another fingerprinting method for microbial community analysis. dHPLC is still a new method and has as yet seldom been applied to complex environmental samples. Considering the huge amount of work that was necessary to bring DGGE (with all its problems) to the level where it is now makes us optimistic concerning the potential of dHPLC.
FIG. 3.
dHPLC (A) and DGGE (B) analysis of 16S rRNA gene fragments amplified with the primer pair 934f/1390GCr using optimized dHPLC running conditions. Lane M, marker.
In Fig. 3A, elution profiles of mixed microbial populations extracted from FS, CT, and SL are depicted. Different peak profiles were obtained for all samples indicating the expected differences in microbial population diversity and abundance but also suggesting overlaps in community composition, since peaks eluting at the same time should represent the same nucleotide sequence (see below). In order to describe the microbial population as represented by the obtained dHPLC peak profile systematically, different approaches for its determination were tried (Fig. 1).
First of all, for the isolation of fragments in the FS sample, the top of each peak was collected separately, reamplified via PCR, and checked by dHPLC (approach A). This collection and amplification procedure was conducted until a single peak was obtained and the resulting PCR product was sequenced. Unfortunately, it turned out that this approach was not suited to describe the peak profile obtained by dHPLC completely. Reamplification of collected peaks did not result in single peaks, although fraction collection was conducted manually as described by Barlaan et al. (1) and a reduced number of PCR cycles (22 instead of 30) was used. In most cases, in addition to the desired fragment, one of the peaks representing what was probably the most abundant species was coamplified, suggesting possible column overload. However, even a dramatically reduced injection volume could not solve this problem. Only if the collected fragment eluted after the highest peak was sequencing possible at all, but it was still problematic. Moreover, the repeated collection and reamplification procedure is susceptible to amplification errors, and the resulting fragments differed considerably in length, so that reliable assignment of collected peaks was hardly possible.
Therefore, fraction collection within time windows was performed as an alternative approach (approach B) for the CT sample; the fractions were reamplified, and the resulting PCR product was cloned into a vector, sequenced, and subsequently assigned to the respective peak on the dHPLC chromatogram of the initial PCR product. This approach is very costly since for each time window one line of competent cloning cells is needed. This is also very time-consuming because many similar fragments are produced that all have to be checked on gels for the appropriate fragment length and on dHPLC to knock out identical clones before sequencing and because not all fragments from the collection window can be found again after the cloning and sequencing procedure.
Therefore, a third alternative was tested for the SL sample to determine the community composition (approach C). The initial PCR product was directly cloned into a vector, sequenced, and reassigned to the chromatogram. Although by this approach the whole community could not be determined after the sequencing/cloning procedure either, it was best suited for the systematic characterization of an environmental sample for several reasons. (i) The major part of the community could be described, since for the SL sample 81.95% of the obtained peaks could be assigned to cloned fragments when using this approach (in contrast to 51.61% for CT with approach B and 12.0% for FS with approach A). (ii) Only one clone library was needed, and therefore costs and time could be saved. (iii) Finally, this approach was less susceptible to PCR errors; since all clones were derived from the initial mixed population PCR product, no additional PCR step was necessary and therefore amplified clone fragments were reliable in their sequence.
The sequencing results and respective RTs of derived fragments on dHPLC are listed in Table 3. Analysis of nucleotide sequences showed that the assignment of a unique peak to a single organism was possible for several peaks with RTs differing by only 0.05 min (or 3 s). Thus, for instance, the pure culture of M. siciliae (DSM 3028) showed an RT of 12.64 min, and for the collected peak Fcm12 (M. siciliae 98%) an RT of 12.65 was obtained. However, similar RTs (RT tolerance < 0.15 min) did not always result in the same identification of the peaks, especially when the sequences of clones derived from different habitats were compared.
TABLE 3.
Sequencing results from collected peaks (FS) or cloned fragments derived from CT and SL and their relationship to sequences in the databasea
| Sample type and ID or cloneb | RT | % GC | Length (bp) | Closest relative | % Identity |
|---|---|---|---|---|---|
| FS | |||||
| F3-2 | 2.58 | 227 | Uncultured euryarchaeote gene | 99 | |
| Fcm12 | 12.60 | 496 | Methanosarcina siciliae | 98 | |
| Fcm15 | 13.77 | 478 | Clostridium sp. | 98 | |
| F3-4 | 14.01 | 369 | Uncultured euryarchaeote gene | 100 | |
| Fa4 | 14.25 | 227 | Uncultured euryarchaeote gene | 99 | |
| Fa6 | 14.25 | 227 | Uncultured euryarchaeote gene | 99 | |
| F3-8 | 14.56 | 496 | No significant results | ||
| F17m13 | 15.02 | 421 | Uncultured bacterium clone M55_D21_H_B_A08 | 97 | |
| F3-2-13 | 16.95 | 751 | Uncultured euryarchaeote isolate DGGE band 672729-MODIFIZIERT | 93 | |
| F3-2-14 | 16.95 | 488 | M. thermoformicicum (SF-4) | 88 | |
| FA1 | 17.27 | 369 | Uncultured euryarchaeote gene | 96 | |
| FA3 | 17.27 | 369 | Uncultured euryarchaeote gene | 100 | |
| F23m | 17.38 | 372 | Uncultured euryarchaeote gene | 99 | |
| CT | |||||
| KA002 | 4.48 | 55.3 | 474 | Uncultured Flavobacterium sp. clone Y17 | 98 |
| KA030 | 15.26 | 55.9 | 478 | Uncultured organism clone ctg_NISA060 | 98 |
| KA039 | 14.35 | 58.9 | 474 | Uncultured bacterium clone PC-PA9-4 | 98 |
| KA061 | 11.80 | 54.6 | 478 | Uncultured bacterium clone l_E07 | 97 |
| KB010 | 10.95 | 493 | Uncultured archaeon gene | 97 | |
| KB024 | 12.29 | 56.1 | 483 | Uncultured archaeon gene | 98 |
| KB029 | 15.07 | 56.3 | 478 | Alphaproteobacterium JL1032 | 98 |
| KB051 | 15.97 | 57.2 | 479 | Bacillus sp. enrichment culture clone 866 ECW 19 | 91 |
| KB073 | 11.03 | 57.5 | 494 | Uncultured archaeon gene | 100 |
| KB104 | 11.77 | 56.7 | 494 | Methanoculleus thermophilus strain JB-1 16S | 99 |
| KB126 | 16.76 | 53.3 | 483 | Streptomyces marokkonensis strain 174443 | 99 |
| KB165 | 19.22 | 57.3 | 496 | Uncultured archaeon gene | 98 |
| KC009 | 12.91 | 53.9 | 498 | Uncultured archaeon gene | 96 |
| KC035 | 19.25 | 60.0 | 493 | Uncultured bacterium clone 101-114 | 98 |
| KC090 | 12.46 | 57.3 | 494 | Uncultured archaeon gene | 99 |
| KC128 | 2.52 | 55.6 | 444 | Uncultured bacterium clone EPR3967-O2-Bc72 | 94 |
| KE033 | 3.00 | 60.0 | 493 | Uncultured bacterium clone 101-114 | 98 |
| KE169 | 19.22 | 60.7 | 476 | Uncultured bacterium clone C6B | 96 |
| SL | |||||
| B008 | 16.10 | 56.7 | 499 | Uncultured planctomycete clone Amb_16S_1421 | 96 |
| B009 | 13.76 | 57.2 | 493 | Uncultured crenarchaeote 03_12a | 99 |
| B014 | 13.44 | 55.8 | 493 | Uncultured crenarchaeote clone OdenE-150iia | 99 |
| B018 | 14.17 | 56.1 | 476 | Uncultured soil bacterium clone 169-2 | 98 |
| B024 | 17.00 | 55.1 | 477 | Uncultured proteobacterium clone Elev_16S_1449 | 98 |
| B032 | 17.64 | 57.5 | 475 | Unidentified bacterium clone 61_H_RHIZO_H1_T7s | 98 |
| B043 | 29.50 | 60.2 | 455 | Uncultured bacterium gene | 96 |
| B046 | 18.91 | 56.0 | 479 | Uncultured bacterium clone NR.1.037 | 97 |
| B103 | 15.64 | 57.2 | 479 | Uncultured bacterium gene | 99 |
| B104 | 16.13 | 55.6 | 478 | Uncultured bacterium clone FCPT661 | 99 |
| B105 | 16.89 | 57.1 | 476 | Acidobacteria bacterium Ellin7137 | 99 |
| B111 | 13.26 | 56.2 | 495 | Uncultured archaeon clone COSAS-F3 | 99 |
| B119 | 17.27 | 56.7 | 476 | Uncultured Verrucomicrobia bacterium clone 172 | 99 |
| B121 | 13.42 | 56.0 | 480 | Uncultured bacterium clone C55_D6_H_B_C12 | 93 |
| B123 | 18.19 | 59.3 | 492 | Uncultured euryarchaeote 03_27e | 99 |
| B127 | 13.74 | 57.6 | 496 | Uncultured crenarchaeote 03_02a | 99 |
| B133 | 13.14 | 56.2 | 495 | Uncultured crenarchaeote 03_06c | 100 |
| B143 | 17.74 | 56.3 | 476 | Uncultured bacterium clone P1mB_062 | 100 |
| B144 | 22.55 | 59.3 | 474 | Uncultured proteobacterium clone Amb_16S_520 | 100 |
| B150 | 12.88 | 57.7 | 494 | Uncultured archaeon gene | 99 |
| B151 | 16.20 | 55.9 | 483 | Uncultured bacterium clone BacB_007 | 99 |
| B158 | 16.97 | 57.4 | 476 | Uncultured alphaproteobacterium clone KF002 | 97 |
| B161 | 20.63 | 60.1 | 494 | Uncultured crenarchaeote 12_30a | 99 |
| B162 | 18.95 | 56.8 | 477 | Uncultured bacterium clone BacC-u_010 | 95 |
| B168 | 17.38 | 57.1 | 476 | Uncultured alphaproteobacterium clone FI-2M_B06 | 99 |
| B169 | 12.95 | 56.2 | 495 | Uncultured crenarchaeote clone OdenB-100iib | 99 |
| B171 | 11.13 | 54.6 | 476 | Uncultured proteobacterium clone Amb | 99 |
| B172 | 12.74 | 57.3 | 494 | Uncultured archaeon gene | 99 |
| B175 | 20.01 | 57.5 | 475 | Uncultured bacterium clone M55_D21_H_B_F10 | 99 |
| B177 | 18.17 | 57.1 | 478 | Uncultured Holophaga sp. | 100 |
| B178 | 17.62 | 57.5 | 477 | Uncultured bacterium clone Elev_16S _1673 | 100 |
| B179 | 12.39 | 54.8 | 476 | Uncultured bacterium clone TSC176 | 99 |
| B180 | 19.12 | 59.7 | 494 | Uncultured crenarchaeote 12_30a | 99 |
| B190 | 17.20 | 57.4 | 477 | Uncultured bacterium clone FCPO684 | 99 |
| B191 | 12.18 | 52.7 | 474 | Uncultured bacterium clone FCPS486 | 99 |
| B205 | 11.45 | 55.9 | 494 | Uncultured archaeon gene | 98 |
| B206 | 12.10 | 57.1 | 494 | Uncultured archaeon gene | 99 |
| B207 | 13.01 | 476 | Uncultured soil bacterium clone 169-2 | 98 | |
| B212 | 14.94 | 57.9 | 496 | Uncultured archaeon clone NAK1-a2 | 97 |
| B217 | 13.22 | 57.5 | 494 | Uncultured archaeon clone GW70-20-11 16S | 100 |
| B218 | 16.29 | 58.6 | 495 | Uncultured crenarchaeote clone E11_ARCH1 | 98 |
| B220 | 17.53 | 57.6 | 479 | Uncultured alphaproteobacterium clone EB1049 | 100 |
| B226 | 12.76 | 55.3 | 474 | Galbibacter mesophilus gene | 98 |
| B229 | 16.28 | 57.0 | 474 | Rhizobiales bacterium Mfc52 gene | 100 |
| B233 | 13.90 | 54.8 | 478 | Uncultured alphaproteobacterium clone lhap30 | 99 |
| B238 | 18.39 | 57.7 | 477 | Uncultured Acidobacteria bacterium clone E05_WMSP1 | 98 |
| B239 | 10.40 | 51.6 | 479 | Uncultured Bacteroidetes bacterium clone AKYG516 | 98 |
| B244 | 16.86 | 57.4 | 479 | Uncultured Hyphomicrobiaceae bacterium clone Elev_16S_1015 | 100 |
| B245 | 17.08 | 58.2 | 478 | Uncultured soil bacterium clone S013 | 100 |
| B260 | 11.63 | 54.0 | 469 | Uncultured archaeon isolate DGGE band 672730-MODIFIZIERT | 87 |
| B261 | 26.29 | 57.2 | 954 | No significant result | |
| B288 | 17.61 | 56.0 | 495 | Uncultured archaeon clone COSAS-F3 16S | 100 |
A total of 208 clones were collected for CT in different time windows, and 289 were collected for SL.
FS, CT, and SL sample types are represented. The identification (ID) is given for the FS samples; the clone is specified for the CT and SL samples.
The great majority of BLAST hits were to uncultured organisms. For instance, in the SL clone library, only one of 54 clones could be assigned to the species level. Thus, a systematic classification was hardly possible, pointing to the difficulties of culture-based methods.
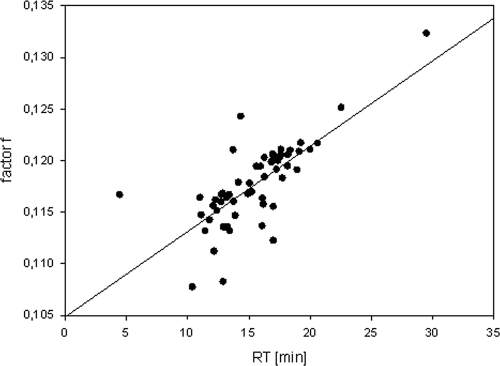
In the SL clone library, archaeal and bacterial amplicons had significantly different fragment lengths (P < 0.001) and GC contents (P < 0.05). The average fragment lengths were 494 bp for clones assigned to the Archaea and 476 bp for clones assigned to the Bacteria, and the GC content was higher in the archaeal amplicons. Barlaan et al. (1) found a correlation of these variables for dHPLC analysis. In the present investigation such a correlation could be confirmed (R2 = 0.418, P < 0.001). When GC content is expressed per base, as f (where f = GC content/fragment length), the correlation is even stronger (R2 = 0.547, P < 0.001) (Fig. 4). This result implies that, unless all fragments in an analysis are the same length (which cannot be guaranteed), a peak with a different RT does not necessarily represent a different organism and vice versa.
FIG. 4.
Influence of fragment length and GC content as represented by factor f plotted against RT (RT [min]). Factor f is calculated by GC content (%)/fragment length. R2 = 0.547, P < 0.001. The plot is derived from the SL clone library with the primer pair 934f/1390GCr.
The highest apparent microbial diversity was found for the CT samples with 31 identified peaks, followed by FS with 25 peaks and the SL sample with 21 peaks (Fig. 3). The RTs of eluting fragments of CT and FS sample were quite similar, whereas the SL sample showed a rather different peak profile: when an RT tolerance of 9 s (or 0.15 min) was taken as a basis, an overlap of 11 peaks was found for CT and FS representing ca. 20% similarity, but only of 5 peaks for CT and SL (ca. 10% similarity) and 7 peaks for SL and FS (ca. 15% similarity). This result is consistent with the origin of the samples since both FS and CT came from a biowaste treatment plant. FS was processed there anaerobically in a digester, and the CT sample represents the FS sample but after an additional aerobic composting for several weeks. However, despite several overlapping peaks in the FS and CT samples, a distinct difference in microbial composition and abundance is obvious, and differences may be due to the influence of the composting process (Fig. 3).
Although dHPLC chromatograms for the CT and SL sample showed one probably highly abundant species, there is more than one in FS. Also, after consideration of the peak areas of the highest peaks differences between the samples were observed. For instance, M. siciliae could be detected as the probably most abundant species in the FS samples, representing ca. 21.3% of total peak area, thus pointing to an anaerobic digester operating with a high acetoclastic methanogen population. In the SL sample Galbibacter mesophilus belonging to the family Flavobacteriaceae was found as the probably most abundant species, a species commonly thought not to be typical for this environment. In contrast, the highest peak for CT could not be classified, representing an uncultured archaeon with 22.3% of total peak area, confirming data of various other studies which also concluded that the abundance of Archaea has often been underestimated thus far.
Conclusion.
dHPLC was shown to work with a conventional HPLC system that is commonly available in microbiological laboratories, using Transgenomic DNASep cartridges. It is thus suitable for broad application in microbial ecology. With minor modification of the HPLC system, there is nearly no limit for its application in molecular (microbial) population analysis. With the creation of clone libraries, the majority of peaks could be related to cloned fragments. Even though the attempt to assign a unique RT to a special amplicon with a defined sequence from a known species, to a cloned amplicon, or to a collected peak failed, a reliable comparison of the microbial communities of various habitats was demonstrated to be possible. We believe that the fast and easy application of conventional PCR products that do not necessarily have to be cleaned up, the high degree of automatization, and the possibility to quantify fragments make the application of dHPLC in microbial ecology superior to other fingerprinting techniques. Apart from that, dHPLC exhibits a comparable sensitivity and even higher resolution than DGGE, if samples from complex and difficult matrices are applied. Possibly, the most striking advantage of dHPLC is the assignment of RTs to different fragments without the need of further graphical processing. The latter is usually inevitable and a source of misinterpretation when analyzing, e.g., DGGE gels. However, since dHPLC is quite new and never before has been applied to such complex and difficult matrices as those described here, there is still potential to improve the method. Considering the work that has been necessary to bring other fingerprinting methods (all of which still have problems) to the level where they are now, we are optimistic concerning the potential of dHPLC. Thus, additional work is needed to optimize not only the dHPLC running conditions but also DNA extraction procedures, as well as PCR.
Acknowledgments
This study was supported by the Austrian FWF in project 193220.
Footnotes
Published ahead of print on 16 December 2008.
REFERENCES
- 1.Barlaan, E. A., M. Sugimori, S. Furukawa, and K. Takeuchi. 2005. Profiling and monitoring of microbial populations by denaturating high-performance liquid chromatography. J. Microbiol. Methods 61:399-412. [DOI] [PubMed] [Google Scholar]
- 2.Coenye, T., and P. Vandamme. 2003. Intragenomic heterogeneity between multiple 16S rRNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 228:45-49. [DOI] [PubMed] [Google Scholar]
- 3.Dahllöf, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frueh, F. W., and M. Noyer-Weidner. 2003. The Use of denaturing high-performance liquid chromatography (DHPLC) for the analysis of genetic variations: impact for diagnostics and pharmacogenetics. Clin. Chem. Lab. Med. 41:452-461. [DOI] [PubMed] [Google Scholar]
- 5.Gjerde, D. T., C. P. Hanna, and D. Hornby. 2002. DNA chromatography. Wiley-VCH, Weinheim, Germany.
- 6.Goldenberg, O., S. Herrmann, G. Marjoram, M. Noyer-Weidner, G. Hong, S. Bereswill, and U. B. Göbel. 2006. Molecular monitoring of the intestinal flora by denaturing high-performance liquid chromatography. J. Microbiol. Methods 68:94-105. [DOI] [PubMed] [Google Scholar]
- 7.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugenholtz, P., C. Pitulle, and K. L. Hershberger. 1998. Novel division level bacteria diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illmer, P., and G. Gstraunthaler. 2008. Effect of seasonal changes in quantities of biowaste on full-scale anaerobic digester performance. Waste Manag. doi: 10.1016/j.wasman.2008.02. [DOI] [PubMed]
- 11.Illmer, P., U. Obertegger, and F. Schinner. 2003. Microbiological properties in acidic forest soils with special consideration of KCl extractable Al. Water Air Soil Pollut. 148:3-14. [Google Scholar]
- 12.Liu, W.-T., T. L. Marsh, H. Cheng, and L. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, W., D. I. Smith, K. J. Rechtzigel, S. N. Thibodeau, and C. D. James. 1998. Denaturing high performance liquid chromatography used in the detection of germline and somatic mutations. Nucleic Acids Res. 26:1396-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malin, C., and P. Illmer. 2007. Ability of DNA-content and DGGE analysis to reflect the performance condition of an anaerobic biowaste fermenter. Microbiol. Res. doi: 10.1016/j.micres.2007.07.004. [DOI] [PubMed]
- 15.Muyzer, G. 1999. Genetic fingerprinting of microbial communities: present status and future perspective, p. 1-10. In C. R. Bell, M. Brylinsky, and P. Johnson-Green (ed.), Proceedings of the 8th International Symposium on Microbial Ecology. Atlantic Canada Society for Microbial Ecology, Halifax, Nova Scotia.
- 16.Muyzer, G., E. C. DeWaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reysenbach, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplicons: formation, consequence, and elimination by “reconditioning PCR.” Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner, A. O., C. Malin, G. Gstraunthaler, and P. Illmer. 2009. Survival of selected pathogens in diluted sludge of a thermophilic waste treatment plant and in NaCl-solution under aerobic and anaerobic conditions. Waste Manag. 29:425-429. [DOI] [PubMed] [Google Scholar]
- 20.Wu, J.-H., W.-T. Liu, I.-C. Tseng, and S.-S. Cheng. 2001. Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiology 147:373-382. [DOI] [PubMed] [Google Scholar]
- 21.Wurzburger, R. J., A. P. Parnassa, S. Jain, J. A. Wexler, J. L. Chu, K. B. Elkon, and R. D. Blank. 2003. Use of GC clamps in dHPLC mutation scanning. Clin. Med. Res. 1:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao, W., and P. J. Oefner. 2001. Denaturating high-performance liquid chromatography: a review. Hum. Mutat. 17:439-474. [DOI] [PubMed] [Google Scholar]