Abstract
In this study, Pseudomonas savastanoi pv. savastanoi isolates were demonstrated to contain two iaaL paralogs, which are both chromosomally located in most strains. Comparative analysis of iaaL nucleotide sequences amplified from these two paralogs revealed that one paralog, iaaLPsn, is 100% identical to iaaL from P. savastanoi pv. nerii, while the other paralog, iaaLPsv, exhibited 93% identity to iaaL from Pseudomonas syringae pv. tomato (iaaLPto). A 3-nucleotide motif (TAC) comprised of 3 to 15 repeats, which remained stable after propagation of the strains in olive plants, was found in iaaLPsv. Based on the observed nucleotide sequence variations, a restriction fragment length polymorphism assay was developed that allowed differentiation among iaaLPsn, iaaLPsv, and iaaLPto. In addition, reverse transcriptase PCR on total RNA from P. savastanoi pv. savastanoi strains demonstrated that both iaaLPsv and iaaLPsn containing 14 or fewer TAC repeats are transcribed. Capillary electrophoresis analysis of PCR-amplified DNA fragments containing the TAC repeats from iaaLPsv allowed the differentiation of P. savastanoi pv. savastanoi isolates.
Infections with Pseudomonas savastanoi pv. savastanoi (46), pv. fraxini, and pv. nerii (13) result in knots and galls on members of the Oleaceae family, and P. savastanoi pv. nerii also causes galls on oleander. Symptoms of infected trees include hypertrophy formation on the stems and branches and occasionally on the leaves and fruits. In the olive, the disease is considered to reduce both olive yield and productivity (41, 42); however, quantitative data on the impact of the disease on crop yield or crop quality are not available (19). Nevertheless, losses can be caused directly by localized infections that inhibit flowering and affect fruit development, as well as taste, and can be caused indirectly by weakening immature main leader branches that result in later damage to the tree frame (48).
Currently, the only molecular P. savastanoi determinants known to be involved in knot development are the phytohormones indoleacetic acid (IAA) and cytokinins (1, 15, 33, 38, 45), as well as biosynthesis of a functional type III secretion system, encoded by the hrp-hrc gene clusters (43, 44). Recently, a global genomic analysis of P. savastanoi pv. savastanoi plasmids allowed the identification of several putative virulence factors in the olive pathogen, including several type III secretion system protein effectors and a variety of genes encoding known Pseudomonas syringae virulence factors (32).
The genetic determinants of P. savastanoi that are involved in the conversion of tryptophan (Trp) to IAA are Trp monooxygenase (encoded by the iaaM gene), which converts Trp to indoleacetamide (IAM), and IAM hydrolase (encoded by the iaaH gene), which catalyzes transformation of IAM to IAA (28). IAA can be further metabolized in P. savastanoi to an amino acid conjugate, 3-indole-acetyl-ɛ-l-lysine (IAA-lysine), through the action of the iaaL gene (14). In P. savastanoi pv. savastanoi and pv. nerii, iaaM and iaaH are organized in an operon with the iaaM locus first in the gene cluster (12). The IAA operon and the iaaL gene reside on a plasmid (pIAA) in P. savastanoi strains isolated from oleander galls (5, 7, 8, 14); however, the direction of iaaL transcription has been determined to be opposite to IAA operon transcription (14). In contrast, most olive isolates have been reported to carry all three IAA genes on the chromosome (5, 8, 32). In a recent study, P. savastanoi pv. savastanoi strains carrying a pIAA plasmid were also suggested to contain chromosomally encoded copies of all three iaa genes (32); however, the exact copy numbers for the three genes in P. savastanoi isolates have not yet been estimated.
Molecular detection of P. savastanoi pv. savastanoi in olive plants is usually performed by an enrichment-PCR assay based on amplification of an internal 454-bp fragment of the iaaL gene followed by HaeIII digestion (29) or by nested PCR followed by dot blot hybridization (2). Although iaaL is widespread among plant-associated bacteria (10, 16), this assay has been designed using the published sequence of iaaL from an oleander isolate (37) and appears to be specific for P. savastanoi (29).
The aim of this study was to develop an improved method for the detection and differentiation of P. savastanoi pv. savastanoi strains based on the existence of nucleotide sequence variations in the iaaL gene. First, we determined the copy numbers of plasmid- and chromosome-borne iaaL sequences for a collection of 30 P. savastanoi pv. savastanoi strains that were isolated in different countries. Then, nucleotide sequence differences, including short tandem repeat sequences (STRs), between iaaL paralogs were analyzed. Based on the observed variations, a PCR-restriction fragment length polymorphism (RFLP) assay was developed that allowed differentiation between iaaL paralogs and transcriptional analysis of paralogs by reverse transcriptase (RT) PCR, followed by RFLP. Capillary electrophoresis analysis of PCR-amplified STRs within the iaaL gene allowed the differentiation of P. savastanoi pv. savastanoi isolates.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and media.
The Pseudomonas strains used in this study (Table 1) were grown at 28°C in Luria-Bertani medium (26) or King's medium B (21), as specified below. P. savastanoi pv. savastanoi identification was performed by LOPAT biochemical tests (22) and olive plant inoculations (30) for all strains included in this study. In addition, amplification and analysis of gyrB and rpoD nucleotide sequences (36, 47) were performed for 10 of the strains (Table 1).
TABLE 1.
Bacterial strains used in this study
| Name and straina | Country (province)b |
iaaL hybridization
|
iaaLPsv size (bp)e | (TAC)nf | Reference or source | |
|---|---|---|---|---|---|---|
| No.c | Classificationd | |||||
| P. syringae pv. tomato | ||||||
| DC3000 | United Kingdom | 1C | 3 | 9 | ||
| P. savastanoi pv. nerii | ||||||
| ITM519 | Italy | 2P | 3 | 45 | ||
| LPVM Psn2 | Italy | 1P | 3 | S. Tegli | ||
| P. savastanoi pv. savastanoi | ||||||
| NCPPB 1506 | Italy | 2C | II | 122 | 3 | 29 |
| ITM 317h | Italy | 2C | 122 | 3 | 45 | |
| PVFi 1 | Italy | 2C | 122 | 3 | 18 | |
| IVIA 2733-1a | Italy | 2C | 122 | 3 | 35 | |
| CFBP 1020 | France | 2C | II | 122 | 3 | 29 |
| CFBP 2074h | Algeria | 2C | 122 | 3 | 29 | |
| NCPPB 1342 | United States | 2C | II | 128 | 5 | 29 |
| NCPPB 1344h | United States | 2C | II | 128 | 5 | 29 |
| NCPPB 3335h | France | 2C | 128 | 5g | 29 | |
| IVIA 1628-3h | Spain (Valencia) | 2C | 128 | 5g | 29 | |
| IVIA 2743-3 | Spain (Badajoz) | 2C | II | 128 | 5 | 35 |
| GUMA Psv160 | Spain (Jaén) | 2P (25-95) | III | 131 | 6 | This study |
| GUMA Psv185 | Spain (Málaga) | 2C | I | 131 | 6 | This study |
| LSV C3.01 | Spain (Sevilla) | 2C | I | 131 | 6g | 32 |
| IVIA 2445-4 | Spain (Jaén) | 1P (70)/1C | II | 131 | 6 | 35 |
| IVIA 1624-b1h | Spain (Valencia) | 1P (75)/1C | II | 131 | 6 | 29 |
| NCPPB 2327 | Italy | 1P (60)/1C | II | 131 | 6 | 29 |
| NCPPB 1479 | Serbia | 1P (65)/1C | II | 131 | 6 | 29 |
| NCPPB 639Th | Serbia | 1P (90)/1C | II | 131 | 6 | 16 |
| LSV C1.01 | Spain (Sevilla) | 2C | II | 137 | 8 | 32 |
| LSV B15.00 | Spain (Sevilla) | 2C | I | 137 | 8 | 32 |
| IVIA 1649-1h | Spain (Castellón) | 2C | I | 137 | 8g | 29 |
| LSV C2.01 | Spain (Sevilla) | 2C | I | 140 | 9 | 31 |
| IVIA 1657-a2 | Spain (Alicante) | 2C | I | 140 | 9 | 29 |
| IVIA 1637-B3h | Spain (Alicante) | 2C | III | 143 | 10g | 29 |
| IVIA 1629-1a | Spain (Valencia) | 2C | I | 149 | 12 | 29 |
| IVIA 1637-C1 | Spain (Alicante) | 2C | I | 151 | 13g | 29 |
| IVIA 1651-c15 | Spain (Alicante) | 2C | I | 155 | 14g | 29 |
| NCPPB 64h | Portugal | 2C | I | 155 | 14 | 29 |
| IVIA 1657-b8h | Spain (Valencia) | 2C | I | 158 | 15g | 29 |
ATCC, American Type Culture Collection, Manassas, VA; CFBP, Collection Francaise des Bacteries Phytopathogenes, Angers, France; IVIA, Instituto Valenciano de Investigaciones Agrarias Collection, Valencia, Spain; LPVM, Laboratorio di Patologia Vegetale Molecolare Collection, Firenze, Italy; LSV, Laboratorio de Sanidad Vegetal Collection, Sevilla, Spain; NCPPB, National Collection of Plant Pathogenic Bacteria, York, United Kingdom; GUMA, Área de Génetica, Universidad de Málaga Collection, Málaga, Spain.
The province of isolation is indicated only for Spanish strains.
Number of chromosomal (C) and plasmid (P)-borne copies of iaaL. The size (in kilobases) of iaaL-hybridizing plasmids is indicated in parentheses.
Classification groups for iaaL hybridization patterns of P. savastanoi pv. savastanoi strains exhibiting a common BglII hybridization band of approximately 8 kb. I, banding pattern exhibits a second hybridization band at 20 kb (Fig. 1, lanes 8 and 10 to 14); II, banding pattern exhibits an additional hybridization band larger than 9.4 kb and smaller than 20 kb (Fig. 1, lanes 6, 7, 16, and 17); and III, banding pattern exhibits an additional hybridization band larger than 23 kb (Fig. 1, lane 9).
Size of PCR-amplified iaaLPsv fragments calculated by capillary electrophoresis.
Estimated number of TAC repeats in the iaaL gene. For P. savastanoi pv. savastanoi strains, the number of TAC repeats refers to the iaaLPsv paralog.
Strain with both iaaL paralogs sequenced. Accession numbers of iaaLPsv fragments are as follows: NCPPB 3335 (EU616802), IVIA 1628-3 (EU616795), LSV C301 (EU616796), IVIA1649-1 (EU616800), IVIA 1637-B3 (EU616799), IVIA 1637-C1 (EU616798), IVIA 1651-c15 (EU616801), and IVIA 1657-b8 (EU616797).
Strain with partially sequenced gyrB and rpoD genes. Nucleotide sequences exhibited 100% identity with those of P. savastanoi pv. savastanoi strain NCPPB 639 (accession numbers AB039514 and AB039469 for rpoD and gyrB, respectively).
General molecular techniques.
Agarose gel electrophoresis and other standard recombinant DNA techniques were performed by standard procedures as described previously (40). Genomic DNA was extracted using the Jet Flex Extraction Kit (Genomed, Löhne, Germany), digested with the appropriate restriction enzymes, and separated by electrophoresis, as described by Pérez-Martínez and coworkers (32). Plasmid DNA minipreps from P. savastanoi were performed and separated through 0.7% agarose gels in 1× Tris-acetate-EDTA, as previously reported (31, 32). A DNA fragment of the iaaL gene, amplified from P. savastanoi pv. savastanoi strain NCPPB 3335 using primers IAAL-F and IAAL-R (29), was labeled with a nonradioactive digoxigenin kit (Roche Diagnostics) and used as a probe, as previously described (32).
PCR-RFLP analysis of iaaL paralogs.
PCR amplification of an internal fragment of the iaaL gene was performed using GoTaq DNA polymerase (Promega Corporation, Madison, WI) and primers IAAL-F and IAAL-R, as previously described (29), with slight modifications. Briefly, the amplification conditions consisted of an initial denaturation step at 94°C for 1 min, followed by 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min and a final extension step at 72°C for 5 min in a Perkin Elmer GeneAmp PCR System 2400 thermocycler. In order to reduce nonspecific DNA products, 5% formamide was added to the reaction mixture. The amplified products were separated by electrophoresis through a 3% Tris-borate-EDTA 1× agarose gel for 4 hours at 80 V. Under these conditions, the target iaaL fragment was amplified not only from P. savastanoi pv. savastanoi and pv. nerii, but also from P. syringae pv. tomato, pv. maculicola, pv. tabaci, and pv. pisi (data not shown). The PCR products were digested with HaeIII, which recognizes GGCC sites, and the resulting fragments were separated by electrophoresis through a 3% agarose gel for 6 hours at 80 V. After electrophoresis, the gels were stained with ethidium bromide.
Sequencing of iaaL fragments.
Primers IAAL-PSN-F (5′-CATATGACTGCCTACGATATGG-3′) and IAAL-PTO-F (5′-CATATGACTGCCTACGATGTA-3′) were designed to specifically amplify a fragment from the iaaL paralog iaaLPsn or iaaLPsv (see below), respectively, in combination with IAAL-R (29). Amplicons obtained using both primer pairs from eight different P. savastanoi pv. savastanoi isolates (Table 1) were sequenced using primers IAAL-F and IAAL-R (29). The amplified DNA fragments were sequenced on both strands by the dideoxy sequencing termination method using an ABI Prism 310 (Applied Biosystem) sequencer. Contiguous DNA sequences were assembled using Seqman (DNAStar) and compared with sequences in public databases using NCBI BLAST.
Capillary electrophoresis to assess the copy numbers and stabilities of tandem repeats.
Determination of the number of tandem TAC repeats in the iaaLPsv allele and stability analysis of repeats in P. savastanoi pv. savastanoi strains was performed by capillary electrophoresis as follows. Total-DNA samples isolated from P. savastanoi pv. savastanoi strains or enriched bacterial cultures recovered from olive knots were used as a template in a PCR with primers IAAL-F (29) and 6-carboxyfluorescein (6-FAM)-labeled primer IAAL-6-FAM (5′-6-FAM-TTGGGCAGCGATCAC-3′). The amplification products, which varied in size from 122 bp to 158 bp depending on the iaaL allele and strain, were analyzed using an ABI3130 genetic analyzer, a LIZ500 size standard, and GeneMapper v. 3.7 software (Applied Biosystems). To determine the stability of the number of tandem TAC repeats in plants, P. savastanoi pv. savastanoi strains were inoculated into olive plants as previously described (31). After a period of 7 (IVIA 1628-3, IVIA 2445-4, IVIA 2733-1a, IVIA 2743-3, and NCPPB 639) or 12 (CFBP 2074, ITM 317, NCPPB 64, and NCPPB 1344) months, P. savastanoi pv. savastanoi strains were recovered from the developed knots, as described by Quesada et al. (34, 35), and grown overnight in LB medium for bacterial enrichment.
Preparation of RNA and RT-PCR-RFLP.
Pure bacterial cultures were grown overnight at 28°C in either LB or minimal SSM medium (25), diluted in the same medium to an optical density at 600 nm of 0.1, and incubated at the same temperature until the cultures reached a turbidity of 0.4 (exponential phase) or approximately 1.0 (stationary phase). Isolation of RNA from bacterial cultures was performed as described by Castillo and coworkers (6). The RNA concentration was determined using a Nanodrop ND-1000 (NanoDrop Technologies, Wilmington, DE), and RNA integrity was assessed by agarose gel electrophoresis. RT-PCR was performed with 100 ng RNA in a final volume of 50 μl using the Tital OneTube RT-PCR system, according to the manufacturer's instructions (Roche Diagnostics), with primers IAAL-F and IAAL-R. A 40-cycle amplification program (94°C for 30 s, 57°C for 30 s, and 68°C for 1 min) was performed, followed by a final extension cycle at 68°C for 7 min. Positive control reactions, containing total DNA isolated from the corresponding bacterial strain, were included in all assays.
RESULTS
P. savastanoi pv. savastanoi contains two paralogs of the iaaL gene.
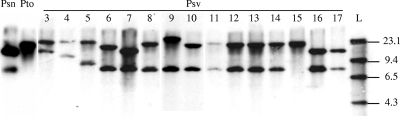
Hybridization analysis of P. savastanoi plasmids with an iaaL probe revealed that 5 of the 30 P. savastanoi pv. savastanoi isolates tested, as well as the P. savastanoi pv. nerii strain LPVM Psn2, contained a native plasmid that hybridized with the probe. In addition, GUMA-Psv160 and P. savastanoi pv. nerii strain ITM 519 contained two different plasmids that hybridized to the probe (Table 1). While BglII-digested total DNA isolated from P. syringae pv. tomato DC3000 resulted in a single band of approximately 20 kb (Fig. 1), as expected from the published genome sequence of the strain (4), total DNA isolated from P. savastanoi yielded two clearly distinguishable hybridization bands for most of the strains tested (28 out of 30 P. savastanoi pv. savastanoi strains and P. savastanoi pv. nerii strain ITM 519) (Table 1 and Fig. 1). Interestingly, a common BglII hybridization band of approximately 8 kb was detected in 24 P. savastanoi pv. savastanoi strains (80%), indicating that most of these strains carry a copy of iaaL in similar or identical locations (Fig. 1). Three hybridization patterns were found within this group of strains: group I, composed of 11 strains isolated in the Iberian Peninsula (Fig. 1, lanes 8 and 10 to 14); group II, including all P. savastanoi pv. savastanoi strains carrying only one plasmid-borne iaaL sequence (Fig. 1, lanes 6, 7, 16, and 17); and group III, composed of two strains (Fig. 1, lane 9). The remaining six strains (20%) exhibited a strain-specific hybridization pattern that was different from those found in these three groups (Fig. 1, lanes 3, 4, 5, and 15, and Table 1).
FIG. 1.
Detection of iaaL gene sequences in P. savastanoi strains. Shown is Southern blot analysis of BglII-digested total DNA using an iaaL probe. Lane 1 (Psn) is P. savastanoi pv. nerii ITM 519, and lane 2 (Pto) is P. syringae pv. tomato DC3000. Lanes 3 to 17 (Psv) are P. savastanoi pv. savastanoi strains. Lanes: 3, CFBP 2074; 4, PVF 1; 5, IVIA 1628-3; 6, NCPPB 1344; 7, IVIA 1624-b1; 8, LSV C2.01; 9, IVIA 1637-B3; 10, IVIA 1637-C1; 11, IVIA 1629-1a; 12, IVIA 1651-c15; 13, NCPPB 64; 14, IVIA 1657-b8; 15, IVIA 2733-1a; 16, IVIA 2743-3; and 17, IVIA 2445-4. L, HindIII-digested λ phage DNA. The positions (in kilobases) of the molecular size markers are indicated on the right of the gel.
Total isolated DNA from P. savastanoi pv. savastanoi strains yielding a single hybridization band (Fig. 1, lane 15) was digested with alternative restriction enzymes (BamHI and HindIII) and hybridized again with the iaaL probe. The hybridization results confirmed the existence of two different DNA fragments that hybridized to the probe for all 30 P. savastanoi pv. savastanoi strains tested (Table 1). All of these results indicate that most P. savastanoi pv. savastanoi isolates (24 out of 30 strains) contain at least two chromosomally located copies of iaaL.
Differentiation of iaaL paralogs in P. savastanoi pv. savastanoi by PCR-RFLP.
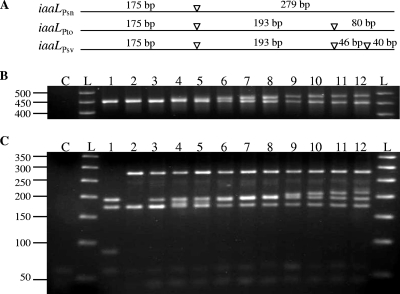
The nucleotide sequences of iaaL genes that were currently annotated included only a plasmid-borne open reading frame from P. savastanoi pv. nerii strain EW2009 (iaaLPsn; accession number M35373) and the chromosomally located ortholog from P. syringae pv. tomato strain DC3000 (iaaLPto; accession number NC_004578). The internal 454-bp fragment of this gene used for PCR identification of P. savastanoi pv. savastanoi (29) contains one and two HaeIII sites in iaaLPsn and iaaLPto, respectively (Fig. 2A). The observed differences between these two ortholog sequences were further confirmed by PCR-RFLP (Fig. 2).
FIG. 2.
PCR-RFLP differentiation of the iaaL gene sequences from P. syringae and P. savastanoi strains. (A) Schematic representation of iaaL DNA fragments amplified by PCR from the indicated strains using primers IAAL-F and IAAL-R. Target positions for HaeIII are indicated by arrowheads. iaaLPsn, P. savastanoi pv. nerii EW-2009; iaaLPto, P. syringae pv. tomato DC3000; and iaaLPsv, P. savastanoi pv. savastanoi ITM 317. (B and C) Gel electrophoresis (3% agarose) of iaaL amplicons and HaeIII-digested amplicons, respectively. Lane 1, P. syringae pv. tomato DC3000; lane 2, P. savastanoi pv. nerii strain LPVM Psn2. Lanes 3 to 12 are P. savastanoi pv. savastanoi strains. Lanes: 3, CFBP 2074; 4, IVIA 1628-3; 5, IVIA 1624-b1; 6, IVIA 1649-1; 7, LSV C2.01; 8, IVIA 1637-B3; 9, IVIA 1629-1a; 10, IVIA 1637-C1; 11, IVIA 1651-c15; and 12, IVIA 1657-b8. Lane C, negative control without template. A fragment of approximately 60 bp, most likely formed by nonspecific primer annealing, is visible in all samples, including C. Lane L, 50-bp DNA step ladder (Promega Co.); the positions (in base pairs) of the molecular size markers are indicated on the left of the gel.
PCR amplification of the 454-bp iaaL fragments from some of the tested P. savastanoi pv. savastanoi isolates yielded an additional fragment of variable size, which was always greater than 454 bp (Fig. 2B, lanes 6 to 12). Further HaeIII digestion of these amplicons revealed RFLP profiles of at least four fragments, with two of them identical to those generated by iaaLPsn. Additionally, all P. savastanoi pv. savastanoi isolates exhibited a 193-bp fragment, also generated by iaaLPto, and a fragment of approximately 45 bp. Moreover, some of the tested strains exhibited an additional fragment of variable size (between 175 bp and 250 bp). Together, these results suggest that the P. savastanoi pv. savastanoi isolates contain two iaaL paralogs, one paralog with a PCR-RFLP profile that is identical to that of iaaLPsn and a second paralog with a variable profile among strains, which shows some similarities to that of iaaLPto and is referred to hereafter as iaaLPsv (Fig. 2).
Identification of a trinucleotide repeat sequence, (TAC)n, in the iaaLPsv paralog of P. savastanoi pv. savastanoi.
DNA fragments from iaaLPsn and iaaLPsv were specifically amplified and sequenced for eight P. savastanoi pv. savastanoi strains (Table 1). While the nucleotide sequences of the iaaLPsn amplicons were 100% identical to that of iaaLPsn from P. savastanoi pv. nerii strain EW2009, iaaLPsv paralogs exhibited a variable number (n) of trinucleotide TAC tandem repeats, which were located in frame and immediately after a (TAC)3 sequence encoding three consecutive tyrosine residues (Tyr78 to Tyr80) in both iaaLPto and iaaLPsn. Except for these additional TAC codons, the nucleotide sequences of all eight iaaLPsv paralogs were 100% identical and exhibited 93% identity to that of iaaLPto. Nucleotide variations found between iaaLPto from P. syringae pv. tomato strain DC3000 and iaaLPsv from these eight P. savastanoi pv. savastanoi strains demonstrated the existence of an additional HaeIII site in iaaLPsv, which is located 40 nucleotides upstream from the 3′ end of this amplicon (Fig. 2A). Accession numbers of iaaLPsv for these eight strains are shown in Table 1.
Determination of the number of tandem TAC repeats and repeat stability in P. savastanoi pv. savastanoi iaaLPsv.
Determination of the number of tandem TAC repeats in the iaaLPsv allele for the remaining 22 P. savastanoi pv. savastanoi strains was performed using capillary electrophoresis. While the primers designed for this purpose, which are identical to the sequence flanking (TAC)n for both iaaLPsn and iaaLPsv, amplified a fragment of 122 bp from iaaLPsn, the sizes of the fragments amplified from iaaLPsv varied between 122 (n = 3) and 158 (n = 15) bp among P. savastanoi pv. savastanoi strains (Table 1). Figure S1A in the supplemental material shows the chromatograms obtained for 10 of the tested P. savastanoi pv. savastanoi strains. Interestingly, P. savastanoi pv. savastanoi strains containing eight or more TAC repeats (59%) were all isolated in the Iberian Peninsula (Spain, 10 strains; Portugal, 1 strain). In comparison, all six strains with a plasmid-borne iaaL gene contain six TAC repeats in iaaLPsv (Table 1). Purification of pIAA plasmids from the five strains containing only one plasmid-borne iaaL sequence, followed by PCR-RFLP analysis, demonstrated that iaaLPsn was found on a plasmid in all five strains (data not shown). Thus, iaaLPsv is located on the chromosome for these five strains.
The stability of the TAC repeats in olive knots was tested for nine P. savastanoi pv. savastanoi strains (see Materials and Methods). The sizes of the iaaL fragments amplified for all these strains were identical to those of the corresponding wild-type strains, indicating that the STR unit remains stable under the conditions tested. Figure S1B in the supplemental material shows the results obtained for four of these strains.
Transcriptional analysis of iaaLPsn and iaaLPsv in P. savastanoi pv. savastanoi.
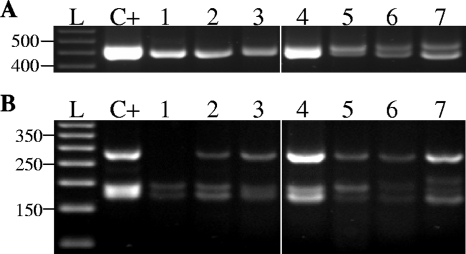
Transcriptional analyses of both iaaLPsv and iaaLPsn were performed for five different P. savastanoi pv. savastanoi strains. The expected product sizes for iaaLPsn (454 bp) and iaaLPsv (from 454 bp to 490 bp, depending on the number of TAC repeats) were obtained for all strains tested, and the transcript levels for each strain in LB medium (data not shown) and minimal medium (Fig. 3) were similar. Digestion of RT-PCR products with HaeIII resulted in the expected RFLP profile for all strains tested, further demonstrating expression of both iaaL paralogs in these strains, regardless of the number of TAC repeats (from n = 3 to n = 14) contained in iaaLPsv. A higher intensity of the amplified RT-PCR product, reflected in the intensity of RFLP fragments corresponding to iaaLPsn (279 bp and 175 bp), was observed for NCPPB 639. As iaaLPsn is plasmid borne in this strain, these results suggest a higher level of expression of iaaLPsn located on the plasmid than for the chromosomally located iaaLPsv. No differential expression between iaaL paralogs was found for strains containing both alleles located on the chromosome (Fig. 3).
FIG. 3.
Transcriptional analysis of iaaLPsn and iaaLPsv in P. savastanoi pv. savastanoi. Gel electrophoresis (3% agarose) of RT-PCR amplicons of iaaL genes (A) and HaeIII-digested iaaL amplicons shown in panel A (B) are shown. Lane 1, P. syringae pv. tomato DC3000. Lanes 2 to 6 correspond to P. savastanoi pv. savastanoi strains. Lanes: 2, ITM 317; 3, NCPPB 3335; 4, NCPBB 639; 5, IVIA 1637-B3; 6, IVIA 1637-C1; and 7, NCPPB 64. Lane C+, positive control RT-PCR amplification reaction with total DNA of NCPPB 3335 as the template. Lane L, 50-bp DNA ladder (Promega Co.); the positions (in base pairs) of the molecular size markers are indicated on the left of the gel.
DISCUSSION
In this study, we demonstrated that P. savastanoi pv. savastanoi isolates contain two iaaL paralogs that are both chromosomally located in most strains analyzed (80%) (Table 1). This high frequency of chromosome-borne iaaL genes in P. savastanoi olive isolates could be a response to the need for stabilization of the virulence factor (24, 49) through mechanisms of integration into the host chromosome. In fact, P. savastanoi pIAA plasmids that have been extensively characterized thus far belong to the pPT23A family of plasmids (32, 49), and the maintenance and evolution of these plasmids is probably based on horizontal transfer and recombination events, which result in either inclusion of plasmid-borne genes on the chromosome or gene duplications (24, 49). Interestingly, iaaL, iaaM, and iaaH seem to coevolve in concert, since all three genes have been found in the same replicon and copy number for some of the P. savastanoi pv. savastanoi strains included in this study (32). Most analyzed P. savastanoi pv. savastanoi strains (80%) contain an iaaL paralog at similar or identical chromosomal locations (Fig. 1 and Table 1). Moreover, purification of DNA fragments (6.5-kb to 9.0-kb) from BglII-digested total DNAs isolated from several of these P. savastanoi pv. savastanoi strains and further PCR-RFLP analysis demonstrated that this allele was iaaLPsv (data not shown). This suggests that iaaLPsn, which was found on the pIAA plasmids of P. savastanoi pv. nerii and pv. savastanoi (Table 1), has stabilized more recently into the P. savastanoi pv. savastanoi genome.
Microsatellites, defined as tracts of DNA that are composed of multiple tandem repeats of a motif with a length between 1 and 6 nucleotides, are ubiquitous in prokaryotes and eukaryotes (11, 17, 23). Capillary electrophoresis analysis of STR numbers in the iaaLPsv allele (Table 1; see Fig. S1 in the supplemental material) allowed the differentiation of P. savastanoi pv. savastanoi strains isolated from different geographical locations. For instance, strains containing eight or more TAC repeats were all isolated in the Iberian Peninsula (11 strains) (Table 1). Moreover, 7 of these 11 strains were isolated from the Valencian Community (Valencia, Alicante, and Castellón Provinces), and three were from the Sevilla Province (n = 8 or 9). However, no correlation between the number of TAC repeats and the cultivar or orchard of isolation (35) was found for strains isolated within the same Spanish province. Determination of the number of STRs in a higher number of P. savastanoi pv. savastanoi strains isolated from different geographical locations, as well as from P. savastanoi pv. savastanoi strains isolated from unknown olive knot samples, would be necessary in order to evaluate the potential of this marker for epidemiological studies of olive knot disease. The stability of the number of STRs at 7 to 12 months after inoculation in olive plants (see Fig. S1B in the supplemental material) further supports the suitability of this marker for strain differentiation. However, STR stability under various stress conditions, which is known to induce STR modification in other bacterial phytopathogens (20, 39), was not analyzed in this study.
The expansion and contraction of the number of STRs in protein-coding regions as a result of slipped-strand mispairing during DNA synthesis (3, 20) is related to bacterial adaptation to environmental changes. In fact, functional repeats have been found in or near antigenic-determinant genes or genes for other virulence factors in bacterial pathogens (27), including bacterial phytopathogens (20). Taking into account that the TAC motif identified in iaaLPsv was found in frame for all strains analyzed and that both iaaL paralogs were transcribed (Fig. 3), translation of the corresponding mRNAs containing this motif would result in IAA-lysine synthetase enzymes that contain 3 to 15 consecutive tyrosine residues. Given that all data on this enzyme are restricted to proteins with only three TAC repeats and crude extracts of P. savastanoi oleander and olive strains (14, 15), which were not included in this study, the activities of IAA-lysine synthetase enzymes carrying a higher number of consecutive tyrosine repeats cannot be determined. Alteration and inactivation of IAA-lysine synthetase activity in P. savastanoi pv. savastanoi would probably have an effect on the amount of IAA and the virulence of this bacterial pathogen, as described for P. savastanoi pv. nerii strains (15). Comparative virulence analysis of P. savastanoi pv. savastanoi strains included in this study and determination of the levels of IAA produced by these strains would be necessary to verify this hypothesis.
Supplementary Material
Acknowledgments
We thank J. Murillo for assistance with Southern hybridizations. We also thank J. Porta and J. M. Porta for help with capillary electrophoresis. We are grateful to M. Duarte and L. Cruzado for excellent technical assistance. A. Barceló-Munóz and S. Tegli are thanked for providing us with olive explants and P. savastanoi pv. nerii strain LPVM Psn2, respectively.
This project was supported by Spanish MCYT grant AGL2005-02090, MICINN grant AGL2008-05311-C02-02, and Junta de Andalucía grant CVI-264. I.M.M. was supported by the Ramón Areces Foundation (Spain).
Footnotes
Published ahead of print on 19 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akiyoshi, D. E., D. A. Regier, and M. P. Gordon. 1987. Cytokinin production by Agrobacterium and Pseudomonas spp. J. Bacteriol. 169:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolini, E., R. Penyalver, A. García, A. Olmos, J. M. Quesada, M. Cambra, and M. M. López. 2003. Highly sensitive detection of Pseudomonas savastanoi pv. savastanoi in asymptomatic olive plants by nested-PCR in a single closed tube. J. Microbiol. Methods 52:261-266. [DOI] [PubMed] [Google Scholar]
- 3.Bichara, M., J. Wagner, and I. B. Lambert. 2006. Mechanisms of tandem repeat instability in bacteria. Mutat. Res. 598:144-163. [DOI] [PubMed] [Google Scholar]
- 4.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van-Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caponero, A., A. M. Contesini, and N. S. Iacobellis. 1995. Population diversity of Pseudomonas syringae subsp. savastanoi on olive and oleander. Plant Pathol. 44:848-855. [Google Scholar]
- 6.Castillo, T., J. L. Ramos, J. J. Rodríguez-Herva, T. Fuhrer, U. Sauer, and E. Duque. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 189:5142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comai, L., and T. Kosuge. 1980. Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J. Bacteriol. 143:950-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comai, L., G. Surico, and T. Kosuge. 1982. Relation of plasmid DNA to indoleacetic-acid production in different strains of Pseudomonas syringae pv. savastanoi. J. Gen. Microbiol. 128:2157-2163. [Google Scholar]
- 9.Cuppels, D. A. 1986. Genetation and caracterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fett, W. F., S. F. Osman, and M. F. Dunn. 1987. Auxin production by plant-pathogenic pseudomonads and xanthomonads. Appl. Environ. Microbiol. 53:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field, D., and C. Wills. 1996. Long, polymorphic microsatellites in simple organisms. Proc. R. Soc. Lond. 263:209-215. [DOI] [PubMed] [Google Scholar]
- 12.Gaffney, T. D., O. da Costa e Silva, T. Yamada, and T. Kosuge. 1990. Indoleacetic acid operon of Pseudomonas syringae subsp. savastanoi: transcription analysis and promoter identification. J. Bacteriol. 172:5593-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardan, L., C. Bollet, M. Abughorrah, F. Grimont, and P. A. D. Grimont. 1992. DNA relatedness among the pathovar strains of Pseudomonas syringae subsp. savastanoi Janse (1982) and proposal of Pseudomonas savastanoi sp. nov. Int. J. Syst. Bacteriol. 42:606-612. [Google Scholar]
- 14.Glass, N. L., and T. Kosuge. 1986. Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas savastanoi subsp. savastanoi. J. Bacteriol. 166:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass, N. L., and T. Kosuge. 1988. Role of indoleacetic acid lysine synthetase in regulation of indoleacetic-acid pool size and virulence of Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 170:2367-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glickmann, E., L. Gardan, S. Jacquet, S. Hussain, M. Elasri, A. Petit, and Y. Dessaux. 1998. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant-Microbe Interact. 11:156-162. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, J. M. 1996. Simple sequences and the expanding genome. Bioessays 18:421-425. [DOI] [PubMed] [Google Scholar]
- 18.Iacobellis, N. S., A. Sisto, and G. Surico. 1993. Occurrence of unusual strains of Pseudomonas syringae subsp. savastanoi on olive in central Italy. Bull. OEPP 23:429-435. [Google Scholar]
- 19.Iacobellis, N. S. 2001. Olive knot, p. 713-715. In O. C. Maloy and T. D. Murray (ed.), Encyclopedia of plant pathology. John Wiley and Sons, New York, NY.
- 20.Jock, S., T. Jacob, W. S. Kim, M. Hildebrand, H. P. Vosberg, and K. Geider. 2003. Instability of short-sequence DNA repeats of pear pathogenic Erwinia strains from Japan and Erwinia amylovora fruit tree and raspberry strains. Mol. Genet. Genom. 268:739-749. [DOI] [PubMed] [Google Scholar]
- 21.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two single media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 22.Lelliot, R. A., E. Billing, and A. C. Hayward. 1966. A determinative scheme for fluorescent plant pathogenic bacteria. J. Appl. Bacteriol. 29:470-478. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y. C., A. B. Korol, T. Fahima, and E. Nevo. 2004. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 21:991-1007. [DOI] [PubMed] [Google Scholar]
- 24.Ma, Z. H., J. J. Smith, Y. Z. Zhao, R. W. Jackson, D. L. Arnold, J. Murillo, and G. W. Sundin. 2007. Phylogenetic analysis of the pPT23A plasmid family of Pseudomonas syringae. Appl. Environ. Microbiol. 73:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, J. M., and M. A. Abdallah. 1978. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physiochemical properties. J. Gen. Microbiol. 107:319-328. [Google Scholar]
- 26.Miller, J. H. (ed.). 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 27.Moxon, R., C. Bayliss, and D. Hood. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40:307-333. [DOI] [PubMed] [Google Scholar]
- 28.Patten, C. L., and B. R. Glick. 1996. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42:207-220. [DOI] [PubMed] [Google Scholar]
- 29.Penyalver, R., A. García, A. Ferrer, E. Bertolini, and M. M. López. 2000. Detection of Pseudomonas savastanoi pv. savastanoi in olive plants by enrichment and PCR. Appl. Environ. Microbiol. 66:2673-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penyalver, R., A. García, A. Ferrer, E. Bertolini, J. M. Quesada, C. I. Salcedo, J. Piquer, J. Pérez-Panadés, E. A. Carbonell, C. del Río, J. M. Caballero, and M. M. López. 2006. Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculations and its use for evaluation of olive cultivar susceptibility. Phytopathology 96:313-319. [DOI] [PubMed] [Google Scholar]
- 31.Pérez-Martínez, I., L. Rodríguez-Moreno, I. M. Matas, and C. Ramos. 2007. Strain selection and improvement of gene transfer for genetic manipulation of Pseudomonas savastanoi isolated from olive knots. Res. Microbiol. 158:60-69. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Martínez, I., Y. Zhao, J. Murillo, G. W. Sundin, and C. Ramos. 2008. Global genomic analysis of Pseudomonas savastanoi pv. savastanoi plasmids. J. Bacteriol. 190:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell, G. K., and R. O. Morris. 1986. Nucleotide sequence and expression of a Pseudomonas savastanoi cytokinin biosynthetic gene—homology with Agrobacterium tumefaciens tmr and tzs loci. Nucleic Acids Res. 14:2555-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quesada, J. M., A. García, E. Bertolini, M. M. López, and R. Penyalver. 2007. Recovery of Pseudomonas savastanoi pv. savastanoi from symptomless shoots of naturally infected olive trees. Int. Microbiol. 10:77-84. [PubMed] [Google Scholar]
- 35.Quesada, J. M., I. Pérez-Martínez, C. Ramos, M. M. López, and R. Penyalver. 2008. IS53: an insertion element for molecular typing of Pseudomonas savastanoi pv. savastanoi. Res. Microbiol. 159:207-215. [DOI] [PubMed] [Google Scholar]
- 36.Rico, A., R. López, C. Asensio, M. Aizpún, S. Asensio, C. Manzanera, and J. Murillo. 2003. Nontoxigenic strains of P. syringae pv. phaseolicola are a main cause of halo blight of beans in Spain and escape current detection methods. Phytopathology 93:1553-1559. [DOI] [PubMed] [Google Scholar]
- 37.Roberto, F. F., H. Klee, F. White, R. Nordeen, and T. Kosuge. 1990. Expression and fine structure of the gene encoding N-indole-3-acetyl-l-lysine synthetase from Pseudomonas savastanoi. Proc. Natl. Acad. Sci. USA 87:5797-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodríguez-Moreno, L., A. Barceló-Muñoz, and C. Ramos. 2008. In vitro analysis of the interaction of Pseudomonas savastanoi pvs. Phytopathology 98:815-822. [DOI] [PubMed] [Google Scholar]
- 39.Ruppitsch, W., A. Stöger, and M. Keck. 2004. Stability of short sequence repeats and their application for the characterization of Erwinia amylovora strains. FEMS Microbiol. Lett. 234:1-8. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schroth, M. N., D. Hildebra, and H. J. Oreilly. 1968. Off-flavor of olives from trees with olive knot tumors. Phytopathology 58:524-529. [Google Scholar]
- 42.Schroth, M. N., J. W. Osgood, and T. D. Miller. 1973. Quantitative assessment of effect of olive knot disease on olive yield and quality. Phytopathology 63:1064-1065. [Google Scholar]
- 43.Sisto, A., M. G. Cipriani, and M. Morea. 2004. Knot formation caused by Pseudomonas syringae subsp. savastanoi on olive plants is hrp-dependent. Phytopathology 94:484-489. [DOI] [PubMed] [Google Scholar]
- 44.Sisto, A., M. Morea, F. Zaccaro, G. Palumbo, and N. S. Iacobellis. 1999. Isolation and characterization of Pseudomonas syringae subsp. savastanoi mutants defective in hypersensitive response elicitation and pathogenicity. J. Phytopathol. 147:321-330. [Google Scholar]
- 45.Surico, G., N. S. Iacobellis, and A. Sisto. 1985. Studies on the role of indole-3-acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. savastanoi. Physiol. Plant Pathol. 26:309-320. [Google Scholar]
- 46.Vivian, A., and J. Mansfield. 1993. A proposal for a uniform genetic nomenclature for avirulence genes in phytopathogenic pseudomonads. Mol. Plant-Microbe Interact. 6:9-10. [Google Scholar]
- 47.Yamamoto, S., H. Kasai, D. L. Arnold, R. W. Jackson, A. Vivian, and S. Harayama. 2000. Phylogeny of the genus Pseudomonas: intragenic structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385-2394. [DOI] [PubMed] [Google Scholar]
- 48.Young, J. M. 2004. Olive knot and its pathogen. Australas. Plant Pathol. 33:33-39. [Google Scholar]
- 49.Zhao, Y., A. K. Hull, N. R. Gupta, K. A. Goss, J. Alonso, J. R. Ecker, J. C. Normanly, and J. L. Celenza. 2002. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16:3100-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.