Abstract
A total of 103 root nodule isolates were used to estimate the diversity of bacteria nodulating Lotus tenuis in typical soils of the Salado River Basin. A high level of genetic diversity was revealed by repetitive extragenic palindromic PCR, and 77 isolates with unique genomic fingerprints were further differentiated into two clusters, clusters A and B, after 16S rRNA restriction fragment length polymorphism analysis. Cluster A strains appeared to be related to the genus Mesorhizobium, whereas cluster B was related to the genus Rhizobium. 16S rRNA sequence and phylogenetic analysis further supported the distribution of most of the symbiotic isolates in either Rhizobium or Mesorhizobium: the only exception was isolate BA135, whose 16S rRNA gene was closely related to the 16S rRNA gene of the genus Aminobacter. Most Mesorhizobium-like isolates were closely related to Mesorhizobium amorphae, Mesorhizobium mediterraneum, Mesorhizobium tianshanense, or the broad-host-range strain NZP2037, but surprisingly few isolates grouped with Mesorhizobium loti type strain NZP2213. Rhizobium-like strains were related to Rhizobium gallicum, Rhizobium etli, or Rhizobium tropici, for which Phaseolus vulgaris is a common host. However, no nodC or nifH genes could be amplified from the L. tenuis isolates, suggesting that they have rather divergent symbiosis genes. In contrast, nodC genes from the Mesorhizobium and Aminobacter strains were closely related to nodC genes from narrow-host-range M. loti strains. Likewise, nifH gene sequences were very highly conserved among the Argentinian isolates and reference Lotus rhizobia. The high levels of conservation of the nodC and nifH genes suggest that there was a common origin of the symbiosis genes in narrow-host-range Lotus symbionts, supporting the hypothesis that both intrageneric horizontal gene transfer and intergeneric horizontal gene transfer are important mechanisms for the spread of symbiotic capacity in the Salado River Basin.
The so-called Salado River Basin in Buenos Aires Province is the most important region devoted to beef and dairy cattle production in Argentina, and natural and cultivated grasslands are the main forage resource in this area (43). The growth and productivity of pastures in this region are limited by alternating cycles of extreme water excess and drought conditions. Soils in the Salado River Basin are heterogeneous and poorly drained and have low nutrient contents, high levels of sodic salts, and alkaline pHs (38, 57).
Legumes have the ability to establish mutualistic symbiotic relationships with soil bacteria collectively known as rhizobia, and these relationships allow the legumes to be independent of nitrogen levels in the soil. Lotus tenuis is a valuable forage legume native to the Mediterranean region which has become naturalized in the Salado River Basin during the last few decades and has shown good potential for adaptation to the soils there (32, 44). Thus, cultivation of L. tenuis could contribute to improvements in forage quality and production in the Salado River Basin.
It is generally accepted that Lotus species establish highly specific nitrogen-fixing symbioses with bacteria belonging to the genera Mesorhizobium and Bradyrhizobium (30, 31, 33). Based on the rhizobial partners, two groups can be distinguished in the genus Lotus. One of these groups includes Lotus corniculatus and L. tenuis, two species that form nitrogen-fixing nodules only with bacteria belonging to the genus Mesorhizobium (particularly Mesorhizobium loti). The second group comprises species like Lotus subbiflorus and Lotus uliginosus, which establish nitrogen-fixing nodules mainly in association with slow-growing Bradyrhizobium-like strains (4, 8, 28, 45). As a general rule, the rhizobia nodulating Lotus species have a narrow host range, but some Mesorhizobium strains (e.g., strain NZP2037) are thought to have a broad host range since they can form nitrogen-fixing nodules on most Lotus species. However, the symbiotic effectiveness of broad-host-range mesorhizobia is low compared with that of narrow-host-range strains (6, 50, 60).
Legume inoculants are usually based on selected highly efficient rhizobia and evaluated in particular environments. However, inoculant success is frequently limited by the presence of native soil rhizobia (14, 64). Inoculation often leads to improved productivity in soils with no previous history of the host legume where native rhizobial populations are small or nonexistent, whereas inoculant success is uncertain in soils where the native rhizobial populations are large. This is frequently due to the superior competitive ability of native strains, which occupy the majority of nodules under field conditions because of their large populations, their distribution throughout the soil profile, or their better adaptation to the local soil environment (14, 64).
Inoculation of L. tenuis with selected rhizobia is expected to increase forage production in the Salado River Basin. However, knowledge about native rhizobia nodulating Lotus spp. in Argentinean soils is sparse. A preliminary study of rhizobia able to nodulate L. tenuis in the Salado River Basin suggested that there is considerable diversity (20). However, detailed information regarding taxonomic and physiological features of native rhizobial strains in this region is not available. The aim of the present work was to characterize bacteria able to nodulate L. tenuis that were obtained from three different soil environments typical of the Salado River Basin.
MATERIALS AND METHODS
Collection of rhizobia and culture conditions.
Samples from three soil environments typical of the Salado River Basin (saline lowlands, nonsaline lowlands, and transitional plains) were composed of mixtures of subsamples taken at several sites located in Chascomús County in Buenos Aires Province. Soils samples were transported from the field to the laboratory, stored at 4°C for 2 days, and then used for recovery of rhizobia by using L. tenuis cultivar Pampa INTA as the trap plant. The isolates obtained are listed in Table 1. Yeast extract-mannitol medium was routinely used for rhizobial isolation, purification, and culture (71). All strains were stored at −80°C in the same medium with 20% (vol/vol) glycerol.
TABLE 1.
Seventy-seven isolates from L. tenuis nodules, their habitats, and the chemical properties of the corresponding soils in the Salado River Basin
| Isolates | Soil properties
|
|||
|---|---|---|---|---|
| Soil type or habitat | pH | Electrical conductivity (mS/cm) | Na+ level (% of total) | |
| BA148, BA146, BA128, BA125, BA124, BA120, BA116, BA144, BA139, BA135, BA115, BA150, BA151, BA152, BA138, BA140, BA121, BA136, BA143, BA149, BA131, BA134, BA113, BA123 | Saline lowlands | 9.7 | 7.93 | 48.3 |
| BD53, BD61, BD46, BD68, BD65, BD66, BD74, BD51, BD67, BD70, BD72, BD56, BD44, BD60, BD50, BD63, BD57, BD58, BD41, BD48, BD43, BD45, BD40, BD47, BD49, BD59, BD55, BD54 | Lowlands | 5.66 | 0.51 | 9.6 |
| ML108, ML106, ML110, ML90, ML79, ML104, ML105, ML91, ML103, ML100, ML93, ML83, ML92, ML77, ML101, ML87, ML98, ML95, ML97, ML85, ML81, ML96, ML88, ML102, ML84 | Transitional plains | 6.2 | 0.33 | 8.2 |
Isolation of genomic DNA.
Total DNA was extracted from 3-ml cultures of the bacterial isolates grown in yeast extract-mannitol broth (71) at 30°C. Cultures were centrifuged at 13,500 × g for 3 min and washed in 0.5 ml of 0.1% (wt/vol) N-lauroylsarcosine in Tris-EDTA buffer (TE buffer) (10 mM Tris, 1 mM EDTA; pH 8). The pellets were suspended in 1 ml of 1 M NaCl and incubated for 1 h at 4°C in an orbital shaker. After this, the cell suspensions were centrifuged at 13,500 × g for 3 min, and the supernatants were removed. The pellets were suspended in 0.25 ml of 20% (wt/vol) sucrose in TE buffer prior to addition of 0.25 ml of a lysozyme solution (5 mg lysozyme in TE buffer) and 0.25 ml of RNase (1 mg/ml). After 30 min of incubation at 37°C, 0.1 ml pronase (10 mg/ml pronase in 5% N-lauroylsarcosine) was added, and the mixtures were incubated for 1 h at 37°C. A 0.07-ml aliquot of 3 M sodium acetate (pH 5.2) was added, and lysates were extracted with 0.4 ml of phenol-chloroform-isoamyl alcohol (25:24:1) saturated with 10 mM Tris-HCl (pH 8). The mixtures were vortexed and centrifuged at 13,500 × g for 3 min. Each aqueous phase was transferred to a clean tube, and 0.3 ml chloroform was added. The tubes were then vortexed and centrifuged at 13,500 × g for 3 min, the aqueous phase was transferred to clean tubes, and the DNA was precipitated with 0.7 ml of isopropanol. The precipitated DNA was washed with 0.5 ml of 70% ethanol, vacuum dried, and subsequently dissolved in 0.1 ml of MilliQ water by incubation for 30 min at 65°C.
Repetitive extragenic palindromic PCR (REP-PCR) genomic fingerprinting.
Primers REP1R-1 (5′-IIIICGICGICATCIGGC-3′) and REP 2I (5′-ICGICTTATCIGGCCTAC-3′) (70) were synthesized by the Service of Oligonucleotide Synthesis of the “Instituto de Parasitología y Biomedicina Lopez Neyra,” CSIC, Spain. PCR amplification was carried out by using a 0.025-ml reaction mixture containing 100 ng of genomic DNA, 2 μM primer REP1R-1, 2 μM primer REP2-1, each deoxynucleoside triphosphate (Roche) at a concentration of 1.25 mM, 1× polymerase reaction buffer (Sigma, United States), 7 mM MgCl2, and 4 U Taq DNA polymerase (Sigma, United States). Amplification was performed with a Px2 thermal cycler (Thermo Electron Corporation, United States) using the following temperature profile: initial denaturation at 95°C for 6 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 40°C for 1 min, and elongation at 65°C for 8 min; and a final extension for 16 min at 65°C (27). Amplified fragments were separated by electrophoresis in 1.5% agarose gels for 2 h at 85 V in 1× Tris-borate-EDTA buffer (100 mM Tris, 83 mM boric acid, 1 mM EDTA; pH 8.5). A 100-bp DNA ladder (Roche, Molecular Biochemicals, New England Biolabs) was used as the size marker. Gels were stained in an aqueous solution containing ethidium bromide (0.1%) and photographed using a UV transilluminator.
Computer-assisted analysis of the REP-PCR genomic fingerprints was performed by using the BioNumerics software program, version 4.0 (Applied Maths, Kortrijk, Belgium). The similarity between a pair of REP-PCR genomic fingerprints was calculated by using the product-moment correlation coefficient (r value) (51) applied to the whole densitometric curves for the gel tracks (52). Cluster analysis of the pairwise similarity values was performed by using the unweighted-pair group method using averages (UPGMA) algorithm (63).
PCR amplification and RFLP analysis of amplified 16S rRNA genes.
Nearly full-length 16S rRNA genes were amplified from isolates using primers 41f (5-GCTCAAGATTGAACGCTGGCG-3) and 1488r (5-CGGTTACCTTGTTACGACTTCACC-3) as previously described (27). The isolates were grouped according to their restriction fragment length polymorphism (RFLP) patterns into rRNA types, compared with 33 reference strains belonging to the genera Rhizobium, Mesorhizobium, Sinorhizobium, and Bradyrhizobium (Table 2), and analyzed further with the PAST software, version 1.30 (26). Levels of similarity for the 16S rRNA gene sequences of isolates and reference strains were estimated by using the proportion of shared restriction fragments. A dendrogram was constructed using the UPGMA (63).
TABLE 2.
Reference strains and genes used in this study
| Strain | Species | Host plant (origin) | 16S rRNA accession no. | nodC accession no. | nifH accession no. | Reference |
|---|---|---|---|---|---|---|
| LMG6133 | Sinorhizobium meliloti | Medicago sativa | X67222.2 | 74 | ||
| IAM12611 | Sinorhizobium meliloti | Medicago sativa | D14509.1 | 49 | ||
| ATCC 35423 | Sinorhizobium fredii | Glycine max | D14516.1 | 49 | ||
| USDA 205 | Sinorhizobium fredii | Glycine max | AY260147.1 | 58 | ||
| NGR234 | Sinorhizobium fredii | Lotus purpureus | AY260149.1 | 58 | ||
| CIAT899 | Rhizobium tropici | Phaseolus vulgaris | U89832.1 | 68 | ||
| IFO15247 | Rhizobium tropici | Phaseolus vulgaris | D11344.1 | 49 | ||
| LMG9517 | Rhizobium tropici | Phaseolus vulgaris | X67234.2 | 74 | ||
| Rhizobium leguminosarum bv. trifolii | Trifolium repens | U31074.1 | 7 | |||
| USDA2671 | Rhizobium leguminosarum bv. phaseoli | Phaseolus vulgaris | U29388.1 | 68 | ||
| Rhizobium leguminosarum bv. viciae | Pisum sativum | U29386.1 | 68 | |||
| CFN 42 | Rhizobium etli | Phaseolus vulgaris | U28916.1 | 68 | ||
| SEMIA384 | Rhizobium etli | Phaseolus vulgaris | AY904730.1 | 42 | ||
| R 602 | Rhizobium gallicum | Phaseolus vulgaris | AF008130.1 | 62 | ||
| ICMP 12856 | Agrobacterium rhizogenes | AY626393.1 | 76 | |||
| DSM 30105 | Agrobacterium tumefaciens | M11223.1 | 75 | |||
| hpig4.1 | Rhizobium sp. | Mimosa pigra | AY691401.1 | 5 | ||
| H152 | Rhizobium giardinii | Phaseolus vulgaris | U86344.1 | 1 | ||
| DSM6450T | Aminobacter aminovorans | AB167232.1 | 21 | |||
| UPM-Ca7 | Mesorhizobium ciceri | Cicer arietinum | U07934.1 | 47 | ||
| A-1Bs | Mesorhizobium tianshanense | Glycyrrhiza pallidiflora | AF041447.1 | 73 | ||
| ORS1096 | Mesorhizobium plurifarium | Acacia tortilis | AJ295079.1 | 2 | ||
| UPM-Ca36 | Mesorhizobium mediterraneum | Cicer arietinum | L38825.1 | 47 | ||
| LMG PR5 | Mesorhizobium chacoense | Prosopis alba | AJ278249.1 | 69 | ||
| ACCC 19665 | Mesorhizobium amorphae | Amorpha fruticosa | AF041442.1 | 73 | ||
| IFO 15243 | Mesorhizobium huakuii | Astragalus sinicus | D13431.1 | 49 | ||
| NZP2037 | Mesorhizobium loti | Lotus divaricatus | Y14159.1 | 12 | ||
| NZP 2213 | Mesorhizobium loti | Lotus tenuis | D14514.1 | 37 | ||
| USDA 110 | Bradyrhizobium japonicum | Glycine max | D13430.1 | 49 | ||
| 1021 | Sinorhizobium meliloti | Medicago sativa | M11268 | 16 | ||
| USDA 257 | Sinorhizobium fredii | Glycine max | M73699 | 34 | ||
| NGR234 | Sinorhizobium fredii | Lotus purpureus | U00090 | 19 | ||
| IIACFN299 | Rhizobium tropici | Phaseolus vulgaris | X98514 | 13 | ||
| USDA2071 | Rhizobium leguminosarum bv. trifolii | Trifolium repens | AF217271 | 35 | ||
| H132 | Rhizobium leguminosarum bv. phaseoli | Phaseolus vulgaris | AF217263 | 35 | ||
| CFN 42 | Rhizobium etli | Phaseolus vulgaris | U80928 | 35 | ||
| FL27 | Rhizobium gallicum | Phaseolus vulgaris | AF217270 | 35 | ||
| SN33 | Mesorhizobium sp. | Oxytropis arctobia | U53327 | 9 | ||
| IC2091 | Mesorhizobium ciceri | Cicer arietinum | AJ457929 | 39 | ||
| IC60 | Mesorhizobium mediterraneum | Cicer arietinum | AJ457928 | 39 | ||
| ACCC 19665 | Mesorhizobium amorphae | Amorpha fruticosa | AF217261 | 35 | ||
| MAFF303099 | Mesorhizobium loti | Lotus corniculatus | BA000012 | 33 | ||
| LMG 6123 (= NZP2037) | Mesorhizobium loti | Lotus divaricatus | X52958.2 | 10 | ||
| R7A | Mesorhizobium loti | Lotus corniculatus | AL672113 | 65 | ||
| CCBAU 43063 | Bradyrhizobium elkanii | Macroptilium atropurpureum | DQ010040 | 24 | ||
| USDA 110 | Bradyrhizobium japonicum | Glycine max | AF322013 | 24 | ||
| 8c-3 | Rhizobium etli | Phaseolus vulgaris | DQ058415 | 18 | ||
| CIAT899 | Rhizobium tropici | Phaseolus vulgaris | M55225 | 15 | ||
| Rch981 | Mesorhizobium ciceri | Cicer arietinum | AY318755 | 39 | ||
| SU329 | Rhizobium leguminosarum bv. trifolii | Trifolium repens | K00490 | 61 | ||
| Ss140 | Mesorhizobium plurifarium | Sesbania sericea | AY688619 | 72 | ||
| NGR234 | Sinorhizobium sp. | Lotus purpureus | U00090 | 19 | ||
| R7A | Mesorhizobium loti | Lotus corniculatus | AL672114 | 65 | ||
| MAFF303099 | Mesorhizobium loti | Lotus corniculatus | BA000012 | 33 | ||
| RCAN13 | Mesorhizobium amorphae | Cicer arietinum | DQ022841 | 54 | ||
| FL27 | Rhizobium leguminosarum bv. phaseoli | Phaseolus vulgaris | M55226 | 15 | ||
| CCBAU 57015 | Rhizobium hainanense | Desmodium sinuatum | AY934876 | 25 | ||
| R602 | Rhizobium gallicum | Phaseolus vulgaris | AF218126 | 35 | ||
| CCBAU 65199 | Bradyrhizobium japonicum | Glycine max | AY934872 | 25 | ||
| CCBAU 23174 | Bradyrhizobium elkanii | Macroptilium atropurpureum | AY934870 | 25 |
PCR amplification of nodC and nifH genes.
Forward primers nodCF, nodCFu, nodCF2, nodCF4, and nodCFn and reverse primer nodCI were used for amplification of nodC genes, as described by Laguerre and coworkers (35). Primers nifHI and nifHF were used for PCR amplification of about 780 bp of the nifH gene (35).
Sequencing of 16S rRNA, nodC, and nifH DNA fragments.
PCR products were purified by electroelution using a dialysis membrane (Spectra/Por; Spectrum Laboratories, Inc.) and were sequenced by The Sequencing Service of the Instituto de Parasitología y Biomedicina “Lopez Neyra,” CSIC, Spain. The 16S rRNA was sequenced using primers 41f and 1488r. In order to sequence the central portion of the amplified 16S rRNA genes, primer U-447 (5-GCGGTAATACGAAGG-3) was used. PCR products of nodC genes were sequenced using primers nodCF and nodCI. PCR products of the nifH gene were sequenced using primers nifHI and nifHF. The sequences obtained were compared with the sequences of reference strains deposited in the GenBank by using the BLASTN program (http://www.ncbi.nlm.nih.gov/blast).
PCR amplification and sequencing of atpD gene.
Approximately 450 bp of the atpD gene was amplified by using forward primer atpD1 or atpD2 and reverse primer atpD3 (22). Purified DNA was added to a 50-μl PCR mixture containing each deoxynucleoside triphosphate at a concentration of 200 nM, 1.5 mM MgCl2, 10 pmol each primer, 1 U Taq DNA polymerase, and 1× reaction buffer (Promega). The atpD gene was amplified as described previously by Gaunt et al. (22). The sequence obtained was compared with the sequences of reference strains deposited in the GenBank by using the BLASTN program (http://www.ncbi.nlm.nih.gov/blast).
Phylogenetic analyses.
Sequence alignment was performed with the ClustalW software from the EMBL server (http://www.ebi.ac.uk/). Aligned sequences were analyzed using the MEGA software, version 4.0 (67). Phylogenetic analyses of the 16S rRNA sequences were performed by the UPGMA (63). Phylogenetic analyses of nodC and nifH sequences were performed by using the neighbor-joining method (56). The phylogenetic distances were computed by using the p-distance method and were calculated based on the proportion of different nucleotides (p-distance), which was obtained by dividing the number of nucleotide differences by the total number of nucleotides compared (46). Statistical support for tree nodes was evaluated by performing a bootstrap analysis (17).
DNA genomic hybridization.
Total genomic DNAs were digested with endonuclease EcoRI, electrophoresed in 0.8% agarose gels, and then transferred to positively charged nylon membranes by the method of Southern (59). DNA hybridization probes were labeled with digoxigenin used according to the manufacturer's instructions (Roche, Barcelona, Spain). Hybridization and membrane washing were carried out under standard conditions. Membranes were prepared for chemiluminescence detection (Roche) and exposed to Kodak X-Omat film (Sigma).
Plant nodulation assays.
Seeds of L. tenuis cv. Pampa INTA or L. uliginosus LE 627 (experimental line; INIA-La Estanzuela, Uruguay) were scarified and surface disinfected with concentrated sulfuric acid for 15 min. After acid treatment, the seeds were soaked for 2 min in a 1% aqueous sodium hypochlorite solution and then thoroughly washed with sterile water. The seeds were distributed on the surface of 1% water agar plates and incubated for 24 to 48 h at 25°C for germination in the dark. In order to evaluate the infectivity of isolates, 10 seedlings were transferred to square petri dishes containing Rigaud-Puppo agar (53). Seedlings were inoculated with individual strains by adding exponentially growing rhizobial cultures (108 cells per seedling). Uninoculated seedlings were used as negative controls. For nodulation assays with L. tenuis seedlings inoculated with M. loti NZP 2213 were used as positive controls, and for nodulation assays with L. uliginosus seedlings inoculated with Bradyrhizobium loti NZP 2309 were used as positive controls. In both assays, root nodules appeared 10 to 20 days after inoculation and were fully developed 2 weeks later.
Nucleotide sequence accession numbers.
Sequences of the 16S rRNA genes obtained in this work have been deposited in the GenBank database under the following accession numbers: EU748908 (BD68), EU748909 (BD60), EU748910 (ML105), EU748911 (BA151), EU748912 (BA134), EU748913 (ML93), EU748914 (BA135), EU748915 (BD56), EU748916 (BA128), EU748917 (ML108), EU748918 (BD55), EU748919 (ML92), EU748920 (ML96), EU748921 (ML98), EU748922 (ML102), EU748923 (ML83), EU748924 (ML81), and EU748925 (ML85). Sequences of the nodC genes obtained in this work have been deposited in the GenBank database under the following accession numbers: EU748926 (ML93), EU748927 (BD56), EU748928 (ML108), EU748929 (BA135), EU748930 (BA128), EU748931 (BD68), EU748932 (BA151), EU748933 (ML105), and EU748934 (BA134). Sequences of the nifH genes obtained in this work have been deposited in the GenBank database under the following accession numbers: EU748935 (BA135), EU748936 (BD68), EU748937 (BA128), EU748938 (BA151), EU748939 (ML93), EU748940 (BA134), EU748941 (BD56), EU748942 (ML105), and EU748943 (ML108).
RESULTS
Analysis of REP-PCR genomic fingerprints.
Purified genomic DNAs from 103 isolates were used as templates for PCR with REP primers to obtain DNA fingerprints. In the first step, REP fingerprints of isolates from each habitat were analyzed separately to determine similarities and to identify siblings (data not shown). As a general rule, each isolate had a unique and complex fingerprint, but some isolates with identical fingerprints were identified. In such cases, only one of the isolates was used for further analysis. A total of 77 fingerprints from all habitats were used to construct a dendrogram with the Bionumerics software (see Fig. S1 in the supplemental material). The REP-PCR analysis revealed a high level of genetic diversity among the isolates, and the degree of relatedness among strains isolated from different habitats ranged from 55 to 80% (see Fig. S1 in the supplemental material).
RFLP analysis of amplified 16S rRNA.
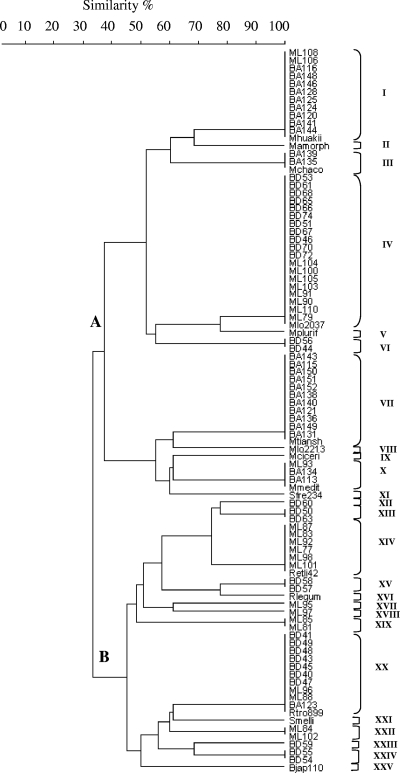
The 16S rRNA genes of 77 nonredundant isolates were PCR amplified, which resulted in a single band at about 1,500 bp in all cases; this size corresponded to the expected size of the 16S rRNA genes of most members of the Rhizobiaceae (37). On the basis of previous work performed by Laguerre et al. (37), endonucleases HinfI and MspI were selected for 16S rRNA RFLP analysis in order to estimate the taxonomic positions of isolates by comparing their restriction profiles with the restriction profiles of 15 reference strains that represented different rhizobial species and genera (Table 2). This analysis resulted in 11 and 15 different restriction patterns for HinfI and MspI, respectively. Restriction patterns were arbitrarily identified by letters (Table 3) and used for classification of isolates into ribogroups. For the 77 isolates analyzed, 17 distinct ribogroups were distinguished, each comprising 1 to 19 isolates. A dendrogram constructed from the distance matrix as described in Materials and Methods confirmed the distribution of the isolates into two distinct clusters, clusters A and B, at a similarity level of 32%, and 17 subclusters that corresponded to the 17 ribogroups identified (Fig. 1). Only 7 of the 17 ribogroups identified matched any of the reference strains included in the analysis.
TABLE 3.
Restriction patterns of amplified 16S rRNA and 16S rRNA genotypes of rhizobial strains isolated from L. tenuis and reference strains
| Isolates and/or reference straina | Restriction pattern of amplified 16S rRNA digested withb:
|
RFLP ribogroup | |
|---|---|---|---|
| HinfI | MspI | ||
| BA148, BA146, BA128, BA125, BA124, BA120, BA116, BA144, ML108, ML106, Mesorhizobium huakuii IFO15243 | B | B | I |
| Mesorhizobium amorphae ACCC19665 | B | D | II |
| BA139, BA135, Mesorhizobium chacoense LMGPR5 | B | F | III |
| BD53, BD61, BD46, BD68, BD65, BD66, BD74, BD51, BD67, BD70, BD72, ML110, ML90, ML79, ML104, ML105, ML91, ML103, ML100, M. loti NZP2037 | E | B | IV |
| Mesorhizobium plurifarium ORS 1096 | E | D | V |
| BD56, BD44 | E | A | VI |
| BA115, BA150, BA151, BA152, BA138, BA140, BA121, BA136, BA143, BA149, BA131, Mesorhizobium tianshanense A-1BS | C | C | VII |
| Mesorhizobium loti NZP2213 | C | A | VIII |
| Mesorhizobium ciceri UPM-Ca7 | A | A | IX |
| BA134, BA113, ML93, Mesorhizobium mediterraneum UMP-Ca36 | A | C | X |
| Sinorhizobium fredii NGR234 | A | J | XI |
| BD60 | F | F | XII |
| BD50, BD63 | D | E | XIII |
| ML83, ML92, ML77, ML101, ML87, ML98, Rhizobium etli CFN42 | F | H | XIV |
| BD57, BD58 | D | F | XV |
| Rhizobium leguminosarum bv. phaseoli USDA2671 | D | H | XVI |
| ML95 | F | N | XVII |
| ML97 | F | K | XVIII |
| ML85, ML81 | H | L | XIX |
| BD41, BD48, BD43, BD45, BD40, BD47, BD49, BA123, ML96, ML88, Rhizobium tropici IIB CIAT899 | D | G | XX |
| Sinorhizobium meliloti LGM 6133 | A | G | XXI |
| ML102, ML84 | J | O | XXII |
| BD59 | I | M | XXIII |
| BD55; BD54 | F | E | XXIV |
| Bradyrhizobium japonicum USDA110 | G | I | XXV |
Isolates that infect and do not infect L. tenuis are indicated by bold and light type, respectively.
Different restriction patterns are indicated by different letters for each endonuclease used.
FIG. 1.
16S rRNA RFLP dendrogram showing genetic relationships among reference strains and rhizobial strains isolated from nodules of L. tenuis in typical soils of the Salado River Basin. The cluster analyses were performed using UPGMA. The endonucleases used were HinfI and MspI. Ribogroups I to XXVI (indicated on the right) are described in Table 3.
Isolates in cluster A, including isolates in ribogroups I, III, IV, VII, and X, comprised 61% of the strains analyzed and clustered with species of the genus Mesorhizobium, and the profiles of these ribogroups were identical to the profiles of reference strains Mesorhizobium huakuii IFO15243, Mesorhizobium chacoense LMGPR5, M. loti NZP2037, Mesorhizobium tianshanense A-1BS, and Mesorhizobium mediterraneum UMP-Ca36, respectively. Two cluster A isolates corresponded to ribogroup VI, which was not represented by any of the reference strains analyzed (Fig. 1 and Table 3). Cluster B comprised 39% of the strains analyzed, which appeared to be related to the genus Rhizobium. Some of the isolates in this cluster corresponded to ribogroups XIV and XX, whose profiles were identical to those of reference strains Rhizobium etli CFN42 and Rhizobium tropici IIB CIAT899, respectively. Other isolates in group B, corresponding to ribogroups XII, XIII, XV, XVII, XVIII, XIX, XXII, XXIII, and XXIV, did not match any of the reference strains used (Fig. 1 and Table 3).
When the environmental distribution of the isolates was examined, less taxonomic diversity was found in saline lowlands. Most the strains from this habitat were restricted to the genus Mesorhizobium; the only exception was isolate BA123, which grouped with reference strain R. tropici CIAT899. On the other hand, strains isolated from the nonsaline lowlands and transitional plains were found to be more diverse in terms of taxonomy, grouping with both the genus Mesorhizobium and the genus Rhizobium (Fig. 1 and Table 3).
Host range of L. tenuis isolates.
The abilities of the 77 isolates to form nodules on the original host, L. tenuis cv. Pampa INTA, were examined. All 47 isolates in cluster A were able to nodulate and fix nitrogen on L. tenuis plants, as shown by the formation of pink nodules and the dark green color of the aerial parts of plants compared with noninoculated plants. In contrast, only isolates BD60, ML83, ML92, ML96, ML98, ML81, ML85, ML102, and BD55 in cluster B (related to the genus Rhizobium) were confirmed to nodulate L. tenuis and formed pink nodules similar to those formed by isolates in cluster A (Table 3). Nodulation of L. tenuis by the remaining isolates could not be verified under our experimental conditions, although it is worth noting that most ribogroups in 16S rRNA RFLP cluster B included both infective and noninfective isolates (Table 3).
On the other hand, several reports have suggested that most rhizobia that nodulate L. tenuis have a narrow host range for nodulation of Lotus spp. and cannot form nitrogen-fixing nodules on L. uliginosus (30, 45); the exceptions are strains, like M. loti NZP2037, that are considered broad-host-range strains since they can form nitrogen-fixing nodules on most, if not all, Lotus species.
Therefore, all isolates that were found to be infective on L. tenuis were tested using L. uliginosus LE627 in order to establish their host ranges for nodulation of Lotus species. Nodulation assays indicated that all the isolates were ineffective on L. uliginosus, as they formed no nodules or small red tumor-like structures that were unable to fix nitrogen (as shown by leaf chlorosis) similar to those on noninoculated plants.
Analysis of 16S rRNA gene sequences.
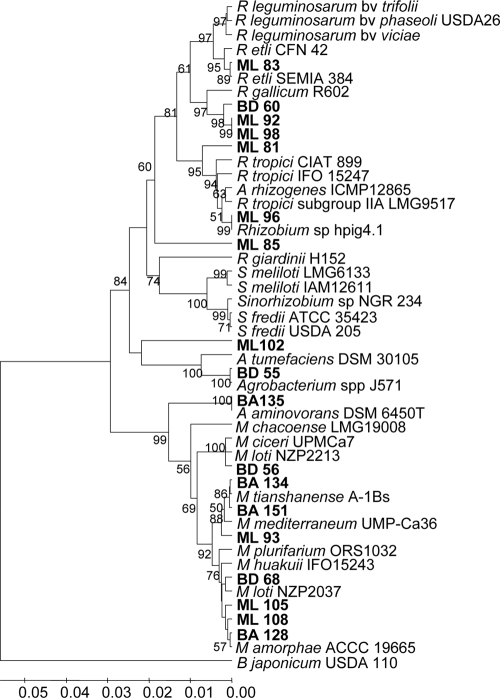
An analysis of the sequences of 16S rRNA genes of representative isolates belonging to each ribogroup that infected L. tenuis (Table 3) was performed to confirm the phylogenetic position estimated using the RFLP analysis results. For this purpose, nearly full-length 16S rRNA sequences were obtained for 18 selected isolates that represented 12 16S rRNA RFLP groups (Table 3 and Fig. 1). Figure 2 shows a phylogenetic tree based on the similarity of the 16S rRNA sequences of the selected isolates and reference strains, which was constructed by using the p-distance model for estimating phylogenetic distances (46) and the UPGMA algorithm (63).
FIG. 2.
16S rRNA gene phylogeny of rhizobial strains isolated from nodules of L. tenuis grown in typical soils of the Salado River Basin. The tree was constructed from the nucleotide sequence data by using the UPGMA algorithm, and phylogenetic distances were calculated by the p-distance method. The numbers at branch points are the significant bootstrap values (expressed as percentages based on 1,000 replicates; only values greater than 50% are shown). The horizontal branch lines are proportional and indicate the p-distances.
The classification of most isolates at the genus level obtained after analysis of 16S rRNA sequences (Fig. 2) was in good agreement with the classification obtained using the 16S rRNA RFLP data; the exceptions were isolates BA135 and BD59 (see below). However, 16S rRNA sequence analyses allowed better discrimination at the species level.
Thus, on the basis of 16S rRNA sequences, isolates BA128 and ML108 clustered with Mesorhizobium amorphae type strain ACCC 19665 (ribogroup II), sharing 99.5% sequence identity (five and six mismatches, respectively).
Isolates BD68 and ML105 exhibited the same ribotype (ribogroup IV) as strain M. loti NZP2037 (Fig. 1 and Table 3), and their 16S rRNA sequences shared 99.4% sequence identity with this reference strain. However, these two isolates appeared to be separated in the phylogenetic tree as isolate ML105 was in an undefined position between NZP2037 and M. amorphae ACCC 19665.
Isolate BD56 (ribogroup VI) was not related to any of the reference strains based on the 16S rRNA RFLP patterns, but its 16S rRNA gene sequence clustered in a branch together with the sequences of Mesorhizobium ciceri UPM Ca7 and M. loti NZP2213, with which it exhibited 99.2 and 98.9% sequence identity, respectively.
In the case of isolate BA151 (ribogroup VII), the 16S rRNA RFLP and sequence analyses provided similar results and grouped this isolate with M. tianshanense A-1BS (Fig. 1 and 2 and Table 3). Isolates ML93 and BA134, which grouped with M. mediterraneum UMP-Ca36 in RFLP ribogroup X, shared more than 99.0% 16S rRNA sequence identity with M. mediterraneum UMP-Ca36 and M. tianshanense A-1BS.
Although 16S rRNA RFLP analyses indicated that isolate BA135 (ribogroup III) grouped with M. chacoense LMG PR5 (Fig. 1 and Table 3), 16S rRNA sequencing revealed that BA135 had greater similarity to the nonsymbiotic bacterium Aminobacter aminovorans strain DSM6450T (99.7% identity and four mismatches) than to M. chacoense LMG PR5 (97.6% identity and a difference of 30 nucleotides). In view of this result, the 16S rRNA sequence of strain A. aminovorans DSM6450T was included in the alignment of 16S rRNA sequences for construction of the phylogenetic tree (Fig. 2).
Based on 16S rRNA sequences, isolates BD60 (ribogroup XII), ML77 (ribogroup XIV), BD63 (ribogroup XIII), and ML98 and ML92 (ribogroup XIV) clustered together in a branch separated from Rhizobium gallicum type strain R602; however, only BD60, ML98, and ML92 could be confirmed to be infective in L. tenuis, and their 16S rRNA sequences were included in the phylogenetic tree. (Fig. 2). The levels of 16S rRNA sequence identity between type strain R602 and the L. tenuis isolates varied from 96.8 to 97.1%.
Isolate ML83, which was one of the three infective isolates that represented RFLP ribogroup XIV (isolates ML98, ML92, and ML83), clustered with R. etli CFN42 on the basis of 16S rRNA sequence analysis. This isolate showed 98.9 and 99.4% sequence identity with R. etli CFN42 and SEMIA 384, respectively (Fig. 2).
Isolates ML88 and ML96 were placed in 16S rRNA RFLP ribogroup XX together with R. tropici IIB CIAT 899 (Table 3), but 16S rRNA sequence analysis showed that both of these isolates were more closely related to Rhizobium sp. strain hpig 4.1 (three and four mismatches, respectively), which was isolated from Mimosa spp. in Costa Rica (5). However, only ML96 could be confirmed to be infective in L. tenuis.
Although isolates ML81 and ML85 both fell into RFLP ribogroup XIX, their 16S rRNA sequences were rather distinct (94.0% identity). ML81 branched out of the R. tropici-Agrobacterium rhizogenes cluster, whereas ML85 formed a differentiated branch within the R. etli-R. leguminosarum-R. gallicum cluster.
Isolates BD55 (ribogroup XXIV) and ML97 (noninfective, ribogroup XVIII) appeared to be closely related to the Agrobacterium tumefaciens cluster, showing 99.9 and 99.7% sequence identity, respectively, with Agrobacterium sp. strain J571, although they were more divergent from A. tumefaciens type strain DSM 30105 (Fig. 2). On the other hand, isolate ML102 was more closely related to Rhizobium taeanense PSB2-6 (a strain isolated from a leguminous sand dune plant; accession number DQ114473.1; not included in the tree), exhibiting higher sequence similarity to this strain (98.6% sequence identity) than to A. tumefaciens 358 (96.2% sequence identity) (Fig. 2).
Analysis of nodC and nifH sequences.
PCR amplification of nodC and nifH gene fragments was carried out with isolates representing each of the 16S rRNA RFLP ribogroups identified for the L. tenuis isolates. A nodC gene fragment about 840 to 890 bp long could be amplified from all representative isolates belonging to ribogroups in 16S rRNA RFLP cluster A by using the nodCF-nodCI primer pair (35). In contrast, even though different combinations of primers described by Laguerre et al. (35) were used, a nodC gene fragment could not be amplified from any of the isolates in cluster B, regardless of their proven ability to nodulate L. tenuis under our experimental conditions (see above and Table 3). Genomic DNAs from nodulating and nonnodulating cluster B strains were hybridized with an M. loti R7A nodC gene probe, but no nodC-hybridizing bands were visualized for either strain, suggesting that the nodulation genes of the cluster B strains isolated from L. tenuis are very divergent from those of M. loti, which would at least partially explain our failure to amplify nodC from these isolates.
Likewise, an 800-bp nifH amplification product was obtained from all isolates in cluster A but from none of the infective representatives of cluster B. In contrast to the findings for nodC, when genomic DNAs from cluster B strains were hybridized with an R. etli CFN42 nifH probe, weak hybridization signals were observed for many of the isolates regardless of their infectivity, suggesting again that the symbiotic genes of these isolates may be rather divergent from those of other rhizobia (data not shown).
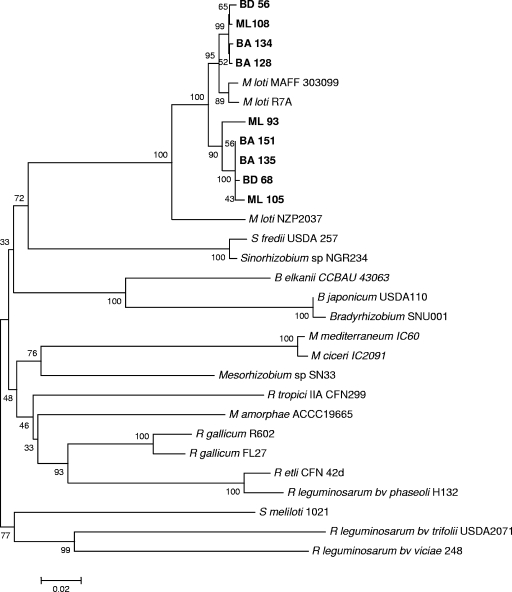
The sequences of nodC and nifH gene fragments were obtained and compared to corresponding rhizobial sequences reported previously. Phylogenetic analysis of nodC and nifH sequences was performed by using the neighbor-joining method (56), and the phylogenetic distances were calculated by using the p-distance method (46). As shown in Fig. 3, nodC sequences from Lotus-nodulating rhizobia clustered separately from nodC sequences from other legume symbiotic rhizobia. Furthermore, within the Lotus group, nodC phylogenies clearly differentiated broad-host-range strains (NZP2037) from narrow-host-range strains (R7A and MAFF303099), suggesting that nodC is a good indicator of host range for Lotus symbionts. All the Argentinian nodC sequences clustered in the narrow-host-range group of Lotus rhizobia, although they formed two subgroups diverging from the reference strains.
FIG. 3.
Phylogenetic analysis of nodC genes from isolates nodulating L. tenuis. Aligned sequences of nodC genes that were 800 bp long (positions 286 −1123 of the nodC nucleotide sequence of M. loti MAFF 303099) were used. The tree was constructed by using the neighbor-joining method. The percentages of replicate trees in which the isolates and reference strains clustered together in the bootstrap test (1,000 replicates) are indicated at branch points (only values greater than 50% are shown). The tree is drawn to scale, and the units for the branch lengths are the same as the units for the phylogenetic distances used to infer the phylogenetic tree.
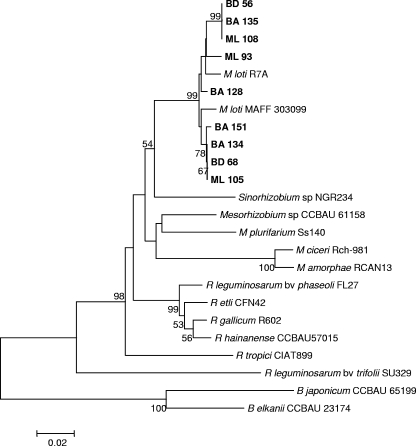
Phylogenetic analysis of nifH sequences also clustered the Lotus bacteria separate from other symbiotic rhizobia (Fig. 4). As observed with nodC, two separate subgroups were distinguished for the Lotus rhizobia, although in this case reference strains R7A and MAFF303099 appeared to be divergent and the Argentinian isolates fell into one of the subgroups regardless of the habitat from which they were isolated.
FIG. 4.
Phylogenetic analysis of nifH genes from isolates nodulating L. tenuis. Aligned sequences of nifH genes that were 500 bp long (positions 317 to 815 of the nifH nucleotide sequence from M. loti MAFF303099) were used. The tree was constructed by using the neighbor-joining method. The percentages of replicate trees in which the isolates and reference strains clustered together in the bootstrap test (1,000 replicates) are indicated at branch points (only values greater than 50% are shown). The tree is drawn to scale, and the units for the branch lengths are the same as the units for the phylogenetic distances used to infer the phylogenetic tree.
DISCUSSION
Analysis of REP-PCR fingerprints revealed a high level of genetic diversity among the L. tenuis nodule isolates, considering that 77 unique and complex genomic fingerprints were obtained. A comparison of REP-PCR profiles for different habitats revealed levels of similarity among isolates that ranged from 80% to less than 60%. Similarly, other authors found considerable genetic diversity among rhizobia nodulating L. corniculatus by using other methodological approaches, like DNA-DNA hybridization (11, 29, 30) or numerical taxonomy (48).
Seventy-seven isolates representing unique REP-PCR genomic fingerprints were further differentiated into two clusters, clusters A and B, after 16S rRNA gene RFLP analysis. A majority of the isolates (61%, cluster A) appeared to be related to the genus Mesorhizobium and were distributed in six ribogroups; the sixth ribogroup was not related to any of our reference strains. In contrast, cluster B, comprising 39% of the isolates, included 11 16S rRNA RFLP ribogroups related to species in the genus Rhizobium. When reinoculated onto the original host, L. tenuis, all of the cluster A isolates formed nitrogen-fixing nodules, in contrast to the cluster B isolates (related to Rhizobium), only nine of which were able to nodulate L. tenuis roots under our experimental conditions. It is possible that the noninfective isolates may have arisen through loss of symbiotic genes during the process of isolation and subsequent culture (36), an explanation supported by the fact that most ribogroups in cluster B included both nodulating and nonnodulating isolates. Furthermore, only Rhizobium-like bacteria were isolated from the nodules formed after reinoculation onto L. tenuis roots. Nevertheless, other explanations, such as the possibility that certain plant-bacterium interactions can occur only under particular soil conditions not met by our experimental protocol, should not be ruled out.
The distribution of most of the symbiotic isolates among the genera Rhizobium and Mesorhizobium is further supported by the results of the analyses of nearly full-length PCR-amplified 16S rRNA genes; the only exception is isolate BA135, whose 16S rRNA sequence was closely related to the A. aminovorans sequence. The phylogenetic relationship of strain BA135 with A. aminovorans is further supported by its atpD gene sequence, which also shows the highest level of similarity with the atpD gene sequence of strain A. aminovorans DSM 10368 (98% identity; accession number EU748944). The genus Aminobacter is phylogenetically related to Mesorhizobium but has never been reported to nodulate legumes (41). Thus, this is the first report of a member of this genus with symbiotic capacity. The genus Aminobacter includes several pesticide- and herbicide-degrading bacteria isolated from agricultural soils that are able to grow on CH3Cl, CH3Br, CH3I, and methylated amines as sole carbon and energy sources (41). Given the high proportion of species with bioremediation activity in this genus, the isolation and further characterization of new symbiotic isolates belonging to this genus in soils of the Salado River Basin could have important practical environmental implications.
All other isolates that infect L. tenuis were related to previously described rhizobial species, particularly members of the genera Mesorhizobium or Rhizobium, as suggested by their 16S rRNA gene sequences. Unexpectedly, an overwhelming majority of these isolates were related to species other than M. loti, which is the type species for L. tenuis and L. corniculatus symbionts (20, 30, 55, 66). Thus, only isolates BD56 and BD44 were phylogenetically closely related to M. loti type strain NZP2213, whereas the remaining isolates in 16S rRNA RFLP cluster A were more closely related to other Mesorhizobium species, like M. amorphae, M. mediterraneum, or M. tianshanense. A significant proportion of isolates were closely related to broad-host-range strain NZP2037; however, according to 16S rRNA phylogenies, this strain is more closely related to other Mesorhizobium spp. than to the M. loti type strain.
Another surprising result was the fact that a significant number of L. tenuis symbionts seem to be closely related to Rhizobium species, like R. gallicum, R. etli, or R. tropici. Although R. etli has previously been shown to nodulate Lotus japonicus (3), this is the first time that Lotus was identified as the primary host of a Rhizobium sp. strain. Intriguingly, the L. tenuis symbionts were closely related to rhizobial species that have been shown to be common symbionts of Phaseolus vulgaris (25, 40). Unfortunately, despite various attempts we were unable to amplify nodC or nifH gene sequences from Rhizobium symbionts of L. tenuis, which prevented us from obtaining further insight into the origin of their symbiotic capacities. However, the fact that no nodC gene sequences could be amplified when various primer sets were used indicates that the nodulation genes of these L. tenuis symbionts may be different from those of other rhizobia. This conclusion is supported by the negative results obtained when genomic hybridization with symbiotic gene probes was performed. In particular, no nodC-hybridizing DNA fragments were found in either nodulating or nonnodulating Rhizobium isolates, which could even suggest the rare but previously proven presence of nodulation in the absence of Nod factors, as recently reported for certain photosynthetic bradyrhizobia (23).
In contrast, nodC and nifH gene fragments from all of the Mesorhizobium and Aminobacter-like isolates could be amplified. The nodC genes were all very similar and closely related to nodC genes from narrow-host-range M. loti strains, such as R7A and MAFF303099. This perfectly correlates with the narrow host range of the Argentinian isolates inferred from their inability to nodulate L. uliginosus. Indeed, nodC gene phylogenies clearly differentiated between narrow- and broad host-range Lotus symbionts, suggesting that nodC is a valid marker for this purpose.
nifH gene sequences in the Argentinian and reference Lotus bacteria were very highly conserved and formed a phylogenetic cluster differentiated from other rhizobial nifH gene sequences. The fact that similar symbiotic genes are shared by distinguishable 16S rRNA species and genera suggests that genes for symbiosis with L. tenuis had a common origin and that horizontal gene transfer has played an important role in the spread of symbiotic capacity among Mesorhizobium species and even to the closely related genus Aminobacter. Although intrageneric lateral transfer of symbiotic determinants has been shown previously for Mesorhizobium (66), our results with isolate BD135 are the first evidence that there has been symbiotic gene exchange between Mesorhizobium and Aminobacter.
Our work indicates that the high level of genetic diversity of L. tenuis symbionts in the Salado River Basin is not evenly distributed in all habitats screened in this work. A comparison of isolates from three typical habitats that characterized the soils in the Salado River Basin revealed that the more stressful environments, the saline lowlands, contain mainly mesorhizobia, in contrast to habitats such as the nonsaline lowlands and transitional plains, where genotypically more diverse symbionts were found, including bacteria belonging to the genera Mesorhizobium, Rhizobium, and Aminobacter. Until now, M. loti has been reported to be the type symbiont for Lotus spp., such as L. japonicus, L. tenuis, or L. corniculatus (55). There have been a few reports of genetic diversity of Lotus symbionts, and it should be noted that much of the previous work on Lotus rhizobia was done in only a few countries, like New Zealand or Uruguay (27, 45, 66). Our work suggests that in environments like the Salado River Basin, M. loti strains represent a minority of the symbionts of L. tenuis. This indicates that Lotus-nodulating rhizobia should be characterized in other countries and environments, and it would not be surprising if species other than M. loti or even genera other than Mesorhizobium were the dominant Lotus symbiotic bacteria.
Supplementary Material
Acknowledgments
This work was supported by grant UE-FP6-517617 (LOTASSA) and by the CONICET (Argentina)-CSIC (Spain) bilateral cooperation program. M.J.E. is a member of the Research Career of Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (Argentina) and was supported in part by a postdoctoral fellowship from MEC (Spain).
F. L. Pieckenstain is particularly acknowledged for his help and for critical reading of the manuscript.
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amarger, N., V. Macheret, and G. Laguerre. 1997. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int. J. Syst. Bacteriol. 47:996-1006. [DOI] [PubMed] [Google Scholar]
- 2.Ba, S., A. Willems, P. de Lajudie, P. Roche, H. Jeder, P. Quatrini, M. Neyra, M. Ferro, J. C. Prome, M. Gillis, C. Boivin-Masson, and J. Lorquin. 2002. Symbiotic and taxonomic diversity of rhizobia isolated from Acacia tortilis subsp. raddiana in Africa. Syst. Appl. Microbiol. 25:130-145. [DOI] [PubMed] [Google Scholar]
- 3.Banba, M., A. M. Siddique, H. Kouchi, K. Izui, and S. Hata. 2001. Lotus japonicus forms early senescent root nodules with Rhizobium etli. Mol. Plant-Microbe Interact. 14:173-180. [DOI] [PubMed] [Google Scholar]
- 4.Baraibar, A., L. Frioni, M. E. Guedes, and H. Ljunggren. 1999. Symbiotic effectiveness and ecological characterization of indigenous Rhizobium loti populations in Uruguay. Pesqui. Agropecu. Bras. 34:1011-1017. [Google Scholar]
- 5.Barrett, C. F., and M. A. Parker. 2006. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 72:1198-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrientos, L., M. Higuera, H. Acuña, J. Guerrero, F. Ortega, and I. Seguel. 2002. Efectividad simbiótica de cepas naturalizadas de Mesoorhizobium loti y Bradyrhizobium sp. (Lotus) en plantas de tres especies del género Lotus. Agric. Tec. (Chile) 62:226-236. [Google Scholar]
- 7.Breil, B., J. Borneman, and E. W. Triplett. 1996. A newly discovered gene, tfuA, involved in the production of the ribosomally synthesized peptide antibiotic trifolitoxin. J. Bacteriol. 178:4150-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockwell, J., D. M. Hebb, and W. M. Kelman. 1994. Symbiotaxonomy of Lotus species and symbiotically related plants and of their root-nodule bacteria, p. 30-35. In P. R. Beuselink and C. A. Roberts (ed.), Proceedings of the First International Lotus Symposium. University Extension, University of Missouri, Columbia.
- 9.Cloutier, J., S. Laberge, D. Prevost, and H. Antoun. 1996. Sequence and mutational analysis of the common nodBCIJ region of Rhizobium sp. (Oxytropis arctobia) strain N33, a nitrogen-fixing microsymbiont of both arctic and temperate legumes. Mol. Plant-Microbe Interact. 9:523-531. [DOI] [PubMed] [Google Scholar]
- 10.Collins-Emerson, J. M., E. A. Terzaghi, and D. B. Scott. 1990. Nucleotide sequence of Rhizobium loti nodC. Nucleic Acids Res. 18:6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow, V. L., B. D. W. Jarvis, and R. M. Greenwood. 1981. Deoxyribonucleic acid homologies among acid-producing strains of Rhizobium. Int. J. Syst. Bacteriol. 31:152-172. [Google Scholar]
- 12.Debelle, F., C. Plazanet, P. Roche, C. Pujol, A. Savagnac, C. Rosenberg, J. C. Prome, and J. Denarie. 1996. The NodA proteins of Rhizobium meliloti and Rhizobium tropici specify the N-acylation of Nod factors by different fatty acids. Mol. Microbiol. 22:303-314. [DOI] [PubMed] [Google Scholar]
- 13.de Lajudie, P., A. Willems, G. Nick, F. Moreira, F. Molouba, B. Hoste, U. Torck, M. Neyra, M. D. Collins, K. Lindstrom, B. Dreyfus, and M. Gillis. 1998. Characterization of tropical tree rhizobia and description of Mesorhizobium plurifarium sp. nov. Int. J. Syst. Bacteriol. 48:369-382. [DOI] [PubMed] [Google Scholar]
- 14.Dowling, D. N., and W. J. Broughton. 1986. Competition for nodulation. of legumes. Annu. Rev. Microbiol. 40:131-157. [DOI] [PubMed] [Google Scholar]
- 15.Eardly, B. D., J. P. Young, and R. K. Selander. 1992. Phylogenetic position of Rhizobium sp. strain Or 191, a symbiont of both Medicago sativa and Phaseolus vulgaris, based on partial sequences of the 16S rRNA and nifH genes. Appl. Environ. Microbiol. 58:1809-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egelhoff, T. T., R. F. Risher, T. W. Jacobs, J. T. Mulligan, and S. R. Long. 1985. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA 4:241-248. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 18.Flores, M., L. Morales, A. Avila, V. Gonzalez, P. Bustos, D. Garcia, Y. Mora, X. Guo, J. Collado-Vides, D. Pinero, G. Davila, J. Mora, and R. Palacios. 2005. Diversification of DNA sequences in the symbiotic genome of Rhizobium etli. J. Bacteriol. 187:7185-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 20.Fulchieri, M. M., M. J. Estrella, and A. A. Iglesias. 2001. Characterization of Rhizobium loti strains from the Salado River Basin. Antonie van Leeuwenhoek 79:119-125. [DOI] [PubMed] [Google Scholar]
- 21.Futamata, H., Y. Nagano, K. Watanabe, and A. Hiraishi. 2005. Unique kinetic properties of phenol-degrading Variovorax strains responsible for efficient trichloroethylene degradation in a chemostat enrichment culture. Appl. Environ. Microbiol. 71:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaunt, M. W., S. L. Turner, L. Rigottier-Gois, S. A. Lloyd-Macgilps, and J. P. W. Young. 2001. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 51:2037-2048. [DOI] [PubMed] [Google Scholar]
- 23.Giraud, E., L. Moulin, D. Vallenet, V. Barbe, E. Cytryn, J. C. Avarre, M. Jaubert, D. Simon, F. Cartieaux, Y. Prin, G. Bena, L. Hannibal, J. Fardoux, M. Kojadinovic, L. Vuillet, A. Lajus, S. Cruveiller, Z. Rouy, S. Mangenot, B. Segurens, C. Dossat, W. L. Franck, W. S. Chang, E. Saunders, D. Bruce, P. Richardson, P. Normand, B. Dreyfus, D. Pignol, G. Stacey, D. Emerich, A. Verméglio, C. Médigue, and M. Sadowsky. 2007. Legume symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316:1307-1312. [DOI] [PubMed] [Google Scholar]
- 24.Gottfert, M. S., S. Rothlisberger, C. Kundig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu, J., E. T. Wang, and W. X. Chen. 2007. Genetic diversity of rhizobia associated with Desmodium species grown in China. Lett. Appl. Microbiol. 44:286-292. [DOI] [PubMed] [Google Scholar]
- 26.Hammer, Ø., D. A. T. Harper, and P. D. Ryan. 2001. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electronica 4:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- 27.Herrera-Cervera, J. A., J. Caballero-Mellado, G. Laguerre, H. V. Tichy, N. Requena, N. Amarger, E. Martinez-Romero, J. Olivares, and J. Sanjuan. 1999. At least five rhizobial species nodulate Phaseolus vulgaris in a Spanish soil. FEMS Microbiol. Ecol. 30:87-97. [Google Scholar]
- 28.Irisarri, P., F. Milnitsky, J. Monza, and E. J. Bedmar. 1996. Characterization of rhizobia nodulating Lotus subbiflorus from Uruguayan soils. Plant Soil 180:39-47. [Google Scholar]
- 29.Jarvis, B. D. W., T. S. Maclean, I. G. C. Robertson, and G. R. Fanning. 1977. Phenetic similarity and DNA base sequence homology of root nodule bacteria from New Zealand native legumes and Rhizobium strains from agricultural plants. N. Z. J. Agric. Res. 20:235-248. [Google Scholar]
- 30.Jarvis, B. D. W., C. E. Pankhurst, and J. J. Patel. 1982. Rhizobium loti, a new species of legume root nodule bacteria. Int. J. Syst. Bacteriol. 32:378-380. [Google Scholar]
- 31.Jordan, D. C. 1982. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 32:136-139. [Google Scholar]
- 32.Kade, M., E. A. Pagani, and R. E. Mendoza. 2003. A morphological study of populations of Lotus glaber Mill. (Fabaceae). Agronomie 23:203-207. [Google Scholar]
- 33.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 34.Krishnan, H. B., and S. G. Pueppke. 1991. Sequence and analysis of the nodABC region of Rhizobium fredii USDA257, a nitrogen-fixing symbiont of soybean and other legumes. Mol. Plant Microb. Interact. 4:512-520. [DOI] [PubMed] [Google Scholar]
- 35.Laguerre, G., S. M. Nour, V. Macheret, J. Sanjuan, P. Drouin, and N. Amarger. 2001. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981-993. [DOI] [PubMed] [Google Scholar]
- 36.Laguerre, G., M. P. Fernandez, V. Edel, P. Normand, and N. Amarger. 1993. Genomic heterogeneity among French Rhizobium strains isolated from Phaseolus vulgaris L. Int. J. Syst. Bacteriol. 43:761-767. [DOI] [PubMed] [Google Scholar]
- 37.Laguerre, G., M. R. Allard, F. Revoy, and N. Amarger. 1994. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl. Environ. Microbiol. 60:56-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavado, R. S. 1992. Río de la Plata grasslands. Soils, p. 377-380. In R. T. Coupland (ed.), Ecosystems of the world, vol. 8A. Natural grasslands. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 39.Maatallah, J., E. B. Berraho, S. Muñoz, J. Sanjuán, and C. Lluch. 2002. Phenotypic and molecular characterization of chickpea rhizobia isolated from different areas of Morocco. J. Appl. Microbiol. 93:531-540. [DOI] [PubMed] [Google Scholar]
- 40.Martínez-Romero, E. 2003. Diversity of Rhizobium-Phaseolus vulgaris symbiosis: overview and perspectives. Plant Soil 252:11-23. [Google Scholar]
- 41.McDonald, I. R., P. Kämpfer, E. Topp, K. L. Warner, M. J. Cox, T. L. Ccnell Hancock, L. G. Miller, M. J. Larkin, V. Ducrocq, C. Coulter, D. B. Harper, J. C. Murrell, and R. S. Oremland. 2005. Aminobacter ciceronei sp. nov. and Aminobacter lissarensis sp. nov., isolated from various terrestrial environments. Int. J. Syst. Evol. Microbiol. 55:1827-1832. [DOI] [PubMed] [Google Scholar]
- 42.Menna, P., M. Hungria, F. G. Barcellos, E. V. Bangel, P. N. Hess, and E. Martinez-Romero. 2006. Molecular phylogeny based on the 16S rRNA gene of elite rhizobial strains used in Brazilian commercial inoculants. Syst. Appl. Microbiol. 29:315-332. [DOI] [PubMed] [Google Scholar]
- 43.Montes, L. 1982. Big trefoil naturalized in southwest of Argentina. Lotus Newslett. 13:22-23. [Google Scholar]
- 44.Montes, L. 1986. Lotus tenuis. Rev. Arg. Prod. Anim. 8:357-376. [Google Scholar]
- 45.Monza, J., E. Fabiano, and A. Arias. 1992. Characterization of an indigenous population of rhizobia nodulating Lotus corniculatus. Soil Biol. Biochem. 24:241-247. [Google Scholar]
- 46.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 47.Nour, S. M., J. C. Cleyet-Marel, P. Normand, and M. P. Fernandez. 1995. Genomic heterogeneity of strains nodulating chickpeas (Cicer arietinum L.) and description of Rhizobium mediterraneum sp. nov. Int. J. Syst. Bacteriol. 45:640-648. [DOI] [PubMed] [Google Scholar]
- 48.Novikova, N. I., E. A. Pavlova, N. I. Vorobjev, and E. V. Limeshenko. 1994. Numerical taxonomy of Rhizobium strains from legumes of the temperate zone. Int. J. Syst. Bacteriol. 44:734. [Google Scholar]
- 49.Oyaizu, H., S. Matsumoto, K. Minamisawa, and T. Gamou. 1993. Distribution of rhizobia in leguminous plants surveyed by phylogenetic identification. J. Gen. Appl. Microbiol. 39:339-354. [Google Scholar]
- 50.Pankhurst, C. E. 1981. Effect of plant nutrient on nodule effectiveness and Rhizobium strain competition for nodulation of Lotus pedunculatus. Plant Soil 60:325-339. [Google Scholar]
- 51.Pearson, K. 1926. On the coefficient of racial likeness. Biometrika 18:105-117. [Google Scholar]
- 52.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1998. Characterization of the diversity of ecologically important microbes by rep-PCR fingerprinting, p. 1-26. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, supplement 3. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 53.Rigaud, J., and A. Puppo. 1975. Indole-3-acetic catabolism by soybean bacteroids. J. Gen. Microbiol. 88:223-228. [Google Scholar]
- 54.Rivas, R., M. Naranjo, P. F. Mateos, S. Oliveira,. E. Martinez-Molina, and E. Velázquez. 2007. Strains of Mesorhizobium amorphae and Mesorhizobium tianshanense, carrying symbiotic genes of common chickpea endosymbiotic species, constitute a novel biovar (ciceri) capable of nodulating Cicer arietinum. Lett. Appl. Microbiol. 44:412-418. [DOI] [PubMed] [Google Scholar]
- 55.Saeki, K., and H. Kouchi. 2000. The Lotus symbiont, Mesorhizobium loti: molecular genetic techniques and application. J. Plant Res. 113:457-465. [Google Scholar]
- 56.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 57.Salazar Lea Plaza, J. C., and G. Moscatelli. 1989. Mapa de suelos de la Provincia de Buenos Aires. Editorial Edipubli, S. A., Buenos Aires, Argentina.
- 58.Saldana, G., V. Martinez-Alcantara, J. M. Vinardell, R. Bellogin, J. E. Ruiz-Sainz, and P. A. Balatti. 2003. Genetic diversity of fast-growing rhizobia that nodulate soybean (Glycine max L. Merr). Arch. Microbiol. 180:45-52. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 60.Scott, D. B., R. Wilson, G. T. Shaw, A. Petit, and J. Tempe. 1987. Biosynthesis and degradation of nodule-specific Rhizobium loti compounds in Lotus nodules. J. Bacteriol. 169:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott, K. F., B. G. Rolfe, and J. Shine. 1983. Biological nitrogen fixation: primary structure of the Rhizobium trifolii iron protein gene. DNA 2:149-155. [DOI] [PubMed] [Google Scholar]
- 62.Sessitsch, A., H. Ramirez-Saad, G. Hardarson, A. D. Akkermans, and W. M. de Vos. 1997. Classification of Austrian rhizobia and the Mexican isolate FL27 obtained from Phaseolus vulgaris L. as Rhizobium gallicum. Int. J. Syst. Bacteriol. 47:1097-1101. [DOI] [PubMed] [Google Scholar]
- 63.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman and Co., San Francisco, CA.
- 64.Streeter, J. G. 1994. Failure of inoculant rhizobia to overcome the dominance of indigenous strains for nodule formation. Can. J. Microbiol. 40:513-522. [Google Scholar]
- 65.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. De Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sullivan, J. T., B. D. Eardly, P. van Berkum, and C. W. Ronson. 1996. Four unnamed species of nonsymbiotic rhizobial isolated from the rhizosphere of Lotus corniculatus. Appl. Environ. Microbiol. 62:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 68.van Berkum, P., D. Beyene, and B. D. Eardly. 1996. Phylogenetic relationships among Rhizobium species nodulating the common bean (Phaseolus vulgaris L.). Int. J. Syst. Bacteriol. 46:240-244. [DOI] [PubMed] [Google Scholar]
- 69.Velazquez, E., J. M. Igual, A. Willems, M. P. Fernandez, E. Munoz, P. F. Mateos, A. Abril, N. Toro, P. Normand, E. Cervantes, M. Gillis, and E. Martinez-Molina. 2001. Mesorhizobium chacoense sp. nov., a novel species that nodulates Prosopis alba in the Chaco Arido region (Argentina). Int. J. Syst. Evol. Microbiol. 51:1011-1021. [DOI] [PubMed] [Google Scholar]
- 70.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. International Biological Programme, Number 15. Blackwell Scientific, Oxford, United Kingdom.
- 72.Vinuesa, P., C. Silva, M. J. Lorite, M. L. Izaguirre-Mayoral, E. J. Bedmar, and E. Martinez-Romero. 2005. Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst. Appl. Microbiol. 28:702-716. [DOI] [PubMed] [Google Scholar]
- 73.Wang, E. T., P. van Berkum, X. H. Sui, D. Beyene, W. X. Chen, and E. Martinez-Romero. 1999. Diversity of rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp. nov. Int. J. Syst. Bacteriol. 49:51-65. [DOI] [PubMed] [Google Scholar]
- 74.Willems, A., and M. D. Collins. 1993. Phylogenetic analysis of rhizobia and agrobacteria based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 43:305-313. [DOI] [PubMed] [Google Scholar]
- 75.Yang, D., Y. Oyaizu, H. Oyaizu, G. J. Olsen, and C. R. Woese. 1985. Mitochondrial origins. Proc. Natl. Acad. Sci. USA 82:4443-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young, J. M., D. C. Park, and B. S. Weir. 2004. Diversity of 16S rRNA sequences of Rhizobium spp.: implications for species determinations. FEMS Microbiol. Lett. 238:125-131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.