Abstract
Supplementation of infant formulas with prebiotic ingredients continues the effort to mimic functional properties of human milk. In this double-blind, controlled, 28-day study, healthy term infants received control formula (control group; n = 25) or control formula supplemented with polydextrose (PDX) and galactooligosaccharide (GOS) (4 g/liter) (PG4 group; n = 27) or with PDX, GOS, and lactulose (LOS) (either 4 g/liter [PGL4 group; n = 27] or 8 g/liter [PGL8 group; n = 25]). A parallel breast-fed group (BF group) (n = 30) was included. Stool characteristics, formula tolerance, and adverse events were monitored. Fecal bacterial subpopulations were evaluated by culture-based selective enumeration (Enterobacteriaceae), quantitative real-time PCR (Clostridium clusters I, XI, and XIV, Lactobacillus, and Bifidobacterium), and fluorescence in situ hybridization (FISH) (Bifidobacterium). Fecal bacterial community profiles were examined by using 16S rRNA gene PCR-denaturing gradient gel electrophoresis. The daily stool consistency was significantly softer or looser in the BF group than in all of the groups that received formula. The formulas were well tolerated, and the incidences of adverse events did not differ among feeding groups. Few significant changes in bacterial subpopulations were observed at any time point. The bacterial communities were stable; individual profiles tended to cluster by subject rather than by group. Post hoc analysis, however, demonstrated that the bacterial community profiles for subjects in the BF, PG4, PGL4, and PGL8 groups that first received formula at a younger age were less stable than the profiles for subjects in the same groups that received formula at an older age, but there was no difference for the control group. These data indicate that formulas containing PDX, GOS, and LOS blends are more likely to influence gut microbes when administration is begun in early infancy and justify further investigation of the age-related effects of these blends on fecal microbiota.
Nondigestible food ingredients called prebiotics pass into the lower gastrointestinal tract and, by definition, may be selectively metabolized by mutualistic microorganisms, such as Lactobacillus spp. and Bifidobacterium spp., which in turn contribute to improved host health (12, 34). After lactose and lipids, oligosaccharides, which have prebiotic activity, are the third largest component of human breast milk (5 to 10 g/liter), and there are as many as 200 distinct molecular structures (5, 26). Lactobacilli and bifidobacteria are the predominant bacteria in the intestinal microbiota of breast-fed infants, whereas infants who receive cow's milk-based infant formulas, which naturally contain low levels of oligosaccharides, often have higher concentrations of potentially pathogenic bacteria, such as Enterobacteriaceae and clostridia, in their intestinal microbiota (4, 15, 17).
Clinical investigations of infant formulas supplemented with galactooligosaccharide (GOS) and fructooligosaccharide (FOS) at a range of concentrations and a range of blends have revealed increases in the numbers of fecal bifidobacteria in preterm infants (7, 18) and in the numbers of both bifidobacteria and lactobacilli in term infants (2, 3, 6, 7, 13, 14, 18, 19, 23, 24, 31) compared with infants receiving unsupplemented formulas. The Bifidobacterium levels (3, 6, 7, 14) and stool consistencies (7, 23, 24) have often been comparable for infants in both prebiotic-supplemented formula groups and human breast milk-fed reference groups. Nondigestible polydextrose (PDX) and lactulose (LOS) also stimulate bifidobacterial growth (8, 16, 29). In a recent safety and tolerance study, healthy term infants who received a formula supplemented with prebiotic blends of PDX, GOS, and LOS when they were 14 to 120 days old exhibited normal growth and stool characteristics more similar to those of breast-fed infants than to those of infants fed an unsupplemented control formula (35).
In the present study, the effects of the same prebiotic blends on the fecal bacterial populations of formula-fed infants were investigated during a 28-day feeding period using 16S rRNA gene-based real-time quantitative PCR (qPCR), 16S rRNA-based fluorescence in situ hybridization (FISH), and PCR-denaturing gradient gel electrophoresis (DGGE). These culture-independent molecular techniques are now commonly used to identify, quantify, and determine the profiles of bacteria in complex microbial communities that are difficult to assess using standard culture methods, such as the communities in the gastrointestinal tract (2, 6, 9, 13-15, 19, 22, 28, 31, 36). In this study, the Clostridium botulinum group (cluster I), the Clostridium lituseburense group (cluster XI), the Clostridium coccoides-Eubacterium rectale group (cluster XIV), and the genera Lactobacillus and Bifidobacterium in infant fecal samples were targeted with 16S rRNA gene group-specific primers, and changes in the subpopulations were evaluated by qPCR. Potential changes in the bifidobacterial subpopulation and total bacteria were also evaluated by using FISH, and Enterobacteriaceae were enumerated on selective media. Specific changes in the sum of the levels of Clostridium clusters I and XI, which include the pathogenic organisms Clostridium perfringens and Clostridium difficile, respectively, were chosen for the primary comparison in this study since consistently lower levels of Clostridium spp. have been found in breast-fed infants than in formula-fed infants (10). Differences in bacterial community composition between feeding groups and between individual participants were analyzed by using DGGE. Infant growth, stool characteristics, and tolerance of formulas were also monitored. The results were compared with the results for a parallel breast-fed reference group.
MATERIALS AND METHODS
Subjects and experimental design.
Healthy term infants who were 13 to 92 days old were recruited for a multicenter, double-blind, controlled, parallel-group feeding trial in the United States. Infants who received antibiotics within 14 days of the beginning of the study were excluded. At the time of enrollment, formula-fed infants (n = 105) started receiving a commercially marketed formula, Enfamil LIPIL with iron (Mead Johnson & Company, Evansville, IN) (control group). Following a 7-day formula run-in phase, all infants participated in a feeding study over a 28-day period. At baseline (on day 1 of the feeding study), formula-fed infants were randomized and placed in one of four groups: (i) the control group (n = 25); (ii) a group that received the control formula supplemented with 4 g/liter of a prebiotic blend containing PDX (Litesse Two PDX; Danisco, Copenhagen, Denmark) and GOS (Vivinal GOS; Friesland Foods Domo, Zwolle, The Netherlands) at a ratio of 1:1 (PG4 group; n = 27); (iii) a group that received the control formula supplemented with 4 g/liter of a prebiotic blend containing PDX, GOS, and LOS (lactulose anhydride; Morinaga Milk Industry Co. Ltd., Tokyo, Japan) at a ratio of 3:2:1 (PGL4 group; n = 27); or (iv) a group that received the control formula supplemented with 8 g/liter of a prebiotic blend containing PDX, GOS, and LOS at a ratio of 3:2:1 (PGL8 group; n = 26). A parallel breast-fed group of infants (BF group) was included as a reference group (n = 30). All of the infants in the reference group were exclusively or predominantly (no more than 120 ml water or juice per day) breast fed prior to the trial, were exclusively breast fed for 7 to 8 days before collection of the baseline stool samples, and then were exclusively breast fed for the remainder of the study. Body weight (in grams), length (in centimeters), and head circumference (in centimeters) were recorded at enrollment and day 28. A parent or caregiver kept a daily record of the formula intake (in ounces), the incidence of diarrhea or constipation, the stool frequency and consistency (using the following scale: 1, hard; 2, formed; 3, soft; 4, loose; 5, watery), the amount of gas (using the following scale: 0, none, 1; small amount, 2; moderate amount; 3, excessive amount), and fussiness (using the following scale: 0, not fussy; 1, slightly fussy; 2, moderately fussy; 3, very fussy; 4, extremely fussy). Adverse events were monitored throughout the study. Institutional review boards for each site reviewed and approved the protocol and procedures, and parents provided written informed consent. The study was conducted in compliance with good clinical practices.
Fecal collection and sampling.
Study personnel collected infant stool specimens at baseline, on day 14, and on day 28 no more than 2 h following defecation by using chemical-free diapers (Tushies; TenderCare International, Eau Claire, WI). Samples were evenly divided and placed into two separate 15-ml transport containers, and the aliquots were treated as follows: (i) shipped under ambient conditions to a central clinical laboratory (North Coast Clinical Laboratory, Sandusky, OH) for enumeration of Enterobacteriaceae or (ii) immediately frozen at ≤−20°C and shipped on cold packs to a second central laboratory (Department of Animal Sciences, University of Illinois, Urbana) for DNA isolation and microbial community analysis. Following storage for approximately 3 years at −20°C, the second set of fecal samples was shipped on dry ice to a third laboratory (Department of Medical Microbiology, University Medical Center, Groningen, The Netherlands) for relative quantification of bifidobacteria by FISH.
Bacterial strains and culture conditions.
Bacterial strains used to obtain the qPCR standard curves are listed in Table 1. All Clostridium strains and Bacteroides fragilis were cultured in reinforced clostridial medium broth, Lactobacillus delbrueckii was cultured in de Man-Rogosa-Sharpe broth, and Bifidobacterium adolescentis was cultured in Belitsky minimal broth supplemented with hemin and menadione. All strains were grown at 37°C and harvested during exponential growth. Enterobacteriaceae were cultured on MacConkey agar at 37°C for at least 48 h, and the results were expressed in CFU.
TABLE 1.
qPCR standards and detection limits
| Bacterial group | Representative strain used for standard curve | Detection limit (log ng/μl) |
|---|---|---|
| Total bacteria | B. fragilis lab strain | −2.0 |
| Lactobacillus | L. delbrueckii lab strain | −3.7 |
| Bifidobacterium | B. adolescentis lab strain | −4.0 |
| Clostridium cluster I | C. paraputrificum 16480 | −3.0 |
| Clostridium cluster XI | C. difficile 17450 | −4.0 |
| Clostridium cluster XIV | C. coccoides ATCC 29236 | −6.0 |
DNA extraction.
Genomic DNA in fecal samples (200 mg) and bacterial cultures was isolated using a previously described method (33).
Real-time qPCR.
Assays were performed using the ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA). The PCR amplification protocols were the protocols described previously for total bacteria (9) and the genera Bifidobacterium (22), Lactobacillus (9), and Clostridium (32). All reaction mixtures contained 1 ng template DNA, a specific primer set (Table 1), and bovine serum albumin (0.01 mg/ml). SYBR green PCR master mixture (2×; Applied Biosystems) was used for detection of total bacteria, Bifidobacterium, and Lactobacillus. For detection of Clostridium, TaqMan PCR master mixture was used, and the internal probe was labeled with 6-carboxyfluorescein as the 5′ reporter dye and 6-carboxytetramethylrhodamine as the 3′ quencher dye. Standard curves were generated using serial dilutions of genomic DNA extracted from pure cultures of representative bacteria (Table 1). The initial data were expressed as target DNA concentrations, which were normalized by expression relative to the total bacterial DNA concentration.
16S rRNA gene PCR-DGGE.
Template DNA (10 ng) was amplified using universal bacterial 16S-V3 primers (Table 2) (a 40-bp GC clamp was added to the forward primer) as described previously (25). PCR amplicons were loaded in 35 to 60% linear DNA-denaturing gradient gels, and DGGE was performed using the Bio-Rad D-code system (Bio-Rad, Los Angeles, CA). The reference ladder contained PCR amplicons from pure cultures of B. fragilis, E. rectale, L. delbrueckii, and B. adolescentis from the top to the bottom (both the fourth and fifth bands were derived from B. adolescentis and consistently appeared on the gel). Following electrophoresis, gels were silver stained and scanned using a GS-710 calibrated imaging densitometer (Bio-Rad). Images were analyzed using Diversity Database software (Bio-Rad). Each band on the gel was identified regardless of the band intensity. Pairwise similarity was determined using the Dice coefficient (Cs), where patterns with no common bands received a score of 0 and identical band patterns received a score of 1.0 (21). Dendrograms were generated by using Ward's algorithm, in which samples were clustered by the degree of similarity to visualize community relatedness.
TABLE 2.
PCR primers and probes used for PCR-DGGE, qPCR, and FISH
| Target bacterial group | Primer or probe | Oligonucleotide sequence (5′→3′) | Annealing temp (°C) | Reference |
|---|---|---|---|---|
| Total bacteria | 341F | CCTACGGGAGGCAGCAGa | 55-65 | 25 |
| 543R | ATTACCGCGGTGCTGG | |||
| Eub338 | GCTGCCTCCCGTAGGAGT | 1 | ||
| Lactobacillus | Lab-0159 | GGAAACAGRTGCTAATACCG | 56 | 9 |
| Univ-0515 | ATCGTATTACCGCGGCTGCTGGCA | |||
| Bifidobacterium | g-Bifid-F | CTCCTGGAAACGGGTGG | 55 | 22 |
| g-Bifid-R | GGTGTTCTTCCCGATATCTACA | |||
| Bif164 | CATCCGGYATTACCACCC | 60.2 | 20 | |
| Clostridium cluster I | CI-F1 | TACCHRAGGAGGAAGCCAC | 63 | 32 |
| CI-R2 | GTTCTTCCTAATCTCTACGCAT | |||
| Clostridium cluster XI | CXI-F1 | ACGCTACTTGAGGAGGA | 58 | 32 |
| CXI-R2 | GAGCCGTAGCCTTTCACT | |||
| Clostridium cluster XIV | CXIV-F1 | GAWGAAGTATYTCGGTATGT | 52 | 32 |
| CXIV-R2 | CTACGCWCCCTTTACAC | |||
| Clostridium | Probe-I | GTGCCAGCAGCCGCGGTAATACG | 32 |
The sequence contained an additional 40-nucleotide GC clamp for DGGE.
FISH.
Quantification of bifidobacteria by FISH was carried out by first suspending 0.5 g of each fecal sample in 4.5 ml filtered phosphate-buffered saline (PBS) with three to five glass beads (diameter, 4 mm) and homogenizing the preparation with a vortex mixer for 3 min. Debris was removed by centrifugation at 1,000 rpm for 1 min, and the supernatant was fixed by diluting it 1:4 in freshly prepared 4% (wt/vol) paraformaldehyde in PBS and incubating it overnight at 4°C. Depending on the probable number, the fixed samples were diluted as necessary in PBS, and 10-μl aliquots were spread completely across the surfaces of 1-cm2 wells in custom-made slides (CBN, The Netherlands) pretreated with a gelatin solution [0.1% gelatin (Oxoid), 0.01% KCr(SO4)2·12H2O (Sigma)], allowed to dry evenly for 30 to 60 min at room temperature, and fixed in 96% (vol/vol) ethanol for 10 min. Bifidobacteria were labeled by hybridization for 16 h at 50.0°C with the genus-specific Bif164 probe (20) that was 5′ end labeled with fluorescein isothiocyanate (Pharmacia) and added at a concentration of 10 ng/μl in buffer (20 mM Tris-HCl, 0.9 M NaCl, 0.1% sodium dodecyl sulfate; pH 7.2). After hybridization, the slides were rinsed in buffer (20 mM Tris-HCl, 0.9 M NaCl) for 30 min at 50.0°C and then with Milli-Q water, dried rapidly using compressed air, and mounted using 6 μl Vectashield (Vector Laboratories, Burlingame, CA) in each well and a coverslip. Total bacteria were labeled in a similar way by hybridization with the Eub338 probe (1). Digital images were captured using a Leica DMRA2 epifluorescence microscope (Wetzlar, Germany), and fluorescent cells were counted either visually (average number in 10 to 25 fields with 30 to 100 positive objects) or automatically using the Quantimet HR600 image analysis software (Leica).
Statistical analysis.
The change in the sum for Clostridium clusters I and XI from baseline to day 28 was the primary outcome used to evaluate study formulas. A sample size of 17 infants per formula group was used to detect a difference of one standard deviation at 80% power (α = 0.05). qPCR data for bacterial subpopulations expressed as percentages of total bacteria were not normally distributed; therefore, data were expressed as median values. Changes from the baseline values among groups of formula-fed infants were analyzed using the Wilcoxon rank sum test; the sign test was used for within-group comparisons. Comparisons of values for all feeding groups at baseline, day 14, and day 28 were performed using the Wilcoxon rank sum test. Comparisons of changes in the levels of Enterobacteriaceae (log10 CFU/g stool) for formula groups compared with the baseline level were performed using analysis of covariance. A repeated-measures analysis that included all feeding groups was performed using data obtained at baseline, day 14, and day 28. Cs and Ward's algorithm calculations were performed using Diversity Database software (Bio-Rad). Cs were analyzed by using analysis of variance (ANOVA). Feeding group and age were factored into the analysis. Anthropometric measurements were converted to z-scores based on U.S. National Center for Health Statistics reference data (http:/www.cdc.gov/nccdphp/dnpa/growthcharts/resources/sas.htm) and were analyzed by using ANOVA. Race, ethnicity, gender, and study discontinuance were analyzed by using Fisher's exact test. Analyses were performed by using SAS, version 8 (Cary, NC).
RESULTS
Infant growth, stool characteristics, and formula tolerance.
There were no statistically significant differences in study discontinuation rates among the feeding groups, and 117 infants completed the study, including 29 infants in the BF group, 21 infants in the control group, 23 infants in the PG4 group, 23 infants in the PGL4 group, and 21 infants in the PGL8 group. One infant (PGL8 group) did not consume the study formula after randomization and was not included in any of the analyses, and one infant (BF group) did not meet the gestational age requirement and was not included in analyses of intestinal bacterial composition, stool characteristics, and tolerance.
The feeding groups were similar with respect to gender distribution and age of the participants. There was a statistically significant difference in race only between the BF group (83% white, 3% black, and 13% more than one race) and the control group (56% white and 44% black) (P = 0.002). The mean formula intake values were similar during the 7-day formula run-in period (195.0 to 214.8 ml/kg per day) and during the 28-day feeding trial (206.3 to 235.0 ml/kg per day). The means for weight-, length-, and head circumference-for-age z-scores at enrollment and day 28 are shown in Table 3. Statistically significant differences in mean weight- and length-for-age z-scores among feeding groups were detected at enrollment and day 28 (Table 3 shows individual P values for all pairwise comparisons of z-scores). Mean weight- and length-for-age z-scores for each group either remained the same or increased between the time of enrollment and day 28. There were no significant differences in mean head circumference-for-age z-scores at enrollment or at study day 28.
TABLE 3.
Mean weight-, length-, and head circumference-for-age z-scores at enrollment and day 28
| Time | Group (n) | Mean z-score for agea
|
||
|---|---|---|---|---|
| Weight | Length | Head circumference | ||
| Enrollment | BF (30) | 0.5 A | 0.3 AC | −0.1 A |
| Control (25) | 0.0 B | 0.0 BC | −0.2 A | |
| PG4 (27) | 0.4 AB | 0.5 A | 0.1 A | |
| PGL4 (27) | 0.0 B | −0.1 B | −0.3 A | |
| PGL8 (25) | 0.8 A | 0.5 A | 0.0 A | |
| Day 28 | BF (29) | 0.6 AB | 0.3 AB | 0.0 A |
| Control (22) | 0.2 A | 0.2 AB | −0.1 A | |
| PG4 (24) | 0.7 BC | 0.6 A | 0.2 A | |
| PGL4 (24) | 0.4 AC | 0.0 B | 0.0 A | |
| PGL8 (21) | 1.0 B | 0.6 A | 0.1 A | |
The standard error for all means was 0.2. In each column means for the same time point (enrollment or day 28) followed by the same letter are not significantly different (P ≥ 0.05).
There were no significant differences among the feeding groups in the numbers of infants who experienced constipation or diarrhea during the run-in period or feeding trial (data not shown). The stool frequency (mean ± standard error) was significantly higher for the BF group (3.6 ± 0.3 stools/day) than for any of the formula groups (1.6 to 2.3 ± 0.3 stools/day) (P ≤ 0.018) during the run-in period; there were no significant differences among feeding groups in average mean stool frequency from baseline to day 28 (1.9 to 2.8 ± 0.3 stools/day). The daily stool consistency scores (mean ± standard error) were significantly higher for the BF group (3.8 ± 0.1) than for all of the formula groups (control group, 3.3 ± 0.1; PG4 group, 3.3 ± 0.1; PGL4 group, 3.2 ± 0.1; PGL8 group, 3.1 ± 0.1) (P ≤ 0.004) during the run-in period; the scores for the BF group (3.8 ± 0.1) and each formula group were also significantly different when the averages from baseline to day 28 were compared (control group, 3.2 ± 0.1; PG4, PGL4, or PGL8 group, 3.4 ± 0.1) (P < 0.05). The only significant difference in the amount of gas (mean ± standard error) was the difference between the BF group (0.9 ± 0.1) and the PGL8 group (1.4 ± 0.1) (P = 0.006) from day 15 to day 21.
There were no significant differences among the feeding groups in the amount of fussiness (mean ± standard error) during the run-in period or feeding trial (1.0 to 1.3 ± 0.1). There were no differences among the study groups in the number of participants who experienced at least one adverse event, and there were no differences in the number of participants who experienced adverse events when body systems or individual events were analyzed. There were no adverse events that had particular clinical significance in any feeding group (data not shown).
Quantification of Enterobacteriaceae.
Selective plating of fecal samples on MacConkey agar was used to enumerate fecal Enterobacteriaceae populations in all feeding groups (log10 CFU/g stool). No significant differences in the mean changes in the Enterobacteriaceae populations from baseline to day 28 were detected among the formula groups (control, PG4, or PGL4 group, 0.2 ± 0.1 log10 CFU/g stool; PGL8 group, 0.3 ± 0.1 log10 CFU/g stool). Within the PGL8 group there was a slight but statistically significant increase in the mean level of Enterobacteriaceae (P = 0.027) from baseline to day 28, but there were no significant changes within the other feeding groups. The means for Enterobacteriaceae for the different feeding groups at baseline, day 14, and day 28 are shown in Table 4. The mean for the BF group was significantly higher than the means for all other groups at baseline (P < 0.001). Significantly higher levels of Enterobacteriaceae were also observed on day 14 for the BF group than for the prebiotic-supplemented groups (P ≤ 0.002). The Enterobacteriaceae levels for the feeding groups at day 28 were similar.
TABLE 4.
Quantification of Enterobacteriaceae in fecal samples from different feeding groups by selective enumeration on MacConkey agar
| Time | Concn of Enterobacteriaceae (log10 CFU/g stool)a
|
||||
|---|---|---|---|---|---|
| BF group | Control group | PG4 group | PGL4 group | PGL8 group | |
| Baseline | 7.9 A | 7.1 B | 7.2 B | 7.1 B | 7.1 B |
| Day 14 | 7.7 A | 7.4 AB | 7.2 B | 7.1 B | 7.2 B |
| Day 28 | 7.5 A | 7.3 A | 7.2 A | 7.3 A | 7.3 A |
The standard error for all means was 0.1. For each time means followed by the same letter are not significantly different (P ≥ 0.05).
Quantification of fecal bacterial groups using real-time qPCR.
To compare the effects of prebiotic blends in infant formulas fed over a 28-day period on fecal bacterial composition, qPCR was used to determine the levels of Clostridium clusters I, XI, and XIV, Lactobacillus spp., and Bifidobacterium spp. expressed as percentages of the total bacteria. Data for eight samples were not included in the statistical analyses due to an unreasonably low density (four samples) or no detection (four samples) of total bacteria by qPCR. No significant differences among the formula groups in changes in the bacterial subpopulations from baseline to day 28 were observed for Clostridium cluster I, cluster XI, or cluster XIV, the sum for Clostridium clusters I and XI, Bifidobacterium spp., or Lactobacillus spp. As determined by using the sign test, a significantly greater number of samples exhibited decreases in the levels of Clostridium cluster XI (P = 0.049) and Lactobacillus spp. (P = 0.013) in the PGL8 group from baseline to day 28 (data not shown).
Median values for Clostridium clusters, Lactobacillus spp., and Bifidobacterium spp. for each feeding group at baseline, day 14, and day 28 are shown in Table 5. No significant differences among feeding groups in Clostridium cluster I values were observed at any time point. The median values for Clostridium cluster XI were significantly lower for the BF group than for the PG4 group (P = 0.006), the PGL4 group (P < 0.001), and the PGL8 group (P = 0.016) at baseline, for all other groups at day 14 (control group, P = 0.011; PG4 group, P < 0.001; PGL4 group, P = 0.003; PGL8 group, P = 0.019), and for the PG4 group (P = 0.005) and the PGL4 group (P = 0.015) at day 28. No significant differences among feeding groups for Clostridium cluster XIV, the sum for Clostridium clusters I and XI, Lactobacillus spp., or Bifidobacterium spp. were observed at any time point.
TABLE 5.
Evaluation of Clostridium clusters I, XI, and XIVa, Lactobacillus spp., and Bifidobacterium spp. at baseline, day 14, and day 28 by qPCR and FISH
| Time | Group (n) | Median % of total bacterial DNAa
|
||||||
|---|---|---|---|---|---|---|---|---|
|
Clostridium clusters
|
Lactobacillus |
Bifidobacterium
|
||||||
| I | XI | XIVa | I and XI | qPCR | FISH | |||
| Baseline | BF (28) | 0.2 A | <0.1 A | 0.2 A | 0.7 A | 3.5 A | 1.5 A | 83.5 A |
| Control (21) | 1.1 A | 0.1 AB | 1.0 A | 2.3 A | 3.9 A | 0.2 A | 34.5 AB | |
| PG4 (24) | 1.4 A | 0.3 B | 0.2 A | 3.0 A | 2.5 A | 0.1 A | 43.7 AB | |
| PGL4 (25) | 0.7 A | 0.3 B | 0.9 A | 1.6 A | 3.1 A | 0.3 A | 42.6 AB | |
| PGL8 (18) | 1.6 A | 0.2 B | 1.1 A | 2.2 A | 3.5 A | 0.1 A | 20.7 B | |
| Day 14 | BF (26) | 1.5 A | <0.1 A | 0.1 A | 2.1 A | 2.4 A | 1.2 A | 53.4 A |
| Control (21) | 1.1 A | 0.1 B | 0.7 A | 3.7 A | 3.2 A | 1.1 A | 45.5 A | |
| PG4 (24) | 1.5 A | 0.4 B | 0.5 A | 2.1 A | 3.2 A | 0.2 A | 42.1 A | |
| PGL4 (25) | 0.1 A | 0.2 B | 0.9 A | 2.6 A | 2.6 A | 0.3 A | 45.5 A | |
| PGL8 (20) | 1.3 A | 0.1 B | 1.1 A | 1.7 A | 2.8 A | 0.3 A | 28.4 A | |
| Day 28 | BF (27) | 0.6 A | <0.1 A | 0.5 A | 0.7 A | 2.7 A | 1.7 A | 70.3 A |
| Control (20) | 4.1 A | 0.2 AC | 0.6 A | 4.5 A | 3.7 A | 1.0 A | 44.9 A | |
| PG4 (23) | 4.0 A | 0.5 BC | 0.6 A | 4.4 A | 2.5 A | 0.5 A | 35.7 A | |
| PGL4 (23) | 0.8 A | 0.2 BC | 1.0 A | 1.6 A | 2.5 A | 0.5 A | 54.3 A | |
| PGL8 (19) | 0.6 A | 0.1 AC | 1.3 A | 0.9 A | 2.0 A | 0.4 A | 57.8 A | |
In each column, values for the same time point (enrollment or day 28) followed by the same letter are not significantly different (P ≥ 0.05).
Repeat quantification of the fecal bifidobacterial subgroup by FISH.
Given the generally low level of bifidobacteria detected by qPCR, fecal samples were reanalyzed by performing FISH using a probe specific for this subpopulation. Compared to qPCR, FISH detected considerably higher median levels of bifidobacteria, expressed as a percentage of the total bacteria. The only significant difference was detected for samples collected during the baseline phase of the study, in which bifidobacteria were found to account for 20.7 and 83.5% of the total bacteria in the PGL8 group and human milk reference group, respectively (Table 5). No other significant differences in levels of bifidobacteria were observed for the various feeding groups at any time.
Profiling of bacterial communities using DGGE.
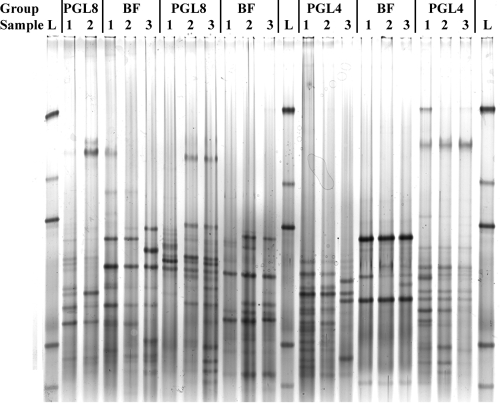
Bacterial community profiles were generated by 16S rRNA gene PCR-DGGE. Samples collected from the same subject at baseline, day 14, and day 28 were run on the same gel to maintain electrophoretic consistency (representative results are shown in Fig. 1). Cs were calculated to quantify the pairwise similarity of DNA banding patterns within (intragroup) and among (intergroup) feeding groups at baseline, day 14, and day 28. The levels of intergroup similarity at baseline (Cs range, 18.0 to 19.2), day 14 (Cs range, 19.2 to 21.7), and day 28 (Cs range, 16.4 to 19.9) were low. The intragroup Cs values were within similar ranges at all time points (data not shown). The intragroup and intergroup Cs values were not significantly different at any time point. A cluster analysis was performed using Ward's algorithm, and dendrograms were constructed to visualize relationships among banding patterns for samples. Samples obtained from the same subject at all time points examined tended to cluster together. No specific clustering patterns were observed for feeding groups, ages, or races (data not shown).
FIG. 1.
Representative DGGE profiles for individual infants at three study time points, baseline (lanes 1), day 14 (lanes 2), and day 28 (lanes 3). Lanes L contained a standard ladder consisting of B. fragilis, E. rectale, L. delbrueckii, and B. adolescentis (fourth and fifth bands) from top to bottom. Each collection of samples is grouped by study diet.
DGGE profiles generated for individual subjects at baseline, day 14, and day 28 were compared across three intervals to index the bacterial stability in response to the feeding regimen during the study period. Individual DGGE profiles obtained at different time points were compared to calculate the Cs value, which was used as an index of bacterial community stability; a higher Cs indicated greater bacterial stability and less population fluctuation during the corresponding feeding period. An analysis of Cs values by using ANOVA with feeding group and age as independent variables indicated that there was a significant age effect but there was not a feeding effect (data not shown). Because of the lack of a feeding effect during the 28-day period, a post hoc analysis was conducted, in which individuals in each feeding group were classified by age to obtain data for younger infants (21 to 50 days old at the beginning of the study) and older infants (51 to 104 days old at the beginning of the study). From baseline to day 28, the mean Cs values were lower for the younger infants in the BF group (P = 0.054) and significantly lower for the younger infants in all three supplemented-formula feeding groups (PG4 group, P = 0.048; PGL4 group, P = 0.040; and PGL8 group, P = 0.006) (Table 6). The mean Cs values for younger and older infants in the control group did not differ from baseline to day 28 (P = 0.511).
TABLE 6.
Cs of individual DGGE profiles between baseline and day 28
| Group | Cs for different agesa
|
P valueb | |
|---|---|---|---|
| 21 to 50 days | 51 to 104 days | ||
| BF | 0.49 ± 0.26 | 0.68 ± 0.23 | 0.054 |
| Control | 0.56 ± 0.25 | 0.62 ± 0.17 | 0.511 |
| PG4 | 0.40 ± 0.16 | 0.60 ± 0.27 | 0.048 |
| PGL4 | 0.41 ± 0.22 | 0.60 ± 0.20 | 0.040 |
| PGL8 | 0.38 ± 0.17 | 0.60 ± 0.11 | 0.006 |
The values are means ± standard deviations.
P values for comparisons of younger and older infants in the same groups. No significant differences among age-matched feeding groups were detected by ANOVA.
DISCUSSION
This study investigated the effects of prebiotic blends containing PDX and GOS (4.0 g/liter) or PDX, GOS, and LOS (4.0 or 8.0 g/liter) on fecal bacterial populations over a 28-day period in healthy term infants. Numerous clinical investigations of preterm and term infants who received formulas supplemented with GOS alone (3) or with GOS and FOS at a 9:1 ratio (2, 6, 7, 14, 18, 19, 23, 24, 31) at a wide range of concentrations (2.0 to 10 g/liter) have shown that the levels of Bifidobacterium spp. in the fecal bacterial population were higher in these infants than in infants who received unsupplemented formula. This is the first clinical evaluation of the effects of new prebiotic blends of PDX, GOS, and LOS on the infant fecal microbiota.
Small but statistically significant differences in mean weight- and length-for-age z-scores were noted among feeding groups at enrollment and study day 28. For all groups, however, the mean weight- and length-for-age z-scores either remained the same or increased between enrollment and day 28, indicating that each group was growing normally. Consequently, differences among feeding groups were not considered clinically significant. The current study was not designed to evaluate growth as a primary clinical outcome. Ziegler et al. (35), however, fed infants from days 14 to 120 old the formulas given to the control, PG4, and PGL8 groups in a study designed specifically to evaluate infant growth according to the American Academy of Pediatrics Task Force recommendations for clinical testing of infant formulas (11). These workers found no differences in the rates of increases in weight or length over the duration of the 120-day study and concluded that infants fed these prebiotic-containing formulas exhibited normal growth (35). Stools from infants in the BF group in the current study were found to be softer and looser than stools from infants in groups that received formula. However, the stool consistency scores changed for the groups of infants who received prebiotic-supplemented formula over the 28-day feeding trial and approached the stool consistency scores for the breast-fed infants. Ziegler et al. (35) also reported that infants fed formula supplemented with prebiotics had stool characteristics more similar to those of breast-fed infants than to those of infants fed unsupplemented formula. Overall, infant formulas containing blends of PDX, GOS, and LOS were generally well tolerated and safe.
Few significant differences in the overall bacterial composition were found from baseline to day 28 within feeding groups or among feeding groups at any time point. Compared to the traditional culture methods used in previous prebiotic feeding studies, the more sensitive molecular methods for quantifying bacteria in complex microbial communities may make broad differences between breast-fed and formula-fed infants less apparent. For example, a recent examination of fecal samples from breast-fed and formula-fed infants using qPCR demonstrated that there were no significant differences in the Bifidobacterium levels (28). Although the levels of bifidobacteria tended to be higher in the BF group throughout the current study, the levels of neither bifidobacteria nor lactobacilli were significantly different in the formula-fed infants and the infants fed breast milk at any time point, with one exception. The exception occurred in the baseline phase, when fecal samples from infants in the PGL8 group contained significantly less bifidobacteria (20.7%) than fecal samples from infants in the human milk reference group (83.5%), as determined by FISH. The general lack of significance could have been due to the great variation in the data, which could be attributed to the wider range of infant ages (21 to 104 days at baseline) and the shorter feeding period (28 days) compared with other similar prebiotic studies.
In addition, subtle differences between the microbiota of breast-fed infants and the microbiota of formula-fed infants might occur only at the species level, where species in certain subpopulations of the gut microbiota shift toward the pattern observed in breast-fed infants (28, 30). Rinne et al. (30) demonstrated that the total number of bifidobacteria was significantly lower in infants who received control formula than in infants who received breast milk or a prebiotic-supplemented formula (GOS and FOS at a ratio of 9:1); additionally, species-specific PCR revealed that the Bifidobacterium species composition obtained for the breast-fed infants was found in infants who received prebiotic-supplemented formula. Using a species-specific qPCR assay, Penders et al. (28) found a significantly lower prevalence and significantly lower numbers of C. difficile, a member of Clostridium cluster XI, in breast-fed infants than in formula-fed infants. In the current study, a significantly greater number of subjects in the PGL8 group exhibited a decrease of the number of Clostridium cluster XI organisms between baseline and day 28. However, no species-specific prebiotic effects, such as a decrease in the level of a pathogen or similarity in species diversity, could be attributed to supplementation of infant formula with GOS, PDX, and LOS with the group-specific primers used.
The level of bifidobacteria observed expressed as a percentage of the total bacterial DNA in this study appeared to be low compared with the common assumption that Bifidobacterium spp. dominate the infant fecal microbiota. However, the results obtained by using a DNA concentration-based standard curve in the qPCR analysis may not have accurately reflected the number of bacterial cells in the complete microbial community. To estimate the number of bifidobacterial cells in the fecal microbiota, we performed post hoc FISH using Bifidobacterium-specific and universal bacterial probes. The percentages of bifidobacterial cells in the original samples indicated by the FISH results were much higher than the percentages estimated by DNA concentration-based qPCR. Consistent with the qPCR results, the percentages of bifidobacteria were higher in BF group than in the other groups, but there were few statistically significant differences among groups due to the great variation among subjects within groups. The difference between the percentages of bifidobacteria estimated by FISH and qPCR can probably be partially explained by inherent differences in methodology. Bifidobacterium spp. have relatively low numbers of 16S rRNA gene copies compared with other major intestinal bacterial species, which leads to potential underestimation by 16S rRNA gene-targeted qPCR. Palmer et al. (27) also observed a relatively low frequency and low abundance of bifidobacteria in infant fecal samples based on the results of molecular methods, including qPCR (as in this study) and microarray analysis, but in the latter analysis the primers used for broad-range amplification of rRNA genes were suboptimal and underestimated bifidobacteria eightfold. Another explanation for the low level of bifidobacteria detected by qPCR in the current study may be the difference in DNA extraction efficiency between gram-positive and gram-negative bacteria, which may result in a bacterial DNA composition bias. The differences in the percentages of bifidobacteria reported in different studies emphasize the importance of reevaluating PCR-related biases, including DNA extraction bias, in order to obtain more accurate estimates of the contribution of this bacterial group to the intestinal microbiota of infants.
An unexpected finding was that the BF group had significantly higher levels of Enterobacteriaceae than the formula-fed groups at the beginning of the study, since the levels of Enterobacteriaceae are reportedly lower in breast-fed infants (28). However, by the end of the study period, the levels of Enterobacteriaceae were comparable for all feeding groups, suggesting that neither prebiotic carbohydrates nor human milk oligosaccharides influenced the growth of these bacteria.
Analysis of the 16S rRNA gene PCR-DGGE results for infant fecal samples revealed little similarity in the bacterial community profiles among subjects regardless of the feeding practice. The calculated Cs for DGGE banding patterns within or among feeding groups were low, and significant differences were not observed. Cluster analysis of DGGE profiles demonstrated that over time the individual bacterial community profiles clustered by subject rather than by feeding group. Significant differences in bacterial community stability were not observed among feeding groups during the 28-day feeding period, whereas a significant age effect was observed. A post hoc analysis of younger and older infants within each group revealed that the shifting microbial populations in younger infants in the PG4, PGL4, and PGL8 groups, as well as the BF reference group, were less stable from baseline to day 28. In contrast, the Cs values for the control group appeared to be relatively high for both younger and older infants, indicating that the overall microbial profile was reasonably stable over the 28-day feeding period regardless of age. These age-related differences might have been due to the presence of prebiotics in the formulas and the presence of human milk oligosaccharides in breast milk, as corresponding age-related differences were not observed for the control group. Although further age-based statistical analyses were not performed, the results suggest that the prebiotic blends had a greater effect on the fecal bacterial communities of younger infants than on the fecal bacterial communities of older infants and imply that infant age is a variable that is more important than originally expected.
With the current study design, few statistically significant differences in the composition of fecal bacterial populations were observed in the infants who received formula supplemented with GOS, PDX, or LOS prebiotic blends compared with the infants who received a control formula or the reference BF group. However, the age-related differences in fecal bacterial community stability observed within the PG4, PGL4, and PGL8 feeding groups indicate that these prebiotic blends may have a greater impact on infant fecal bacterial populations in younger infants than in older infants. Further analysis is necessary before definitive conclusions concerning this possibility can be reached.
Acknowledgments
This research was funded by Mead Johnson & Company, Evansville, IN.
We thank study investigators and their staffs for their cooperation, including Michael Brown (Hill Top Research, St. Petersburg, FL), Ann Grandjean (The Center for Human Nutrition, Omaha, NE), William Geffen (Hillcrest Medical Group, Tulsa, OK), Robert Lewine (Hill Top Research, Scottsdale, AZ), Derek Lewis (Little Rock, AR), Susanne Matthews (Metro Pediatrics, Birmingham, AL), William Parker (Portico Pediatrics, Shreveport, LA), Ekhard Ziegler (Children's Hospital of Iowa, Iowa City, IA), and Stacy Jones (Arkansas Children's Hospital, Little Rock, AR). The participation of parents and infants in this study is gratefully acknowledged. Gerwin C. Raangs and Gjalt W. Welling (University Medical Center, Groningen, The Netherlands) are acknowledged for conducting the FISH analysis of bifidobacteria. Julia Boettcher is acknowledged for assistance with the manuscript.
Footnotes
Published ahead of print on 16 December 2008.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakker-Zierikzee, A. M., M. S. Alles, J. Knol, F. J. Kok, J. J. Tolboom, and J. G. Bindels. 2005. Effects of infant formula containing a mixture of galacto- and fructo-oligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br. J. Nutr. 94:783-790. [DOI] [PubMed] [Google Scholar]
- 3.Ben, X. M., X. Y. Zhou, W. H. Zhao, W. L. Yu, W. Pan, W. L. Zhang, S. M. Wu, C. M. Van Beusekom, and A. Schaafsma. 2004. Supplementation of milk formula with galacto-oligosaccharides improves intestinal micro-flora and fermentation in term infants. Chin. Med. J. (Engl. Ed.). 117:927-931. [PubMed] [Google Scholar]
- 4.Benno, Y., K. Sawada, and T. Mitsuoka. 1984. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28:975-986. [DOI] [PubMed] [Google Scholar]
- 5.Bode, L. 2006. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J. Nutr. 136:2127-2130. [DOI] [PubMed] [Google Scholar]
- 6.Boehm, G., J. Jelinek, B. Stahl, K. van Laere, J. Knol, S. Fanaro, G. Moro, and V. Vigi. 2004. Prebiotics in infant formulas. J. Clin. Gastroenterol. 38:S76-S79. [DOI] [PubMed] [Google Scholar]
- 7.Boehm, G., M. Lidestri, P. Casetta, J. Jelinek, F. Negretti, B. Stahl, and A. Marini. 2002. Supplementation of a bovine milk formula with an oligosaccharide mixture increases counts of faecal bifidobacteria in preterm infants. Arch. Dis. Child. Fetal Neonatal Ed. 86:F178-F181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouhnik, Y., A. Attar, F. A. Joly, M. Riottot, F. Dyard, and B. Flourie. 2004. Lactulose ingestion increases faecal bifidobacterial counts: a randomised double-blind study in healthy humans. Eur. J. Clin. Nutr. 58:462-466. [DOI] [PubMed] [Google Scholar]
- 9.Collier, C. T., M. R. Smiricky-Tjardes, D. M. Albin, J. E. Wubben, V. M. Gabert, B. Deplancke, D. Bane, D. B. Anderson, and H. R. Gaskins. 2003. Molecular ecological analysis of porcine ileal microbiota responses to antimicrobial growth promoters. J. Anim. Sci. 81:3035-3045. [DOI] [PubMed] [Google Scholar]
- 10.Conway, P. L. 1997. Development of intestinal microbiota, p. 3-38. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology, vol. 2. Chapman and Hall, New York, NY. [Google Scholar]
- 11.Finberg, L., E. F. Bell, R. J. Cooke, S. J. Fomon, R. E. Kleinman, P. B. Pencharz, J. W. Reynolds, R. J. Schanler, A. L. Forbes, and American Academy of Pediatrics Task Force on Clinical Testing of Infant Formulas. 1988. Clinical testing of infant formulas with respect to nutritional suitability for term infants. U.S. Food and Drug Administration Center for Food Safety and Applied Nutrition, College Park, MD.
- 12.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 13.Haarman, M., and J. Knol. 2006. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 72:2359-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haarman, M., and J. Knol. 2005. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 71:2318-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 16.Jie, Z., L. Bang-Yao, X. Ming-Jie, L. Hai-Wei, Z. Zu-Kang, W. Ting-Song, and S. A. Craig. 2000. Studies on the effects of polydextrose intake on physiologic functions in Chinese people. Am. J. Clin. Nutr. 72:1503-1509. [DOI] [PubMed] [Google Scholar]
- 17.Kleessen, B., H. Bunke, K. Tovar, J. Noack, and G. Sawatzki. 1995. Influence of two infant formulas and human milk on the development of the faecal flora in newborn infants. Acta Paediatr. 84:1347-1356. [DOI] [PubMed] [Google Scholar]
- 18.Knol, J., G. Boehm, M. Lidestri, F. Negretti, J. Jelinek, M. Agosti, B. Stahl, A. Marini, and F. Mosca. 2005. Increase of faecal bifidobacteria due to dietary oligosaccharides induces a reduction of clinically relevant pathogen germs in the faeces of formula-fed preterm infants. Acta Paediatr. Suppl. 94:31-33. [DOI] [PubMed] [Google Scholar]
- 19.Knol, J., P. Scholtens, C. Kafka, J. Steenbakkers, S. Gro, K. Helm, M. Klarczyk, H. Schopfer, H. M. Bockler, and J. Wells. 2005. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 40:36-42. [DOI] [PubMed] [Google Scholar]
- 20.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magurran, A. 1988. Ecological diversity and its measurement, p. 8-45. Princeton University Press, Princeton, NJ.
- 22.Matsuki, T., K. Watanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moro, G., I. Minoli, M. Mosca, S. Fanaro, J. Jelinek, B. Stahl, and G. Boehm. 2002. Dosage-related bifidogenic effects of galacto- and fructooligosaccharides in formula-fed term infants. J. Pediatr. Gastroenterol. Nutr. 34:291-295. [DOI] [PubMed] [Google Scholar]
- 24.Moro, G. E., F. Mosca, V. Miniello, S. Fanaro, J. Jelinek, B. Stahl, and G. Boehm. 2003. Effects of a new mixture of prebiotics on faecal flora and stools in term infants. Acta Paediatr. Suppl. 91:77-79. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ninonuevo, M. R., Y. Park, H. Yin, J. Zhang, R. E. Ward, B. H. Clowers, J. B. German, S. L. Freeman, K. Killeen, R. Grimm, and C. B. Lebrilla. 2006. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 54:7471-7480. [DOI] [PubMed] [Google Scholar]
- 27.Palmer, C., E. M. Bik, D. B. Digiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penders, J., C. Vink, C. Driessen, N. London, C. Thijs, and E. E. Stobberingh. 2005. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol. Lett. 243:141-147. [DOI] [PubMed] [Google Scholar]
- 29.Probert, H. M., J. H. Apajalahti, N. Rautonen, J. Stowell, and G. R. Gibson. 2004. Polydextrose, lactitol, and fructo-oligosaccharide fermentation by colonic bacteria in a three-stage continuous culture system. Appl. Environ. Microbiol. 70:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinne, M. M., M. Gueimonde, M. Kalliomaki, U. Hoppu, S. J. Salminen, and E. Isolauri. 2005. Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota. FEMS Immunol. Med. Microbiol. 43:59-65. [DOI] [PubMed] [Google Scholar]
- 31.Schmelzle, H., S. Wirth, H. Skopnik, M. Radke, J. Knol, H. M. Bockler, A. Bronstrup, J. Wells, and C. Fusch. 2003. Randomized double-blind study of the nutritional efficacy and bifidogenicity of a new infant formula containing partially hydrolyzed protein, a high beta-palmitic acid level, and nondigestible oligosaccharides. J. Pediatr. Gastroenterol. Nutr. 36:343-351. [DOI] [PubMed] [Google Scholar]
- 32.Song, Y., C. Liu, and S. M. Finegold. 2004. Real-time PCR quantitation of clostridia in feces of autistic children. Appl. Environ. Microbiol. 70:6459-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai, Y. L., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward, R. E., M. Ninonuevo, D. A. Mills, C. B. Lebrilla, and J. B. German. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 72:4497-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegler, E., J. A. Vanderhoof, B. Petschow, S. H. Mitmesser, S. I. Stolz, C. L. Harris, and C. L. Berseth. 2007. Term infants fed formula supplemented with selected blends of prebiotics grow normally and have soft stools similar to those reported for breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 44:359-364. [DOI] [PubMed] [Google Scholar]
- 36.Zoetendal, E. G., C. T. Collier, S. Koike, R. I. Mackie, and H. R. Gaskins. 2004. Molecular ecological analysis of the gastrointestinal microbiota: a review. J. Nutr. 134:465-472. [DOI] [PubMed] [Google Scholar]