Abstract
Type II thioesterases (TE IIs) were shown to maintain the efficiency of polyketide synthases (PKSs) by removing acyl residues blocking extension modules. However, the substrate specificity and kinetic parameters of these enzymes differ, which may have significant consequences when they are included in engineered hybrid systems for the production of novel compounds. Here we show that thioesterase ScoT associated with polyketide synthase Cpk from Streptomyces coelicolor A3(2) is able to hydrolyze acetyl, propionyl, and butyryl residues, which is consistent with its editing function. This enzyme clearly prefers propionate, in contrast to the TE IIs tested previously, and this indicates that it may have a role in control of the starter unit. We also determined activities of ScoT mutants and concluded that this enzyme is an α/β hydrolase with Ser90 and His224 in its active site.
Modular polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) are complexes of multifunctional enzymes with exceptionally high molecular masses (300 to 1,600 kDa) that produce a variety of biologically active compounds. Despite substantial structural differences between the two groups of natural products (polyketides and nonribosomal peptides), the multienzymes responsible for their biosynthesis have important functional analogies (7, 37, 39). Enzymatic domains responsible for one round of chain extension are grouped in modules. During synthesis all substrates and intermediates are covalently bound to the enzyme by a thioester linkage with a 4′-phosphopantetheine (4′PP) arm of an acyl carrier protein (ACP) domain in PKSs or a peptidyl carrier protein (PCP) domain in NRPSs. The full-length chain is usually released by a thioesterase domain located at the C terminus of the last module. Both polyketides and peptides are usually modified further before they become final biologically active compounds.
In bacteria, genes encoding main PKS or NRPS subunits are clustered with genes encoding enzymes that modify the polyketide or peptide chain, as well as regulatory and resistance genes. Many modular PKS and NRPS clusters also include genes for type II thioesterases (TE IIs). These discrete, 25- to 29-kDa proteins were first identified in some mammalian fatty acid synthase (FAS) complexes, where they are alternative chain-terminating enzymes with affinities for medium-chain-length fatty acids (38). In PKS and NRPS systems they appear to have an “editing” role, removing nonreactive acyl residues blocking the multienzyme (16, 36, 50), removing amino acids that cannot be elongated (48), and/or controlling starter unit selection (20). However, recent reports on polyether ionophore antibiotic biosynthesis have shown that TE IIs are responsible for the release of this class of polyketides by hydrolysis (15, 28, 29).
Biosynthesis of polyketides involves repeated cycles of condensation and reduction of short carboxylic acids in a process similar to fatty acid biosynthesis (39). The first step of each biosynthetic cycle is decarboxylative condensation of a starter unit (or the growing chain from the previous module) with a malonyl-coenzyme A (malonyl-CoA)-derived extender unit. This results in chain elongation by a dicarbon unit and generates a β-carbonyl group, which can be further reduced depending on the presence of reducing domains in a given module. The chain is then transferred to the subsequent module, which performs the next elongation cycle. Sometimes, extender units attached to the ACP are decarboxylated without condensation. Products of such aberrant decarboxylation (acetyl, propionyl, and butyryl residues) remain covalently attached to the ACP and block the “assembly line.” They must be removed by a TE II to restore the PKS activity (16).
Since the nonribosomal peptide biosynthesis process does not involve decarboxylation, a different mechanism that gives rise to nonreactive acyl residues has been proposed (36). After synthesis of the NRPS protein, each of the PCP domains is converted into its holo form by attachment of 4′PP in a process called priming catalyzed by a 4′PP transferase. 4′PP transferases use CoA as a source of 4′PP, but they have also been shown to accept different acyl-CoA derivatives. Transfer of acyl-4′PP to the PCP domain is called mispriming and results in inactive NRPS. The role of TE II has been proposed to be hydrolysis of these acyl residues, and this step has been called deblocking after mispriming (36). The ACP domains of modular PKSs also require priming with 4′PP; therefore, it is very likely that deblocking after mispriming is also the role of TE II in polyketide synthesis.
Biochemical studies have shown that TE IIs are able to hydrolyze acyl residues, which may result from aberrant decarboxylation of extender units or from mispriming of ACP/PCP domains with acyl-4′PP (16, 23, 36, 48, 50). These findings are consistent with results of disruption experiments, in which a lack of TE II resulted in drastically reduced levels of production of polyketide (6, 10, 50) or peptide (11, 35, 49) compounds.
In contrast, deletion of the TE II gene (pikAV) from the picromycin/methymycin synthase gene cluster of Streptomyces venezuelae was found to have no effect on the level of polyketide production (8). This finding is in contrast to the findings of Xue et al. (47), who found that less than 5% of the glycosylated products and no intermediates were accumulated by a mutant with a disrupted pikAV gene. Therefore, the role of PikAV thioesterase remains unclear. Overexpression of the pikAV genes in S. venezuelae resulted in 50 to 70% decreases in polyketide production, which can be attributed to hydrolysis of correct extender units (23). On the other hand, polyketide production by picromycin/methymycin synthase expressed in Streptomyces lividans was enhanced up to sevenfold by coexpression of PikAV thioesterase (42).
Disruption of the TE II gene associated with 6-deoxyerythronolide B synthase (DEBS) of Saccharopolyspora erythraea resulted in a notable increase in shunt products formed by utilization of acetate instead of propionate as a starter unit, while the combined level of all polyketides produced was reduced by less than 20% (20). The DEBS TE II exhibited a strong in vitro preference for the acetyl group bound to the ACP from the loading module of DEBS. Therefore, it was proposed that removal of acetate from the ACP of the loading module is the main function of this TE II (20).
This paper deals with the TE II ScoT associated with the type I modular PKS Cpk from Streptomyces coelicolor A3(2) (32). ScoT was shown previously to restore polyketide production in a mutant of Streptomyces fradiae lacking the native TE II (encoded by tylO in the tylosin synthase cluster) (25). It was not possible to study the role of this enzyme in the natural context, since the polyketide produced by the Cpk synthase has not been identified yet. Here we describe kinetic measurements of the activity of the thioesterase ScoT with synthetic substrates mimicking acyl chains blocking the PKS. We also describe the activity of ScoT active-site mutants.
MATERIALS AND METHODS
Materials.
N-Acetylcysteamine (NAC) acetate was synthesized by reacting NAC with acetic anhydride. NAC propionate and NAC butyrate were prepared by reacting NAC with the appropriate acids in the presence of dicyclohexylcarbodiimide. The solvents were removed in vacuo. The products were purified by chromatography using a silica gel column impregnated with anhydrous copper sulfate (13). The compounds obtained were verified by 1H nuclear magnetic resonance (500 MHz) and electrospray mass spectrometry.
p-Nitrophenyl (p-NP) acetate and p-NP propionate were obtained from Sigma, and p-NP butyrate was obtained from Fluka. Other chemical reagents were obtained from Sigma and Roth. Restriction enzymes and polymerases, as well as DNA and protein mass markers, were obtained from MBI Fermentas.
Escherichia coli strains were cultured in LB medium (34).
Construction of the TE II expression plasmids.
DNA manipulations were performed by using standard procedures (34). The TE II gene scoT of S. coelicolor A3(2) was amplified by PCR using plasmid pMK15 as the template; pMK15 is a pBluescriptSK(+) construct containing two BamHI fragments from the T3 terminus of cosmid 1G7 (accession number AF109727). Primers TE-P-Nde (5′ TTT TTT TTT CAT ATG GGA AGT GAC TGG TT 3′) and TE-K-Hind (5′ TTT TTT TAA GCT TGT CGT ACG TAC ACG GA 3′) were designed to introduce restriction sites (underlined). The PCR product was cloned in the pGEM-T Easy vector (Promega), digested with NdeI and HindIII, and cloned into the corresponding sites of pET-28a(+) to obtain plasmid pOS22.
Mutated versions of the ScoT thioesterase were constructed by overlap extension (34). Upstream and downstream portions of the scoT gene were amplified using Pfu polymerase, which generated blunt ends with appropriate primer pairs that introduced mutations (Table 1). The upstream part of the scoT gene was amplified with TE-P-Nde and a mutagenic primer whose designation ends with “R,” and the downstream part of gene was amplified with TE-K-Hind and a mutagenic primer whose designation ends with “F.” In the subsequent step upstream and downstream parts of the scoT gene with mutations (approximately 100 ng each) were mixed, and the whole mutated gene was amplified with primers TE-P-Nde and TE-K-Hind to allow cloning in pET-28a(+) digested with NdeI and HindIII. For cloning in pGEX-6P-1, the NdeI and HindIII adhesive ends were filled with the Klenow enzyme, and the fragment was ligated with an SmaI-digested vector.
TABLE 1.
Oligonucleotides used for site-directed mutagenesis
| Primer | Sequence (5′-3′)a |
|---|---|
| S90A-F | C TTC GGG CAC GCC ATG GGC GCG |
| S90A-R | CGC GCC CAT GGC GTG CCC GAA G |
| S90C-F | C TTC GGG CAC TGC ATG GGC GCG |
| S90C-R | CGC GCC CAT GCA GTG CCC GAA G |
| H210A-F | G TGG CGC GAG GCC ACC ACC GCC |
| H210A-R | GGC GGT GGT GGC CTC GCG CCA C |
| H210Y-F | G TGG CGC GAG TAT ACC ACC GCC |
| H210Y-R | GGC GGT GGT ATA CTC GCG CCA C |
| H224A-F | G CCG GGC GGT GCC TTC TAT CTC |
| H224A-R | GAG ATA GAA GCG ACC GCC CGG C |
| H224Y-F | G CCG GGC GGT TAT TTC TAT CTC |
| H224Y-R | GAG ATA GAA ATA ACC GCC CGG C |
| D194N-F | C CTG GTC GGG AAT CAG GAC CCC |
| D194N-R | GGG GTC CTG ATT CCC GAC CAG G |
| D196N-F | GGG GAC CAG AAT CCC GTC GTT C |
| D196N-R | G AAC GAC GGG ATT CTG GTC CCC |
| DD194/196NN-F | C CTG GTC GGG AAT CAG AAT CCC GTC GTT C |
| DD194/196NN-R | G AAC GAC GGG ATT CTG ATT CCC GAC CAG G |
Nucleotides that are different than the nucleotides in the wild-type scoT sequence are underlined.
Cloned DNA fragments generated by PCR were verified by sequencing. E. coli strain DH5α was used for the cloning steps. Expression plasmids were used to transform E. coli strains BL21(DE3)/pLysS and BL21Star(DE3).
Purification of the recombinant proteins.
Wild-type ScoT protein and the ScoT_H224Y mutant were isolated from E. coli BL21(DE3)/pLysS cells. The ScoT_S90A and ScoT_S90C proteins were isolated from E. coli strain BL21Star(DE3). The ScoT_H210Y protein was isolated from E. coli BL21Star(DE3) cells additionally transformed with plasmid pGroESL encoding chaperones (14).
Cultures (200 ml) of E. coli cells transformed with expression plasmids were grown in 1-liter flasks in LB medium with appropriate antibiotics [30 μg/ml kanamycin for pET28a(+), 100 μg/ml ampicillin for pGEX-6P-1, and 34 μg/ml chloramphenicol for pLysS and pGroESL] at 37°C until the required optical density at 600 nm (OD600) was reached (0.6 for ScoT_H210Y and 1.0 for the remaining proteins). Then the cultures were cooled for 20 min, and protein expression was induced with isopropyl-thio-β-d-galactoside (IPTG) (0.1 mM for ScoT_H224Y and 0.01 mM for the remaining proteins). After overnight incubation on a shaker (at 22°C for ScoT_H210Y and at 9°C for the remaining proteins), the cells were harvested by centrifugation (5,000 × g, 10 min, 4°C) and frozen at −70°C.
Recombinant proteins with an N-terminal His6 tag were isolated by nickel affinity chromatography. Cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl; pH 8.0) (4 ml of lysis buffer/g [wet weight] of cells) and disrupted by sonication on ice (two 5-min treatments; cycle 0.5; 60% amplitude) using a UP200S ultrasonic disintegrator (Dr. Hielscher GmbH). The cell lysate was clarified by centrifugation (10,000 × g, 30 min, 4°C). Two milliliters of HIS-Select nickel affinity gel (Sigma) previously equilibrated with lysis buffer was added to 50 to 100 ml of the lysate and allowed to bind for 1 h with gentle shaking at 4°C. The resin was then collected by centrifugation (5,000 × g, 5 min, 4°C) and packed into a column. The column was washed with lysis buffer containing 10 mM imidazole, and the recombinant protein was eluted with the same buffer containing 50 mM imidazole. Fractions containing the recombinant protein were pooled and desalted using a PD-10 column (GE Healthcare). The buffer was changed to 50 mM Tris-HCl (pH 7.5). Next, 1 ml of Q Sepharose (GE Healthcare) equilibrated with the same buffer was added, allowed to bind for 30 min, packed into a column, and washed. The protein of interest was eluted with 100 mM NaCl in 50 mM Tris-HCl (pH 7.5).
ScoT_H224Y expressed as a fusion protein with glutathione S-transferase was purified by affinity chromatography. Cells collected from a 2-liter culture were resuspended in 100 ml of ATP-containing buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgSO4, 2 mM ATP) and disrupted by sonication as described above. One milliliter of glutathione-Sepharose 4B (GE Healthcare) equilibrated with the same buffer was added to the cleared lysate and allowed to bind for 30 min with gentle shaking at 4°C. The resin was then collected by centrifugation (5,000 × g, 5 min, 4°C), packed into a column, washed with ATP buffer, and equilibrated with PreS buffer (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol). The column-bound fusion protein was digested overnight at 4°C with 80 U of PreScission protease (GE Healthcare). Purified ScoT_H224Y protein was eluted with PreS buffer.
Samples from different purification steps were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were visualized by staining with Coomassie brilliant blue. Protein concentrations were determined by the method of Bradford (3). Fractions containing purified protein were pooled, glycerol was added to a concentration of 10% (vol/vol), and aliquots were shock frozen in liquid nitrogen and stored at −70°C.
Kinetic assays.
The conditions used for the enzymatic activity assays were the conditions described by Heathcote et al. (16). Hydrolysis rates were determined at 30°C using 1 ml (total volume) of assay buffer containing 200 mM potassium phosphate (pH 7.5), 2.5 mM Tris-HCl (pH 7.5), and 50 μM EDTA. Variable amounts of substrates were added from stock solutions in dimethyl sulfoxide (DMSO), and the DMSO concentration was adjusted to 3% (vol/vol). The ScoT concentration used for hydrolysis of p-NP butyrate, NAC butyrate, and NAC acetate was 225 nM, and the ScoT concentration used for hydrolysis of the remaining substrates was 75 nM. The concentrations of mutant proteins ScoT_S90C, ScoT_H210Y, and ScoT_H224Y were 800, 75, and 150 nM, respectively. All samples were examined in triplicate, and the results were corrected for background hydrolysis in the absence of the enzyme. The rate of hydrolysis was proportional to the enzyme concentration; therefore, so that the results could be compared on the same plot, all data were recalculated so that they corresponded to the data obtained with 75 nM enzyme.
Hydrolysis of p-NP esters was visible due to formation of the yellow p-nitrophenolate anion and was monitored by spectroscopy (λmax, 400 nm; ɛ = 8,570 M−1 cm−1). Stock solutions of p-NP esters consisted of 100 mM ester in DMSO.
Hydrolysis of NAC thioesters was monitored by including 0.2 mM 5,5′-dithio-2-nitrobenzoic acid (Ellman's reagent) in the reaction mixture. The yellow product of the reaction of 5,5′-dithio-2-nitrobenzoic acid with free thiol groups (5-thio-2-nitrobenzoic acid) was monitored by using spectroscopy (λmax, 412 nm; ɛ = 13,600 M−1 cm−1). Stock solutions of NAC thioesters consisted of 2 M thioester in DMSO.
ESI-MS analysis.
For electrospray ionization-mass spectrometry (ESI-MS) experiments a micrOTOF-Q mass spectrometer (Bruker) was used. The buffer for the ScoT and ScoT_S90A proteins was changed to 20 mM NH4HCO3 when a PD-10 column (GE Healthacare) was used. Ten microliters of a protein solution (8.5 μM) was diluted with 300 μl of a solution containing acetonitrile, water, and formic acid (500:500:1, vol/vol/vol). In order to determine the presence of an acylated thioesterase, 1 μl of a substrate solution (10 mM p-NP ester in acetonitrile) was added to 10 μl of a protein sample, incubated for 20 s at room temperature, and diluted with 300 μl of acetonitrile-water-formic acid. Samples were introduced into the ion source at a flow rate of 4 μl min−1. Spectra were scanned from m/z 200 to m/z 2200 and were analyzed using the Bruker Daltonics DataAnalysis 3.4 software. The mass scale was calibrated with Tune Mix (Agilent).
RESULTS
Heterologous expression and purification of thioesterase ScoT.
The TE II ScoT was obtained as an N-terminal His6-tagged protein expressed in E. coli. The recombinant protein was found mostly in the insoluble fraction of cell lysates under all induction conditions tested (with IPTG concentrations of 0.1 and 0.01 mM, induction at OD600 of 0.6 and 1.0, and incubation temperatures of 37, 28, and 9°C). A low incubation temperature and a low IPTG concentration slightly increased the amount of ScoT protein in the soluble fraction of the cell lysate. Induction with 0.01 mM IPTG at an OD600 of 1.0 and overnight incubation at 9°C were used for large-scale protein purification. Since enzymatic measurements were obtained in this study, isolation under native conditions was used in spite of the low yield in order to avoid the denaturation step required for isolation of proteins from inclusion bodies. ScoT protein was purified from crude cell extract by nickel affinity chromatography, followed by anion-exchange chromatography on Q Sepharose. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels are shown in Fig. S1 to S5 in the supplemental material. Approximately 100 μg of purified protein was obtained from 1 liter of culture. Problems with insolubility and low yields of recombinant TE IIs have been reported previously by Schwarzer et al. (36).
Substrate specificity of thioesterase ScoT.
Aberrant decarboxylation of polyketide extender units, such as malonate, methylmalonate, and ethylmalonate, gives rise to acetyl, propionyl, and butyryl residues that are attached to ACP domains of PKS proteins and block the activity of the entire multienzyme. The hydrolytic activity of the TE II ScoT was investigated using substrates representing these acyl chains. Two groups of substrates were used: p-NP esters and NAC thioesters. The hydrolysis reaction has two steps, acylation and deacylation. NAC thioesters are good structural analogues of natural thioesterase substrates (acyl groups attached to 4′-PP of ACP). They reflect the acylation step of the hydrolysis reaction well. The more-reactive p-NP esters are useful for studying the deacylation step and have been widely used in studies of other thioesterases (16, 44, 46). The progress of hydrolysis was monitored by spectroscopy. The kinetic constants determined are shown in Table 2.
TABLE 2.
Kinetic constants for synthetic substrate hydrolysis by recombinant TE II ScoT
| Substrate | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| p-NP esters | |||
| Acetate | 788 | ||
| Propionate | 3,567 | ||
| Butyrate | 485 | ||
| NAC thioesters | |||
| Acetate | 109 ± 7 | 56 ± 6 | 33 |
| Propionate | 450 ± 67 | 34 ± 10 | 221 |
| Butyrate | 54 ± 8 | 52 ± 14 | 17 |
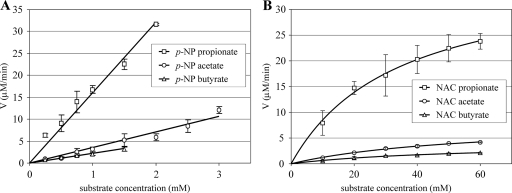
Plots of the initial reaction velocity for p-NP ester hydrolysis versus substrate concentration are shown in Fig. 1A. Due to the limited solubility of p-NP esters in water, it was not possible to obtain substrate-saturating conditions and to determine the kcat and Km values. The specificity constant kcat/Km was used to compare the activities of ScoT with different substrates. The kcat/Km value for each substrate was calculated from the linear slope of each plot at a low substrate concentration. Of the three p-NP esters, propionate was the best substrate for the thioesterase. It was hydrolyzed 4.5 and 7 times more efficiently than acetate and butyrate, respectively, as shown by a comparison of the specificity constants (Table 2).
FIG. 1.
Plots of rate (V) versus substrate concentration for the hydrolysis of p-NP esters (A) and NAC thioesters (B) by thioesterase ScoT. The data are the means ± standard deviations.
Plots of the initial reaction rates of hydrolysis of NAC thioesters versus substrate concentration are shown in Fig. 1B. The kinetic constants kcat and Km were calculated by fitting experimental data to the Michaelis-Menten equation by nonlinear regression using the SigmaPlot software (Table 2). The results obtained for NAC thioesters were consistent with those obtained for p-NP esters. The higher specificity constants observed for the p-NP esters than for the NAC thioesters (approximately 20-fold higher) can be attributed to the high reactivity of p-nitrophenol as a leaving group. NAC propionate was hydrolyzed much more readily than the other substrates. The specificity constant for propionate was 7 and 13 times higher than the specificity constants for acetate and butyrate, respectively. There were no big differences between the Km values for acetate and butyrate, which suggests that the initial binding of these substrates involves similar interactions. The lower Km for propionate indicates that the affinity of the enzyme for this compound is higher. There were significant differences between kcat constants, which reflected differences in the rates of the chemical steps subsequent to substrate binding.
Active site investigation.
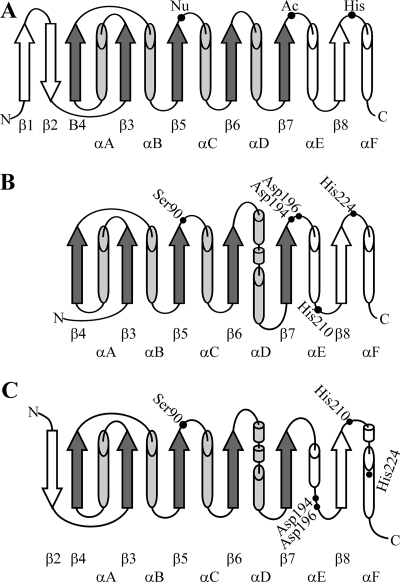
On the basis of the results of a sequence comparison and predicted secondary structure homology search, the ScoT protein was assigned to the family of thioesterases (PF00975) belonging to the clan of α/β hydrolases (CL0028) (Pfam database). The proteins belonging to the α/β hydrolase group (lipases, esterases, dehalogenases, epoxide hydrolases, and others) have conserved structural features despite low or even no sequence homology (17, 19, 31). The canonical α/β hydrolase fold has been described as an eight-strand, mostly parallel β sheet surrounded on both sides by α helices (Fig. 2A). The active sites of α/β hydrolases consist of the catalytic triad Nu-Ac-His (where Nu is a nucleophile [usually Ser] and Ac is an acid [usually Asp]). The nucleophile is located in a tight turn between strand β5 and helix αC called the “nucleophilic elbow” in the consensus sequence Sm-X-Nu-X-Sm (where Sm is a small residue [usually Gly] and X is any residue). Histidine is located on a loop after strand β8. The location of the acidic residue is the most variable; in the canonical structure this residue is after strand β7.
FIG. 2.
Topology of α/β hydrolases. Cylinders, α helices; arrows, β strands. Shading indicates the minimal set of α and β structures. (A) Canonical α/β hydrolase fold. Catalytic triad amino acids are indicated by dots. (B) Predicted topology of secondary structures of ScoT thioesterase representing model 1. (C) Predicted topology of secondary structures of ScoT tioesterase representing model 2. In panels B and C amino acids selected for site-directed mutagenesis are indicated by dots.
Amino acid residues which may form the catalytic triad of the ScoT thioesterase were proposed on the basis of identification of conservative sequence motifs (Fig. 3) and analysis of two structural models. Model 1 was created by using the Phyre software (protein homology/analogy recognition engine; an improved version of 3D-PSSM; www.sbg.bio.ic.ac.uk/phyre/), which performs predicted secondary structure matching and is able to find distant structural homologues even with no sequence similarity (22). In model 2, proteins with the highest sequence homology were selected by searching databases on the PDB and FUGUE servers, and their structures were used to construct the model template. Secondary structure topology diagrams representing models 1 and 2 are shown in Fig. 2B and C, respectively.
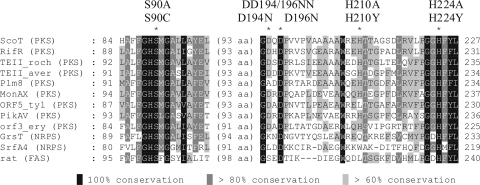
FIG. 3.
Alignment of the ScoT amino acid sequence with the sequences of TE IIs associated with bacterial PKSs, NRPSs, and rat FAS. Amino acids selected for mutagenesis are indicated by asterisks, and the corresponding substitutions are shown above the sequences. RifR (PKS), TE II associated with rifamycin synthase of Amycolatopsis mediterranei (accession number AAG52991); TEII_roch (PKS), predicted TE II of Streptomyces rochei (accession number NP_851508); TEII_aver (PKS), predicted TE II of Streptomyces avermitilis (accession number NP_821582); Plm8 (PKS), TE II associated with phoslactomycin B synthase of Streptomyces sp. strain HK803 (accession number AAQ84143); MonAX (PKS), TE II associated with monensin synthase of Streptomyces cinnamonensis (accession number AAO65810); ORF5_tyl (PKS), TE II associated with tylosin synthase of S. fradiae (accession number AAA21345); PikAV (PKS), TE II associated with pikromycin synthase of S. venezuelae (accession number AAC69333); ORF3_ery (PKS), TE II associated with DEBS of Saccharopolyspora erythraea (accession number CAA42928); GrsT (NRPS), TE II associated with gramicidin S-synthetase of Brevibacillus brevis (accession number AAA58717); SrfA4 (NRPS), TE II associated with surfactin synthetase of Bacillus subtilis (accession number CAA49819); rat (FAS), TE II associated with rat FAS (accession number P08635).
Both models include the minimal set of α helices and β strands of the α/β hydrolase fold (β3 to β7 and αA to αD). The nucleophilic residue of ScoT is probably Ser90 in the conservative sequence G-X-S-X-G typical of thioesterases and acyltransferases; in both models this residue is located on the turn between strand β5 and helix αC. The two models indicate that different histidine residues are located on a loop after strand β8, which could be involved in the catalytic triad. These residues are His224 (model 1) and His210 (model 2). After the predicted strand β7 there are two acidic residues, Asp194 and Asp196. One of these residues could potentially be the acidic component of the catalytic triad. These five amino acids were selected for site-directed mutagenesis (Fig. 3).
Eight mutants with single substitutions (S90A, S90C, H210A, H210Y, H224A, H224Y, D194N, and D196N) and one double mutant (DD194/196NN) were constructed by overlap extension. The double mutant was designed to exclude potential functional replacement of one Asp residue by the other Asp residue. ScoT_S90A, ScoT_S90C, and ScoT_H210Y were isolated using the same procedure that was used for the wild-type ScoT protein. ScoT_ H224Y was expressed as a fusion protein with glutathione S-transferase. The remaining mutant proteins were found in the insoluble fraction of an E. coli lysate. The mutations probably had a negative effect on the protein folding process.
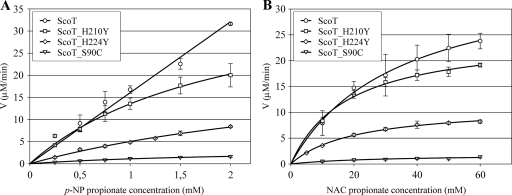
Since propionate derivatives were the best substrates for the wild-type ScoT protein, p-NP propionate and NAC propionate were used to test the enzymatic activity of the mutants. Plots of the initial reaction rates versus substrate concentrations are shown in Fig. 4. Calculated catalytic constants are shown in Table 3. Replacement of serine 90 by alanine completely abolished the hydrolytic activity of ScoT. The mutant with cysteine instead of serine exhibited some residual activity (the kcat/Km value was approximately 10% of the wild-type kcat/Km value). This led to the conclusion that Ser90 is indeed the nucleophilic residue of the catalytic triad. The results obtained indicate that the second element of the catalytic triad is histidine 224 and not histidine 210. The activity of the protein with the H210Y mutation was close to that of the wild-type ScoT protein, and the activity of the ScoT_H224Y mutant was much lower (the kcat/Km value was 40% of the kcat/Km value for p-NP propionate and 55% of the kcat/Km value for NAC propionate).
FIG. 4.
Plots of rate (V) versus substrate concentration for the hydrolysis of p-NP propionate (A) and NAC propionate (B) by ScoT thioesterase mutants. The data are the means ± standard deviations.
TABLE 3.
Summary of ScoT mutant activities
| Substrate | ScoT | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| p-NP | Wild type | 3,567 | ||
| propionate | S90A | Inactive | Inactive | Inactive |
| S90C | 47 ± 8 | 2.09 ± 0.54 | 378 | |
| H210Y | 540 ± 79 | 1.99 ± 0.48 | 4,523 | |
| H224Y | 313 ± 41 | 3.65 ± 0.66 | 1,428 | |
| NAC | Wild type | 450 ± 67 | 34 ± 10 | 221 |
| propionate | S90A | Inactive | Inactive | Inactive |
| S90C | 24 ± 5 | 24 ± 12 | 17 | |
| H210Y | 320 ± 12 | 18 ± 2 | 303 | |
| H224Y | 150 ± 6 | 20 ± 2 | 122 |
Acylation of thioesterase.
In order to capture the acyl-enzyme intermediate, the ESI-MS technique was used. Reaction mixtures containing ScoT with p-NP acetate, propionate, and butyrate were prepared, diluted with an acetonitrile-water-formic acid solution, and introduced into the ion source.
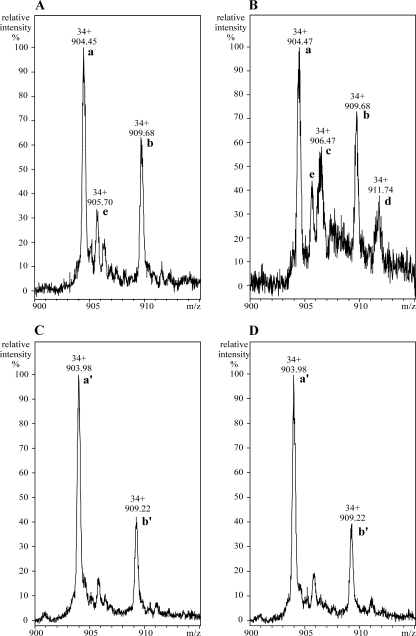
The initial analysis of the mass spectrum of ScoT protein without any substrates added revealed the presence of two major compounds, compounds a and b, whose molecular masses differed by 178 Da (Fig. 5A). The molecular mass of the main compound (compound a) was 30,718.48 ± 0.44 Da (molecule [M+H]+), which is in agreement with the expected molecular mass of recombinant ScoT protein lacking the N-terminal methionine (30,717.98 Da). The second peak (compound b) represented the same protein with a gluconic acid residue attached to its N terminus. Spontaneous α-N-gluconoylation is a common posttranslational modification of His-tagged proteins expressed in E. coli and results in an increase in the molecular mass of 178 Da (12). A small amount of acetylated ScoT was also present in the protein sample (compound e, with molecular mass 42 Da greater than the molecular mass of compound a).
FIG. 5.
Detection of acyl-enzyme intermediate of the hydrolysis reaction catalyzed by thioesterase ScoT. (A) ScoT protein. (B) ScoT protein incubated with p-NP butyrate. (C) ScoT_S90A protein. (D) ScoT_S90A protein incubated with p-NP butyrate. Both ScoT and ScoT_S90A lack the N-terminal methionine. Peaks labeled with letters represent the following compounds: peak a, ScoT; peak b, gluconoylated ScoT; peak c, butyrylated ScoT; peak d, butyrylated and gluconoylated ScoT; peak e, acetylated ScoT; peak a′, ScoT_S90A; peak b′, gluconoylated ScoT_S90A.
When p-NP esters were added, formation of acyl-enzyme was visible, as shown by the reaction with p-NP butyrate (Fig. 5B). Two additional peaks were observed, representing increases in the molecular masses of 70 Da for the two forms of ScoT (native and gluconoylated; compounds c and d, respectively).
Mutated thioesterase with serine 90 replaced by alanine was also subjected to ESI-MS analysis. As in the experiment with the wild-type protein, two forms of the ScoT_S90A protein were found (Fig. 5C): compound a′ (lacking the N-terminal methionine) and compound b′ (demethionylated and gluconoylated). When the ScoT_S90A protein was incubated with p-NP butyrate, formation of the acyl-enzyme was not observed (Fig. 5D). This experiment confirmed that the hydrolysis reaction proceeds through acylation of Ser90, which is the active site of thioesterase ScoT.
DISCUSSION
Thioesterase ScoT hydrolyzes acyl chains and has a preference for propionate.
As shown by our study, thioesterase ScoT from S. coelicolor A3(2) is able to hydrolyze acetyl, propionyl, and butyryl residues which can block ACP domains. The recombinant protein hydrolyzed all substrates tested at significant rates. This is consistent with its editing function suggested by a complementation study of S. fradiae (25). The enzyme showed a clear preference for propionate. The specificity constants for propionate hydrolysis were several times higher than those for acetate hydrolysis. The butyrate residue was hydrolyzed the slowest (the kcat/Km for butyrate was approximately twofold lower than that for acetate).
Michaelis constants and catalytic constants could be determined only for NAC thioesters. The Km for propionate is slightly lower than those for acetate and butyrate, which suggests that ScoT binds this substrate with higher affinity. However, greater differences in the reaction rates were associated with other steps, as indicated by the differences between kcat values (Table 2).
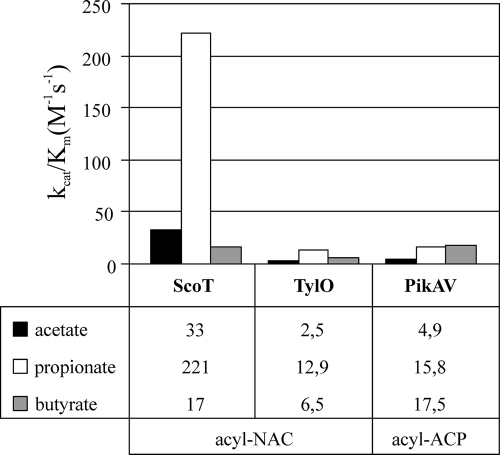
The kinetic constants determined can be directly compared to the parameters for TylO thioesterase associated with the tylosin synthase of S. fradiae, which was tested with the same substrates (16). ScoT was found to be more active than TylO. For both groups of substrates (p-NP and NAC derivatives) the kcat/Km values of ScoT for acetate and propionate were an order of magnitude greater than the corresponding values for TylO (Fig. 6). By comparing the parameters determined for NAC propionate hydrolysis by ScoT (kcat = 450 min−1 and Km = 34 mM) and by TylO (kcat = 29.2 min−1 and Km = 37.9 mM), we concluded that the differences are associated mostly with the deacylation step during hydrolysis (there are significant differences between the kcat values). As suggested by Kim et al. (23), high Km values (>100 μM) indicate that TE II operates in vivo under nonsaturating conditions and is mostly dissociated from the PKS, which enables it to scan all modules in search of stalled residues.
FIG. 6.
Comparison of specificity constants (kcat/Km) of TE IIs ScoT (S. coelicolor), TylO (S. fradiae), and PikAV (S. venezuelae) for different acyl residues attached to NAC (acyl-NAC) or ACP (acyl-ACP). The kcat/Km (in M−1 s−1) values are shown below the graphs. Data for TylO and PikAV were obtained from references 16 and 23, respectively.
The same acyl chains attached to the phosphopanthetheine of an ACP domain were tested as substrates for the TE II (PikAV) associated with the picromycin biosynthetic cluster of S. venezuelae (23). The differences in the specificities of thioesterase ScoT for substrates with different chain lengths were greater than the differences in the specificities of the TylO and PikAV thioesterases (Fig. 6). The order of substrate preferences was also different for the three thioesterases. PKS Cpk from S. coelicolor A3(2) utilizes only malonyl-CoA as extender units, as predicted by sequence analysis of AT domains (32); therefore, theoretically, only acetyl residues can be generated as products of aberrant decarboxylation. However, keeping in mind that mispriming of ACP domains can also give rise to blocking residues, activity with other acyl chains is not surprising. The substrate specificities of ScoT that are different from the substrate specificities of TylO and PikAV are probably associated with the different structures, especially the different shapes of the substrate binding pocket. The active site of ScoT seems to have some specific features which make it fit the two-carbon chain of propionate particularly well; however, detailed structural studies are necessary to understand these features.
Acyl residues which are starter units for polyketide biosynthesis are also possible substrates for TE IIs. The loading module of the Cpk synthase includes a KS domain with glutamine in the active site, which probably decarboxylates a malonyl residue incorporated by the AT domain, resulting in an acetyl starter unit (1). The results obtained caused us to speculate that ScoT may have a role in control of the starter unit by preventing accidental propionate incorporation. To prove this hypothesis, it is necessary to solve the chemical structure of the polyketide produced by the Cpk synthase and to identify the potential shunt product arising from the propionate starter.
High activity of ScoT for propionate can explain the results of our previous experiments. When the scoT gene from S. coelicolor A3(2) was used to complement a tylO disruption mutant of S. fradiae, polyketide production was restored to a level that was up to 50% of the level of the wild-type strain (25), while complementation with the native thioesterase gene (tylO) restored polyketide production to a level that was nearly 100% of the level of the wild-type strain (6). The starter unit for tylosin is propionate. One of the reasons that the level obtained with the strain complemented with the scoT gene was lower may be that there was excessive removal of the propionate starter unit from tylosin synthase. Genes used for complementation were expressed from a strong, constitutive promoter, ermE*p. Perhaps decreasing the expression of ScoT thioesterase would result in a higher level of polyketide production.
Chromosomes of Streptomyces and some other bacteria often contain several PKS and NRPS gene clusters. The mechanisms that prevent unwanted results of interactions between synthases and TE IIs associated with different clusters may include “fine-tuning” of kinetic parameters of the enzymes, substrate specificity, specificity for different ACP (PCP) domains, and temporal and spatial regulation of expression.
The balance resulting from evolution is disturbed by “man-made” modifications of megasynthases (PKS and NRPS), construction of hybrid enzymes, and heterologous expression of the enzymes. Inclusion of a proper editing enzyme in the system is one of the important aspects of such experiments. It is also possible that there are interactions between the introduced synthase and native TE IIs of the host organism. Brown et al. (4) observed that the main product of the first subunit of DEBS expressed in S. coelicolor was the compound resulting from incorporation of acetate instead of propionate. All three DEBS subunits expressed in S. coelicolor produced 6-deoxyerythronolide B with significant addition of 15-nor-6-deoxyerythronolide B (from the acetate starter), even when the TE II associated with DEBS was included (20). It seems probable that high activity of ScoT in the host cells could contribute to this effect by removal of propionate from the loading module of DEBS.
Active site of thioesterase ScoT.
On the basis of the results of a theoretical sequence analysis, we suggest that the ScoT protein belongs to the α/β hydrolase group. Recently, the three-dimensional structure of the TE II involved in the biosynthesis of nonribosomal peptide surfactin of Bacillus subtilis was resolved based on nuclear magnetic resonance titration experiments (24). Surfactin TE II was found to have a typical α/β hydrolase fold. Several crystal structures that have been resolved show that TE I domains of PKS (43) and NRPS (5) also have the α/β hydrolase fold. However, there are important structural differences when TE Is and TE IIs are compared, which explain their different roles. Thioesterase domains form deep substrate channels which accommodate the whole polyketide or peptide products and facilitate their cyclization, while the open shallow active-site cavity of TE II can accommodate only small acyl substrates.
Amino acids which may constitute the catalytic triad of the thioesterase ScoT (Ser-His-Asp) were proposed and changed by site-directed mutagenesis. Our results show that Ser90 is the active site of the enzyme. Replacement of this amino acid with alanine resulted in complete inactivation of the enzyme. The same effect was observed when serine was replaced with alanine in TE IIs (associated with rat FAS [41], surfactin NRPS [27], and nanchangmycin PKS [29]), thioesterase domains associated with the pikromycin PKS (30) and chicken FAS (33), and many other α/β hydrolases.
Replacement of Ser90 with cysteine resulted in a protein with low activity compared to the wild-type enzyme (the kcat/Km value was not more than 10% of the kcat/Km value of the wild-type enzyme). Different results for such a mutation were obtained previously. For example, the S101C mutation in the thioesterase domain of FAS resulted in only a 50% decrease in activity (33). Similarly, rat TE II with the S101C mutation and TE II associated with the surfactin synthetase with the S86C mutation exhibited relatively high activities (27, 41). On the other hand, the activities of many α/β hydrolases decreased by several orders of magnitude when serine was replaced with cysteine (18, 21, 26). The sulfhydryl group is a stronger nucleophile than the hydroxyl group; therefore, often the observed decrease in activity associated with a Ser-to-Cys substitution is associated with deacylation becoming the rate-limiting step (26). The atomic radius of sulfur is larger than that of oxygen. This probably leads to changes in the geometry of the active site and in some cases disables the charge relay system.
The role of Ser90 as the active site was further confirmed by the ESI-MS experiment. Acyl-enzyme formation was observed when p-NP substrates were added to wild-type ScoT protein and not when the mutated protein ScoT_S90A was used.
Two different histidine residues (residues 210 and 224) were identified by two structural models of the ScoT protein as possible elements of the catalytic triad, and each of them was mutated. Initially, they were replaced with alanine, but it was impossible to isolate the mutated proteins from the soluble fraction of E. coli cell lysate. In the second round of the experiment, the histidine residues were replaced with tyrosine. Unexpectedly, the enzymatic activities of both the H210Y and H224Y mutants were fairly high (the kcat values determined for p-NP propionate were 77 and 33% of kcat of wild-type ScoT, respectively). The fact that the effect of the His-to-Tyr substitution at position 224 was more pronounced than the effect of the substitution at position 210 indicates that His224 is part of the catalytic triad. Previously described mutations of histidines in catalytic triads resulted in inactivation or large decreases in activity even if the histidines were replaced with another basic amino acid or tyrosine (2, 9, 27, 29, 45). A typical pKa of the Tyr side chain is 10.0 (40). Theoretically, this residue should remain uncharged at pH 7.5. Perhaps local conditions in the active site of ScoT lead to a decrease in the pKa and allow ionization of tyrosine. The resultant negative charge might function as a proton acceptor and in this way replace histidine.
The third element of the catalytic triad remains unknown, since we failed to isolate Asp194 and Asp196 mutants under native conditions from the soluble fraction of E. coli cell lysate.
Thioesterase ScoT from S. coelicolor A3(2) as characterized here is unique among the TE IIs (associated with macrolide-producing PKSs) tested so far due to its exceptionally high activity for propionate. This feature could be exploited if propionate residues posed a problem and should also be kept in mind when S. coelicolor is used as a host for hybrid PKS expression. We propose that thioesterase ScoT has an editing function similar to that of TylO and also prevents inclusion of the propionate starter unit by the Cpk PKS. We also concluded that this enzyme is an α/β hydrolase with Ser90 and His224 in the active site.
Supplementary Material
Acknowledgments
We thank P. Jakimowicz for help with model construction and analysis and for advice concerning protein isolation. We are grateful to M. Jon for performing mass spectroscopy.
This work was supported by the Ministry of Scientific Research and Information (grant 3P04B 004 29).
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bisang, C., P. F. Long, J. Cortés, J. Westcott, J. Crosby, A. L. Matharu, R. J. Cox, T. J. Simpson, J. Staunton, and P. F. Leadlay. 1999. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502-505. [DOI] [PubMed] [Google Scholar]
- 2.Blee, E., S. Summerer, M. Flenet, H. Rogniaux, A. Van Dorsselaer, and F. Schuber. 2005. Soybean epoxide hydrolase: identification of the catalytic residues and probing of the reaction mechanism with secondary kinetic isotope effects. J. Biol. Chem. 280:6479-6487. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. J. B., J. Cortes, A. L. Cutter, P. F. Leadlay, and J. Staunton. 1995. A mutant generated by expression of an engineered DEBS1 protein from the erythromycin-producing polyketide synthase (PKS) in Streptomyces coelicolor produces the triketide as a lactone, but the major product is the nor-analogue derived from acetate as starter acid. J. Chem. Soc. Chem. Commun. 15:1517-1518. [Google Scholar]
- 5.Bruner, S. D., T. Weber, R. M. Kohli, D. Schwarzer, M. A. Marahiel, C. T. Walsh, and M. T. Stubbs. 2002. Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure 10:301-310. [DOI] [PubMed] [Google Scholar]
- 6.Butler, A. R., N. Bate, and E. Cundliffe. 1999. Impact of thioesterase activity on tylosin biosynthesis in Streptomyces fradiae. Chem. Biol. 6:287-292. [DOI] [PubMed] [Google Scholar]
- 7.Cane, D. E., and C. T. Walsh. 1999. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 6:R319-R325. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S., J. B. Roberts, Y. Xue, D. H. Sherman, and K. A. Reynolds. 2001. The Streptomyces venezuelae pikAV gene contains a transcription unit essential for expression of enzymes involved in glycosylation of narbonolide and 10-deoxymethynolide. Gene 263:255-264. [DOI] [PubMed] [Google Scholar]
- 9.David, F., A. M. Bernard, M. Pierres, and D. Marguet. 1993. Identification of serine 624, aspartic acid 702, and histidine 734 as the catalytic triad residues of mouse dipeptidyl-peptidase IV (CD26), a member of a novel family of nonclassical serine hydrolases. J. Biol. Chem. 268:17247-17252. [PubMed] [Google Scholar]
- 10.Doi-Katayama, Y., Y. J. Yoon, C. Y. Choi, T. W. Yu, H. G. Floss, and C. R. Hutchinson. 2000. Thioesterases and the premature termination of polyketide chain elongation in rifamycin B biosynthesis by Amycolatopsis mediterranei S699. J. Antibiot. (Tokyo) 53:484-495. [DOI] [PubMed] [Google Scholar]
- 11.Geoffroy, V. A., J. D. Fetherston, and R. D. Perry. 2000. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 68:4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geoghegan, K. F., H. B. Dixon, P. J. Rosner, L. R. Hoth, A. J. Lanzetti, K. A. Borzilleri, E. S. Marr, L. H. Pezzullo, L. B. Martin, P. K. LeMotte, A. S. McColl, A. V. Kamath, and J. G. Stroh. 1999. Spontaneous alpha-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: the cause of extra mass of 258 or 178 Da in fusion proteins. Anal. Biochem. 267:169-184. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, I. H., M. Ginty, J. A. O'Neill, T. J. Simpson, J. Staunton, and C. L. Willis. 1995. Synthesis of β-keto and αβ-unsaturated N-acetylcysteamine thioesters. BioMed. Chem. Lett. 5:1587-1590. [Google Scholar]
- 14.Goloubinoff, P., A. A. Gatenby, and G. H. Lorimer. 1989. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337:44-47. [DOI] [PubMed] [Google Scholar]
- 15.Harvey, B. M., H. Hong, M. A. Jones, Z. A. Hughes-Thomas, R. M. Goss, M. L. Heathcote, V. M. Bolanos-Garcia, W. Kroutil, J. Staunton, P. F. Leadlay, and J. B. Spencer. 2006. Evidence that a novel thioesterase is responsible for polyketide chain release during biosynthesis of the polyether ionophore monensin. Chembiochem 7:1435-1442. [DOI] [PubMed] [Google Scholar]
- 16.Heathcote, M. L., J. Staunton, and P. F. Leadlay. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207-220. [DOI] [PubMed] [Google Scholar]
- 17.Heikinheimo, P., A. Goldman, C. Jeffries, and D. L. Ollis. 1999. Of barn owls and bankers: a lush variety of alpha/beta hydrolases. Structure 7:R141-R146. [DOI] [PubMed] [Google Scholar]
- 18.Holm, C., R. C. Davis, T. Osterlund, M. C. Schotz, and G. Fredrikson. 1994. Identification of the active site serine of hormone-sensitive lipase by site-directed mutagenesis. FEBS Lett. 344:234-238. [DOI] [PubMed] [Google Scholar]
- 19.Holmquist, M. 2000. Alpha/beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr. Protein Pept. Sci. 1:209-235. [DOI] [PubMed] [Google Scholar]
- 20.Hu, Z., B. A. Pfeifer, E. Chao, S. Murli, J. Kealey, J. R. Carney, G. Ashley, C. Khosla, and C. R. Hutchinson. 2003. A specific role of the Saccharopolyspora erythraea thioesterase II gene in the function of modular polyketide synthases. Microbiology 149:2213-2225. [DOI] [PubMed] [Google Scholar]
- 21.Huhtinen, K., J. O'Byrne, P. J. Lindquist, J. A. Contreras, and S. E. Alexson. 2002. The peroxisome proliferator-induced cytosolic type I acyl-CoA thioesterase (CTE-I) is a serine-histidine-aspartic acid alpha/beta hydrolase. J. Biol. Chem. 277:3424-3432. [DOI] [PubMed] [Google Scholar]
- 22.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 23.Kim, B. S., T. A. Cropp, B. J. Beck, D. H. Sherman, and K. A. Reynolds. 2002. Biochemical evidence for an editing role of thioesterase II in the biosynthesis of the polyketide pikromycin. J. Biol. Chem. 50:48028-48034. [DOI] [PubMed] [Google Scholar]
- 24.Koglin, A., F. Löhr, F. Bernhard, V. V. Rogov, D. P. Frueh, E. R. Strieter, M. R. Mofid, P. Güntert, G. Wagner, C. T. Walsh, M. A. Marahiel, and V. Dötsch. 2008. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature 454:907-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotowska, M., K. Pawlik, A. R. Butler, E. Cundliffe, E. Takano, and K. Kuczek. 2002. Type II thioesterase from Streptomyces coelicolor A3(2). Microbiology 148:1777-1783. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., R. Szittner, Z. S. Derewenda, and E. A. Meighen. 1996. Conversion of serine-114 to cysteine-114 and the role of the active site nucleophile in acyl transfer by myristoyl-ACP thioesterase from Vibrio harveyi. Biochemistry 35:9967-9973. [DOI] [PubMed] [Google Scholar]
- 27.Linne, U., D. Schwarzer, G. N. Schroeder, and M. A. Marahiel. 2004. Mutational analysis of a type II thioesterase associated with nonribosomal peptide synthesis. Eur. J. Biochem. 271:1536-1545. [DOI] [PubMed] [Google Scholar]
- 28.Liu, T., D. You, C. Valenzano, Y. Sun, J. Li, Q. Yu, X. Zhou, D. E. Cane, and Z. Deng. 2006. Identification of NanE as the thioesterase for polyether chain release in nanchangmycin biosynthesis. Chem. Biol. 13:945-955. [DOI] [PubMed] [Google Scholar]
- 29.Liu, T., X. Lin, X. Zhou, Z. Deng, and D. E. Cane. 2008. Mechanism of thioesterase-catalyzed chain release in the biosynthesis of the polyether antibiotic nanchangmycin. Chem. Biol. 15:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, H., S. C. Tsai, C. Khosla, and D. E. Cane. 2002. Expression, site-directed mutagenesis, and steady state kinetic analysis of the terminal thioesterase domain of the methymycin/picromycin polyketide synthase. Biochemistry 41:12590-12597. [DOI] [PubMed] [Google Scholar]
- 31.Nardini, M., and B. W. Dijkstra. 1999. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9:732-737. [DOI] [PubMed] [Google Scholar]
- 32.Pawlik, K., M. Kotowska, K. F. Chater, K. Kuczek, and E. Takano. 2007. A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Arch. Microbiol. 187:87-99. [DOI] [PubMed] [Google Scholar]
- 33.Pazirandeh, M., S. S. Chirala, and S. J. Wakil. 1991. Site-directed mutagenesis studies on the recombinant thioesterase domain of chicken fatty acid synthase expressed in Escherichia coli. J. Biol. Chem. 266:20946-20952. [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Schneider, A., and M. A. Marahiel. 1998. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch. Microbiol. 169:404-410. [DOI] [PubMed] [Google Scholar]
- 36.Schwarzer, D., H. D. Mootz, U. Linne, and M. A. Marahiel. 2002. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. USA 99:14083-14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarzer, D., R. Finking, and M. A. Marahiel. 2003. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20:275-287. [DOI] [PubMed] [Google Scholar]
- 38.Smith, S. 1994. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB J. 8:1248-1259. [PubMed] [Google Scholar]
- 39.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]
- 40.Stryer, L. 1995. Biochemistry, 4th ed. Freeman, New York, NY.
- 41.Tai, M. H., S. S. Chirala, and S. J. Wakil. 1993. Roles of Ser101, Asp236, and His237 in catalysis of thioesterase II and of the C-terminal region of the enzyme in its interaction with fatty acid synthase. Proc. Natl. Acad. Sci. USA 90:1852-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang, L., H. Fu, M. C. Betlach, and R. McDaniel. 1999. Elucidating the mechanism of chain termination switching in the picromycin/methymycin polyketide synthase. Chem. Biol. 6:553-558. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, S. C., H. Lu, D. E. Cane, C. Khosla, and R. M. Stroud. 2002. Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases. Biochemistry 41:12598-12606. [DOI] [PubMed] [Google Scholar]
- 44.Weissman, K. J., C. J. Smith, U. Hanefeld, R. Aggarwal, M. Bycroft, J. Staunton, and P. F. Leadlay. 1998. The thioesterase of the erythromycin-producing polyketide synthase: influence of acyl chain structure on the mode of release of substrate analogues from the acyl enzyme intermediates. Angew. Chem. Int. Ed. 37:1437-1440. [DOI] [PubMed] [Google Scholar]
- 45.Witkowski, A., J. Naggert, B. Wessa, and S. Smith. 1991. A catalytic role for histidine 237 in rat mammary gland thioesterase II. J. Biol. Chem. 266:18514-18519. [PubMed] [Google Scholar]
- 46.Witkowski, A., H. E. Witkowska, and S. Smith. 1994. Reengineering the specificity of a serine active-site enzyme. Two active-site mutations convert a hydrolase to a transferase. J. Biol. Chem. 269:379-383. [PubMed] [Google Scholar]
- 47.Xue, Y., L. Zhao, H. W. Liu, and D. H. Sherman. 1998. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 95:12111-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh, E., R. M. Kohli, S. D. Bruner, and C. T. Walsh. 2004. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. Chembiochem 5:1290-1293. [DOI] [PubMed] [Google Scholar]
- 49.Yu, F. M., B. Qiao, F. Zhu, J. C. Wu, and Y. J. Yuan. 2006. Functional analysis of type II thioesterase of Streptomyces lydicus AS 4.2501. Appl. Biochem. Biotechnol. 135:145-158. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, Y., Q. Meng, D. You, J. Li, S. Chen, D. Ding, X. Zhou, H. Zhou, L. Bai, and Z. Deng. 2008. Selective removal of aberrant extender units by a type II thioesterase for efficient FR-008/candicidin biosynthesis in Streptomyces sp. strain FR-008. Appl. Environ. Microbiol. 74:7235-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.