Abstract
Dermatophytes are highly specialized filamentous fungi which cause the majority of superficial mycoses in humans and animals. The high secreted proteolytic activity of these microorganisms during growth on proteins is assumed to be linked to their particular ability to exclusively infect keratinized host structures such as the skin stratum corneum, hair, and nails. Individual secreted dermatophyte proteases were recently described and linked with the in vitro digestion of keratin. However, the overall adaptation and transcriptional response of dermatophytes during protein degradation are largely unknown. To address this question, we constructed a cDNA microarray for the human pathogenic dermatophyte Trichophyton rubrum that was based on transcripts of the fungus grown on proteins. Profiles of gene expression during the growth of T. rubrum on soy and keratin protein displayed the activation of a large set of genes that encode secreted endo- and exoproteases. In addition, other specifically induced factors potentially implicated in protein utilization were identified, including heat shock proteins, transporters, metabolic enzymes, transcription factors, and hypothetical proteins with unknown functions. Of particular interest is the strong upregulation of key enzymes of the glyoxylate cycle in T. rubrum during growth on soy and keratin, namely, isocitrate lyase and malate synthase. This broad-scale transcriptional analysis of dermatophytes during growth on proteins reveals new putative pathogenicity-related host adaptation mechanisms of these human pathogenic fungi.
Dermatophytes are highly specialized fungi which are the most common agents of superficial mycoses in humans and animals (13, 34). Among the human pathogenic dermatophytes, the anthropophilic species Trichophyton rubrum is clinically the most commonly observed. In contrast to most other medically important fungi, dermatophytes are not opportunists but are obligate pathogens, infecting exclusively the skin stratum corneum, nails, or hair. The ability of dermatophytes to degrade and utilize compact hard keratin within such host structures is presumably related to their common secreted keratinolytic activity, which is therefore a major putative virulence attribute of these fungi. Individual endo- and exoproteases secreted by dermatophytes are similar to those of species of the genus Aspergillus. However, in contrast to those of Aspergillus spp., dermatophyte-secreted endoproteases are multiple and members of two large protein families, the subtilisins (Subs) S8 (serine proteases) and the fungalysins M36 (metalloproteases) (see the MEROPS peptidase database, http://merops.sanger.ac.uk) (10, 11). More than 20 genes that encode secreted proteases have been identified in dermatophytes, some of which have been characterized in detail on the molecular level (reviewed in reference 23).
In order to better understand the basic mechanisms of protein degradation by dermatophytes, the secretome of different dermatophyte species was previously analyzed during in vitro growth on protein-containing media. By two-dimensional polyacrylamide gel electrophoresis and a shotgun mass spectrometry approach, secreted proteins from T. rubrum and T. violaceum were identified in soy culture supernatants, including endo- and exoproteases and other hydrolases (8). Such analyses of proteolytic dermatophyte supernatants, however, are limited to the identification of secreted proteins. In addition, the possibility could not be excluded that many secreted proteins were degraded because of the high proteolytic activity of the supernatant and hence were not detectable by this approach. Genetic manipulation in dermatophyte research has been hampered by the limited number of available genetic tools and the lack of full genome sequences. Only a small number of genes have therefore been studied, and functional analysis by targeted gene disruption has been demonstrated only in a very few selected cases (6, 7, 35). High-throughput gene discovery by expressed sequence tag (EST) sequencing and cDNA-based microarrays provide additional valuable methodologies for the analysis of biological systems. In dermatophytes, recent applications of such techniques revealed the transcriptional response of T. rubrum cells in distinct developmental growth phases and in the presence of novel fatty acid synthase inhibitors (16, 33). Differential cDNA screening allowed first insights into the response of Trichophyton mentagrophytes and T. rubrum during growth on protein substrates (12, 19); nevertheless, the basic mechanisms of adaptation during growth under such conditions need further investigations. The aim of the present study was to analyze a broad gene expression profile by cDNA microarray analysis in T. rubrum cells during keratin utilization. Our research was devised not only to monitor the expression of protease genes in T. rubrum during protein utilization but also to reveal other dermatophyte-specific mechanisms which are involved in this putative pathogenicity-related process.
MATERIALS AND METHODS
Strain and growth conditions.
T. rubrum strain Lau1673 (14, 37) was routinely grown on Sabouraud dextrose agar (Bio-Rad, Hercules, CA), a medium containing 1% peptone. For liquid cultures, T. rubrum was grown in Sabouraud dextrose medium (hereafter referred to as Sabouraud medium) and, to promote proteolytic activity, in soy liquid medium and keratin-soy medium (10). Soy medium was prepared by dissolving 2 g soy protein (Supro 1711; Protein Technologies International) in 1 liter distilled water. Keratin-soy medium aliquots of 100 ml were prepared by adding 0.2 g keratin (Merck, Dietikon, Switzerland) and 5 ml soy medium to 95 ml distilled water. Growth media were sterilized by autoclaving at 120°C for 15 min. For growth of liquid cultures for subsequent RNA isolation, 100-ml volumes of each medium were poured into 800-ml tissue culture flasks and inoculated with a plug of fresh mycelium grown on Sabouraud agar. T. rubrum liquid cultures in Sabouraud, soy, and keratin-soy media were incubated for 5, 10, and 28 days, respectively, at 30°C without shaking. Longer incubation times were necessary in the case of keratin-soy medium compared to soy medium and Sabouraud medium cultures; however, the fungus was actively growing in all three media at the selected incubation times, i.e., before the stationary phase was reached because of nutrient depletion. In addition, the time points for cultures in soy and keratin-soy media were chosen by a substantial proteolytic activity along with a clarification of the media and, in the case of keratin-soy medium, also by progressive dissolution of the water-insoluble keratin granules.
cDNA library construction and EST sequencing.
A T. rubrum cDNA library was previously constructed from RNA derived from 10-day-old T. rubrum soy cultures (10), and the cloned inserts were sequenced to generate a collection of 3,804 ESTs, representing a total of 2,145 clusters composed of 514 contigs and 1,631 singletons (37). In the present study, we identified the EST sequences by use of the annotations from the T. rubrum Expression Database (www.mgc.ac.cn/TrED/). The T. rubrum Expression Database contains more than 40,000 EST sequences (January 2008) representing 10,224 clusters.
Microarray construction.
Plasmid inserts of the cDNA library were amplified by PCR in a 100-μl PCR mixture with primer SP6 and 3′ poly(T) primers each starting with 15 T's followed by an A, C, or G. PCR mixtures contained 50 ng of plasmid DNA, 5 U of Taq polymerase (Sigma, Buchs, Switzerland), 0.4 μM primers, 0.25 μM deoxynucleoside triphosphates, and Sigma buffer. An initial 2-min denaturation step at 94°C was followed by 35 cycles of 30 s at 94°C, 60 s at 40°C, and 7 min at 70°C. The reaction ended with an additional incubation step of 10 min at 70°C. The longest and shortest plasmid inserts corresponding to ESTs of the same contig and all singletons were used as templates. PCR products were visually analyzed on a 2% agarose Invitrogen E-gels 96 and classified as “single band,” “weak or multiple bands,” or “no band.” The names of clones classified in the latter two categories were given an “f” or a “d” suffix, respectively. The collection is composed of 2,626 PCR products plus 11 full-length cDNA sequences of previously described T. rubrum genes (Table 1). The PCR products were resuspended in 30 μl water and transferred into duplicate 384-well plates with a Tecan liquid-handling robot (Tecan, Männedorf, Switzerland). Afterwards, the PCR products were dried, resuspended in 20 μl 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1.5 M betaine and printed in triplicate on aldehydesilane-coated slides (Nexterion Slide AL; Schott Nexterion, Jena, Germany) with an Omnigrid 300 contact-printing robotic microarrayer (Genomic Solutions, Ann Arbor, MI) equipped with SMP3 pins (TeleChem International Inc.). Spike controls (Lucidea Universal Scorecard; GE Healthcare) were included in each subgrid of the microarray. Spot and printing quality was assessed visually after printing, and the DNA was cross-linked to the slides by baking at 80°C for 1 h. The slides were then processed with NaBH4 (Fluka, Buchs, Switzerland) by the protocol recommended by the manufacturer. Including the controls and spots from empty wells, the microarray contained a total of 9,408 spots.
TABLE 1.
Primers for amplification of full-length protease cDNA sequences
| Gene | Sequence | Primer | Reference |
|---|---|---|---|
| MEP2 | 5′-GTTGAATTCGGTCTTCCAGCCCGTCAACAA-3′ | P1 | This study |
| 5′-GTTTCTAGATTAGCAGTCAGCGGGCATGTCG-3′ | P2 | ||
| NPII-1 | 5′-GTTCTCGAGCCTCTATCCCAGCTGCTGCTC-3′ | P3 | This study |
| 5′-CTTGGATCCTTTAGCAGCCAAGGTAGAT-3′ | P4 | ||
| NPII-2 | 5′-GTTCTCGAGCTCCAGCCCTTGGCTTCTCCAT-3′ | P5 | This study |
| 5′-GTTGGATCCGTTTAGCAGCCAACGTAGATAG-3′ | P6 | ||
| SCPA | 5′-GTTGTCGACTTCAAGGCTTCCCTCCACCCGTT-3′ | P7 | 37 |
| 5′-CTTGTCGACGCGGCCGCCTACAAGAAGAAAGCAAG-3′ | P8 | ||
| SCPB | 5′-CTTCTCGAGCTCAGTTCCCACCAAAACCGG-3′ | P11 | 37 |
| 5′-CTTGGATCCTTACATTGCCAGCTCTATAAC-3′ | P12 | ||
| SUB2 | 5′-GGTTCTCGAGACCTCGCTCCACAGCCTGAGCCG-3′ | P13 | 11 |
| 3′-CTTGGATCCTCAGTAATACTTGGGCAGTTTGC-5′ | P14 | ||
| SUB6 | 5′-GTGCTCGAGATGGTGCTAGAATCCTTGAGGCCGGT-3′ | P15 | 11 |
| 3′-GTTGCGGCCGCTTATTTGCCGCTGCCGTTGTA-5′ | P16 | ||
| SUB7 | 5′-GTTGCTCGAGCTGAGATCTTGGAGACTCGCGCT-3′ | P17 | 11 |
| 3′-GTTGGATCCTTACATGCCAGATCGGTTGTTGATGAGCTTGC-5′ | P18 | ||
| DPPIV | 5′-CTTAGATCTGTTCCTCCTCGTGAGCCCCG-3′ | P19 | This study |
| 5′-CTTGCGGCCGCTCATTCCTCTGCCCTCTCACC-3′ | P20 | ||
| PAP | 5′-GTTGGTACCATGCAAGCAGCAAAATTGTTGAGC-3′ | P21 | This study |
| 5′-CTTGCGGCCGCCTAGTCAATCGTATCATCACGAAG-3′ | P22 | ||
| AMPP | 5′-GTTGAATTCATGCCGCCACCACCGCTTCACACG-3′ | P23 | This study |
| 5′-CTTGCGGCCGCTTAAATAGGTTGTGTCTCGCGCTT-5′ | P24 |
RNA isolation, cDNA synthesis, and microarray hybridization.
For RNA isolation, the fungal cells were ground into a powder with a mortar and pestle in liquid nitrogen to facilitate cell disruption. Total RNA was isolated with the Qiagen RNeasy plant mini kit (Qiagen, Hombrechtikon, Switzerland) according to the instructions of the manufacturer. Three micrograms of total RNA was amplified with the MessageAmp aRNA II kit (Ambion, NY). Three micrograms of amplified RNA was reverse transcribed into cyanin 3 (Cy3)- or Cy5-labeled cDNA with Superscript II reverse transcriptase (Invitrogen) and random primers (Invitrogen), purified with QIAquick columns (Qiagen, Netherlands), and hybridized on the microarrays described above (Gene Expression Omnibus accession number GPL6857). Hybridizations were performed overnight at 65°C in microarray hybridization cassettes (TeleChem International Inc.). Slides were sequentially washed in 2× SSC-0.1% sodium dodecyl sulfate, 0.2× SSC, 0.1× SSC, and 0.1× SSC-0.1% Triton and scanned on an Agilent DNA microarray scanner (Agilent Technologies).
Data analysis.
Tagged image file format images corresponding to the Cy5 and Cy3 fluorescence emission channels were extracted with GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA). Statistical analysis of the data was performed with open-source R software packages (http://www.r-project.org/ and http://www.BioConductor.org/). Gene expression levels were quantified with the limma package by using global loess normalization without background subtraction (30, 36). The resulting expression measurements for each array are the log2 ratios (M values) and the average log2 intensities (A values) of the Cy3 and Cy5 signals. The median M values of the triplicate cDNA probes on each microarray were subsequently used for the analysis. Adjustment for multiple testing was performed by Benjamini and Hochberg's method to control the false-discovery rate (FDR). Statistical analyses were done by pairwise comparisons of cultures grown on soy medium or keratin-soy medium versus Sabouraud medium with the Bioconductor limma package (29). To further assess the differences in gene expression of cultures grown on keratin-soy medium versus cultures grown on soy medium, a linear model was used in limma which took into account all soy medium versus Sabouraud medium, keratin-soy medium versus Sabouraud medium, and direct keratin-soy medium versus Sabouraud medium hybridizations. A metabolic pathway analysis was performed by use of tools provided by the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.ad.jp/kegg/).
Quantitative real-time RT-PCR.
In order to validate the microarray results, we performed quantitative real-time reverse transcriptase PCR (RT-PCR) analysis of a selection of eight genes (Table 2). Specific primers and 5′-6-carboxyfluorescein-3′ Black Hole Quencher probes were designed with PrimerExpress 2.0 software (Applied Biosystems); all of the primers and probes used (Table 2) were ordered from Microsynth (Balgach, Switzerland) and Eurogentec (Seraing, Belgium), respectively. Expression of target genes was quantified by two-step quantitative RT-PCR analysis. Briefly, 200 ng of total RNA was mixed with 0.25 ng of random hexamers (Invitrogen) and reverse transcribed with 200 U of Superscript Reverse Transcriptase II (Invitrogen) and RNasin (Promega). Quantitative PCR conditions were as follows. One nanogram of freshly diluted cDNA was mixed with TaqMan Universal PCR Master Mix (Applied Biosystems) and primers and probes with final concentrations of 900 and 200 nM, respectively. The cycling conditions were as follows: 50°C for 2 min, initial denaturation at 95°C for 10 min, and 45 cycles of 15 s at 95°C and 1 min at 60°C. All samples were tested in triplicate, and for each probe-primer set, reaction efficiency estimates were derived from standard curves generated with serial dilutions of a cDNA sample. The threshold cycle values obtained with the SDS software (Applied Biosystems) were exported into qBase version 1.3.5, a Visual Basic Excel-based script for the management and automated analysis of quantitative PCR data for further analysis (9). Threshold cycle values were transformed to relative quantities and analyzed with geNorm 3.4 software (31). This Microsoft Excel application identifies the most stable reference genes from a set of candidate normalization genes in a given panel of cDNA samples. To correct for any variation in mRNA content and variation in enzymatic efficiency, the relative quantities of the genes of interest were normalized with the geometric mean of the two most stable housekeeping genes. These genes, which encode T. rubrum chitin synthase (TrMZG10ACO) and ADP-ribosylation factor (TrMZC10ACH), respectively, were selected by a sequential pairwise comparison. Briefly, the two genes with the lowest intragroup variation were selected and a gene stability measure, M, was defined as the average pairwise variation. The stability measure M was below the value defined as threshold acceptance (M < 0.5). All PCR plates were assembled with a Tecan Freedom Evo (Männedorf, Switzerland) liquid handler, and the quantitative PCRs were performed with an ABI Prism 7900 sequence detection system.
TABLE 2.
Primers and fluorescent probes used for quantitative real-time RT-PCR
| Primer and probea | Sequence |
|---|---|
| Sub3TrubF | 5′-TATGCTTGCCCAGGGTGTTAG-3′ |
| Sub3TrubR | 5′-CCTGGGTTGCGGATAACG-3′ |
| Sub3TrubTaq | 5′-AGGCCTGCAACCGCTTGAAGCA-3′ |
| Sub4TrubF | 5′-GGGCCCAGTTGTTGACATCT-3′ |
| Sub4TrubR | 5′-GATCATGTAGGCACCCATACCA-3′ |
| Sub4TrubTaq | 5′-CCTCCATGGCCTCTCCCCACG-3′ |
| Sub6TrubF | 5′-GCGGCAGCACTGACACTCT-3′ |
| Sub6TrubR | 5′-CCGATGAGATAGGCACCAAGAC-3′ |
| Sub6TrubTaq | 5′-ACTTCCATGGCTTCTCCTCACGTTTGC-3′ |
| McpATrubF1 | 5′-GCATTGAAGGCGGTGCAT-3′ |
| McpATrubR1 | 5′-GTCAACACTGTCTCCATTAACTTGGT-3′ |
| McpATrubTaq1 | 5′-CCGGACCCATCTGCAACACCATC-3′ |
| AcuDTrubF1 | 5′-TGGAAAGATTCAAGATGGCGATA-3′ |
| AcuDTrubR1 | 5′-TCTGTGCATTTGATGGGTAATCA-3′ |
| AcuDTrubTaq1 | 5′-ACCAAACGTCCATTCACCGCAGAGC-3′ |
| AcuETrubF1 | 5′-GAACATGGTAAATGGTCAGGTGAA-3′ |
| AcuETrubR1 | 5′-CGCCGAGGGTGAAGTCAA-3′ |
| AcuETrubTaq1 | 5′-CTTTACGATGCCATCCGCCGTCA-3′ |
| ChitinSTrubF | 5′-CGAAGTCTCCGGCTACTCGTAT-3′ |
| ChitinSTrubR | 5′-CCGGGAAGCGGAGTATCC-3′ |
| ChitinSTrubTaq | 5′-CCACCAAGACTCACTACGCTCCTGCTACTG-3′ |
| ADPrfTrubF | 5′-CAGAGAAGAGTTGCAAAAGATGCT-3′ |
| ADPrfTrubR | 5′-TGCTTGTTGGCGAAAACGA-3′ |
| ADPrfTrubTaq | 5′-CGAGGATGAACTACGGGATGCTTTGC-3′ |
Primer names ending with F, R, and Taq indicate forward and reverse primers and the TaqMan probe, respectively.
Nucleotide sequence accession numbers.
The EST sequences reported here have been deposited in the GenBank data library under accession numbers AJ880769 to AJ884477.
RESULTS
Individual transcripts are overrepresented in a cDNA library based on T. rubrum cells grown on proteins.
For microarray construction, we used a T. rubrum cDNA library generated from mRNA derived from fungal cells grown in soy medium (10), thus ensuring that genes implicated in protein utilization are present on the array. Soy protein is known to induce the secretion of similar keratinases by dermatophytes compared to keratin but, in contrast to the latter, allows higher fungal growth rates and increased levels of total RNA (24). Previous EST sequencing of this cDNA library identified a collection of 3,804 ESTs, representing a total of 2,145 clusters (37). Sequence analysis in the present study indicated that individual genes which were represented by multiple ESTs (Table 3) were found to be related to stress response, e.g., sequences that encode putative heat shock proteins Hsp70 (TrMZA12ACM), Hsp90 (TrMZH11AAA), and Hsp98 (TrMZC07AAH). Other overrepresented ESTs encode putative elongation factors, e.g., Ef-1-α (TrMZF11ACI) and Ef-3 (TrMZA02ABD); a putative HacA transcription factor (TrMZB07ACL); and metabolic enzymes such as a putative 4-hydroxyphenylpyruvate dioxygenase (TrMZF01ACG), a glutamine synthetase (TrMZH05ACQ), an oxalate decarboxylase (TrMZE11ACL), and an aspartate aminotransferase (TrMZG07ACN). As expected, many of the abundant ESTs encode known secreted proteases such as Sub3 and Sub5, the fungalysins metalloprotease 1 (Mep1) and Mep3, and leucine aminopeptidase 2 (Lap2). Notably, a number of sequences that encode hypothetical proteins were identified (Table 3). These sequences could be dermatophyte specific since no homology with any sequences deposited in the public databases was found by BLAST search.
TABLE 3.
Clusters with the most abundant ESTs (five or more)a
| Cluster | EST accession no. | No. of ESTs/cluster | Tentative annotation | Accession no. (GenBank, GO) |
|---|---|---|---|---|
| TrMZA12ACM | AJ884048 | 46 | T. rubrum HSP70 mRNA | AF052391 |
| TrMZF01ACG | AJ883548 | 39 | 4-Hydroxyphenylpyruvate dioxygenase activity | 0003868 |
| TrMZF11ACI | AJ883743 | 38 | T. rubrum elongation factor 1-α mRNA | AY115575 |
| TrMZA02ABD | AJ882741 | 29 | Hypothetical protein | |
| TrMZE01ABZ | AJ883070 | 19 | A. fumigatus Af293 translation elongation factor eEF-3, putative mRNA | XM_743850 |
| TrMZC12ABZ | AJ883058 | 18 | Hypothetical protein | |
| TrMZH05ACQ | AJ884475 | 18 | Glutamine synthetase activity | 0004356 |
| TrMZF12AAE | AJ881221 | 18 | ADP/ATP carrier protein pattern | 0005471 |
| TrMZC08AAV | AJ882197 | 16 | A. fumigatus Af293 eukaryotic translation initiation factor 4, putative mRNA | XM_749705 |
| TrMZH11AAA | AJ880862 | 15 | A. fumigatus Af293 molecular chaperone and allergen Mod-E/Hsp90/Hsp1 mRNA | XM_742833 |
| TrMZH12ABZ | AJ883111 | 15 | Neosartorya fischeri NRRL 181 woronin body protein HexA, putative | XP_001259804 |
| TrMZH04AAV | AJ882252 | 13 | T. rubrum Sub-like protease SUB3 gene | AY343501 |
| TrMZD08ACB | AJ883155 | 13 | Hypothetical protein | |
| TrMZC08ACP | AJ884338 | 13 | T. rubrum elongation factor 1-α mRNA | AY115575 |
| TrMZD09AAI | AJ881576 | 12 | A. fumigatus Af293 homogentisate 1,2-dioxygenase (HmgA), putative mRNA | XM_744461 |
| TrMZE11ACL | AJ884003 | 12 | Oxalate decarboxylate KEGG K01569 | |
| TrMZF12ABY | AJ883001 | 11 | A. fumigatus Af293 thiazole biosynthesis enzyme mRNA | XM_745632 |
| TrMZE07AAB | AJ880918 | 11 | A. fumigatus Af293 60S ribosomal protein L3 mRNA | XM_750424 |
| TrMZC03ACG | AJ883514 | 11 | Hypothetical protein | |
| TrMZG07ACN | AJ884209 | 10 | A. fumigatus Af293 aspartate aminotransferase, putative mRNA | XM_746692 |
| TrMZE09ACI | AJ883730 | 10 | A. fumigatus Af293 glutamate carboxypeptidase, putative mRNA | XM_749963 |
| TrMZG05AAG | AJ881418 | 10 | A. fumigatus Af293 conserved lysine-rich protein, putative mRNA | XM_746493 |
| TrMZD10ACI | AJ883719 | 10 | A. fumigatus Af293 mitochondrial phosphate carrier protein (Mir1), putative mRNA | XM_747790 |
| TrMZG02ABZ | AJ883092 | 10 | A. fumigatus Af293 glycosylphosphatidylinositol-anchored cell wall organization protein Ecm33 mRNA | XM_747051 |
| TrMZD03AAJ | AJ881665 | 10 | T. rubrum Sub-like protease SUB5 gene | AY344482 |
| TrMZH12AAT | AJ882165 | 10 | A. fumigatus Af293 calcineurin binding protein, putative mRNA | XM_750545 |
| TrMZA10ACG | AJ883497 | 9 | T. rubrum actin (ACT) gene | AY525329 |
| TrMZB07AAB | AJ880882 | 9 | A. fumigatus Af293 mitochondrial Hsp70 chaperone (Ssc70), putative mRNA | XM_750235 |
| TrMZF08ACF | AJ883460 | 9 | A. fumigatus Af293 GTP-binding protein EsdC mRNA | XM_741646 |
| TrMZA02ABC | AJ882645 | 9 | A. fumigatus Af293 Hsp70 chaperone (HscA), putative mRNA | XM_742107 |
| TrMZE02ACO | AJ884268 | 9 | T. rubrum Mep MEP3 gene | AY283569 |
| TrMZB07ACK | AJ883872 | 9 | A. fumigatus Af293 Hsp70 chaperone Hsp88 mRNA | XM_747538 |
| TrMZD07ACI | AJ883716 | 9 | A. fumigatus Af293 integral membrane protein mRNA | XM_745546 |
| TrMZC12AAR | AJ882009 | 9 | A. fumigatus Af293 IMP dehydrogenase, putative mRNA | XM_744401 |
| TrMZB03AAR | AJ881988 | 9 | A. fumigatus Af293 C-4 methyl sterol oxidase Erg25, putative mRNA | XM_741575 |
| TrMZE11ACP | AJ884365 | 8 | A. fumigatus Af293 translation elongation factor eEF-1 subunit γ, putative mRNA | XM_742528 |
| TrMZD04AAA | AJ880807 | 8 | Hypothetical protein | |
| TrMZB07ABB | AJ882566 | 8 | Cytochrome P-450 pattern | PF00067 |
| TrMZB07ACL | AJ883963 | 8 | A. fumigatus Af293 bZIP transcription factor HacA mRNA | XM_743634 |
| TrMZC11ACE | AJ883335 | 7 | A. fumigatus Af293 indoleamine 2,3-dioxygenase family protein mRNA | XM_741638 |
| TrMZG01ACD | AJ883280 | 7 | A. fumigatus Af293 nucleolar GTP-binding protein (Nog1), putative mRNA | XM_750390 |
| TrMZH04AAQ | AJ881965 | 7 | A. fumigatus Af293 ATP synthase F1, β subunit, putative mRNA | XM_748496 |
| TrMZA06AAJ | AJ881633 | 7 | T. rubrum putative secreted Mep1 (MEP1) gene | AF407185 |
| TrMZG06ACJ | AJ883840 | 7 | A. fumigatus Af293 translation elongation factor EF-Tu, putative mRNA | XM_747492 |
| TrMZH11ABD | AJ882834 | 7 | A. fumigatus Af293 mitochondrial F1 ATPase subunit α, putative mRNA | XM_742241 |
| TrMZF10ACI | AJ883742 | 7 | A. fumigatus Af293 fumarylacetoacetate hydrolase FahA mRNA | XM_744462 |
| TrMZB01ABZ | AJ883036 | 7 | A. fumigatus Af293 conserved hypothetical protein mRNA | XM_746193 |
| TrMZG05AAD | AJ881130 | 7 | A. fumigatus Af293 RNA helicase (Dbp), putative mRNA | XM_750315 |
| TrMZG07AAL | AJ881800 | 7 | A. fumigatus Af293 acetyl coenzyme A acetyltransferase, putative mRNA | XM_746201 |
| TrMZD05AAR | AJ882014 | 7 | Hypothetical protein | |
| TrMZH11ACP | AJ884394 | 7 | A. fumigatus Af293 flotillin domain protein mRNA | XM_750060 |
| TrMZB05AAH | AJ881454 | 7 | A. fumigatus Af293 sphinganine hydroxylase Sur2, putative mRNA | XM_747960 |
| TrMZC07AAH | AJ881468 | 7 | A. fumigatus Af293 carbamoyl-phosphate synthase, small subunit mRNA | XM_748862 |
| TrMZA11ABB | AJ882558 | 7 | A. fumigatus Af293 heat shock protein Hsp98/Hsp104/ClpA, putative mRNA | XM_747803 |
| TrMZD06ACB | AJ883153 | 7 | A. fumigatus Af293 protein kinase, putative mRNA | XM_741763 |
| TrMZF09AAC | AJ881027 | 6 | A. fumigatus Af293 glyceraldehyde 3-phosphate dehydrogenase GpdA mRNA | XM_743052 |
| TrMZH11ACG | AJ883582 | 6 | A. fumigatus Af293 stearic acid desaturase (SdeA), putative mRNA | XM_743825 |
| TrMZE10AAA | AJ880825 | 6 | Hypothetical protein | |
| TrMZD04ACK | AJ883893 | 6 | A. fumigatus Af293 fructose-bisphosphate aldolase, class II mRNA | XM_749359 |
| TrMZB07ABX | AJ882853 | 6 | A. fumigatus Af293 conserved hypothetical protein mRNA | XM_748162 |
| TrMZH03AAB | AJ880950 | 6 | A. fumigatus Af293 malate dehydrogenase, NAD-dependent mRNA | XM_743843 |
| TrMZC03AAD | AJ881080 | 6 | A. fumigatus Af293 aspartate transaminase, putative mRNA | XM_750205 |
| TrMZE12AAR | AJ882033 | 6 | A. fumigatus Af293 5-aminolevulinic acid synthase HemA mRNA | XM_748913 |
| TrMZA09ACM | AJ884045 | 6 | A. fumigatus Af293 translation initiation factor SUI1, putative mRNA | XM_749887 |
| TrMZA01ACJ | AJ883764 | 6 | A. fumigatus Af293 NAD-dependent formate dehydrogenase AciA/Fdh mRNA | XM_742493 |
| TrMZF11AAF | AJ881316 | 6 | A. fumigatus Af293 telomere- and ribosome-associated protein Stm1, putative mRNA | XM_749434 |
| TrMZF02AAL | AJ881783 | 6 | A. fumigatus Af293 isocitrate dehydrogenase, NAD-dependent mRNA | XM_747557 |
| TrMZH09ACE | AJ883392 | 6 | A. fumigatus Af293 conserved hypothetical protein mRNA | XM_748162 |
| TrMZA10ABY | AJ882939 | 6 | T. rubrum carboxypeptidase Y (TruScpC) | AY497024 |
| TrMZC04ACK | AJ883881 | 5 | A. fumigatus Af293 histone H4 arginine methyltransferase RmtA mRNA | XM_745275 |
| TrMZE03ACM | AJ884087 | 5 | A. fumigatus Af293 extracellular serine-threonine-rich protein mRNA | XM_749218 |
| TrMZD02AAA | AJ880805 | 5 | A. fumigatus Af293 glycosyl hydrolase, putative mRNA | XM_743297 |
| TrMZG05ACF | AJ883469 | 5 | A. fumigatus Af293 RAB GTPase Ypt5, putative mRNA | XM_746929 |
| TrMZH11ACM | AJ884131 | 5 | A. fumigatus Af293 eukaryotic translation initiation factor 3 subunit EifCb, putative mRNA | XM_744860 |
| TrMZB10ACD | AJ883229 | 5 | A. fumigatus Af293 aspartic endopeptidase Pep2 mRNA | XM_749386 |
| TrMZG04ACO | AJ884293 | 5 | Hypothetical protein | |
| TrMZF01AAH | AJ881497 | 5 | A. fumigatus Af293 MFS toxin efflux pump (AflT), putative mRNA | XM_747539 |
| TrMZD05AAD | AJ881094 | 5 | A. fumigatus Af293 biotin synthase, putative mRNA | XM_742617 |
| TrMZB02AAE | AJ881163 | 5 | Hypothetical protein | |
| TrMZE04AAE | AJ881201 | 5 | A. fumigatus Af293 metacaspase CasA mRNA | XM_745326 |
| TrMZF08ABC | AJ882711 | 5 | A. fumigatus Af293 NAD+-isocitrate dehydrogenase subunit I mRNA | XM_745436 |
| TrMZB10ACN | AJ884154 | 5 | A. fumigatus Af293 conserved hypothetical protein mRNA | XM_746150 |
| TrMZH04AAI | AJ881619 | 5 | A. fumigatus Af293 60S ribosomal protein L5, putative mRNA | XM_747566 |
| TrMZG06ACM | AJ884114 | 5 | A. fumigatus Af293 cytochrome c peroxidase Ccp1, putative mRNA | XM_746821 |
| TrMZE03AAL | AJ881772 | 5 | A. fumigatus Af293 conserved hypothetical protein mRNA | XM_745144 |
| TrMZF09AAL | AJ881790 | 5 | A. fumigatus Af293 phosphoribosyl diphosphate synthase isoform 4 mRNA | XM_746655 |
| TrMZH07AAL | AJ881812 | 5 | A. fumigatus Af293 translation elongation factor EF-2 subunit, putative mRNA | XM_750593 |
| TrMZG02ACI | AJ883745 | 5 | T. rubrum Lap2 | AY496930 |
| TrMZE03ACH | AJ883633 | 5 | A. fumigatus Af293 60S ribosomal protein L4, putative mRNA | XM_742948 |
| TrMZB06ABD | AJ882757 | 5 | A. fumigatus Af293 eukaryotic translation initiation factor 5, putative mRNA | XM_750140 |
| TrMZD05ACG | AJ883528 | 5 | A. fumigatus Af293 RNA polymerase I and III transcription factor complex component Tbp, putative mRNA | XM_749515 |
The ESTs were derived from a cDNA library constructed by the use of transcripts isolated from a 10-day-old T. rubrum culture grown in soy protein medium (for details, see Materials and Methods).
Recently, a larger EST library was obtained from T. rubrum cultures grown on standard laboratory media, i.e., spores and hyphae from yeast extract peptone glucose medium (33) (www.mgc.ac.cn/TrED/). Interestingly, despite the smaller size of our collection, several hundred of our ESTs were found to be absent from the library of Wang et al. (see Table S1 in the supplemental material) (33). Many of these unique sequences, which may be specifically related to the growth of T. rubrum on proteins, encode proteases and hypothetical proteins with unknown functions. A sequence in this collection that shows homology to heat shock-related genes in other filamentous fungi (TrMZE08ACQ) was identified by our microarray analysis as the most upregulated gene during the growth of T. rubrum cells in keratin-soy medium compared to Sabouraud medium (see also Table 5). The results of the EST sequence analysis already indicate that proteolysis by dermatophytes represents an important process for these fungi and that appropriate cDNA libraries are necessary to unravel the underlying transcriptional response.
TABLE 5.
T. rubrum genes commonly upregulated in both soy and keratin-soy mediac
| Cluster | Expression change (n-fold)a
|
Tentative annotation | KEGG orthology | |
|---|---|---|---|---|
| Soy vs Sab | Keratin-soy vs Sab | |||
| TrMZE08ACQ | 32.0 | 73.8 | Hsp70 family chaperone, putative | |
| TrMZA11AAT | 14.0 | 3.5 | Hypothetical protein | |
| TrMZA08ACP | 13.8 | 6.7 | Oxidoreductase, zinc-binding dehydrogenase family | K08070 |
| TrMZC09ACG | 13.5 | 19.4 | No significant hits found | |
| TrMZG08AAC | 12.4 | 6.1 | Plant homeo domain zinc finger protein | |
| TrMZC02ACH | 11.7 | 18.0 | Hypothetical protein | |
| TrMZC11ABC | 7.3 | 8.2 | Hypothetical protein | |
| TrMZA01AAT | 6.6 | 7.5 | Isocitrate lyase AcuD | K01637b |
| TrMZG11ACB | 6.2 | 2.1 | Isochorismatase family hydrolase | |
| TrMZD06ACI | 5.9 | 10.3 | Malate synthase A AcuE | K01638b |
| TrMZH11AAX | 6.0 | 4.1 | No significant hits found | |
| TrMZF07AAX | 5.6 | 9.1 | Hypothetical protein | |
| TrMZD01AAC | 5.0 | 4.1 | General amino acid permease (Agp2) | K03293 |
| TrMZE09ACJ | 4.2 | 10.9 | No significant hits found | |
| TrMZE04ACM | 3.8 | 9.8 | Guanine nucleotide-binding protein α subunit | |
| TrMZB02ACB | 3.0 | 6.6 | Hypothetical protein | |
| TrMZF05AAX | 2.7 | 5.4 | 5′-3′ exoribonuclease Dhp1 | K01146 |
| TrMZB01ACL | 3.4 | 5.1 | Cytochrome P450 monooxygenase | |
| TrMZB02ABY | 3.4 | 3.2 | C6 transcription factor PRO1 | |
| TrMZA10AAT | 3.1 | 2.8 | HLH transcription factor, putative | |
Only genes with an expression change of fivefold or greater are indicated, except for genes that encode putative transcription factors C6Pro1 and HLH.
Entries belonging to the pathway ko00630, i.e., glyoxylate and dicarboxylate metabolism.
Protease genes are indicated in Table 6. All of the probes in this table were significantly regulated with an FDR of <5%.
Construction of a T. rubrum cDNA microarray for the specific analysis of protein digestion.
A T. rubrum microarray was constructed as described in Materials and Methods. Including control sequences, the cDNA microarray contained a total set of 2,626 PCR products that were spotted in triplicate on glass slides so that the microarray contains 9,408 spots. Because secreted proteases are considered particularly important for protein degradation by dermatophytes and for their pathogenicity, known protease genes absent from our EST collection were added as full-length cDNAs to the microarray (Table 1). Only sequences from dipeptidyl protease gene DPPIV are present as both EST sequences and an additional full-length cDNA. This approach allowed the analysis of at least 22 distinct T. rubrum protease genes for their possible involvement in protein digestion (see also Table 6).
TABLE 6.
Changes in T. rubrum protease gene expression in soy and keratin-soy media compared to Sabouraud mediumd
| Genea | EST clusterb | Expresson change (n-fold)c
|
|
|---|---|---|---|
| Soy vs Sabouraud | Keratin-soy vs Sabouraud | ||
| Endoprotease genes | |||
| MEP1 | TrMZA06AAJ | 11.5 | 40.8 |
| MEP2 | PCR | 5.5 | |
| MEP3 | TrMZB07ACH | 16.9 | 38.8 |
| MEP4 | TrMZD09AAV | 21.7 | 63.7 |
| SUB1 | TrMZA12AAE | −2.9 | 4.0 |
| SUB2 | PCR | ||
| SUB3 | TrMZB09AAX | 33.2 | 72.4 |
| SUB4 | TrMZE10AAT | 40.3 | 31.1 |
| SUB5 | TrMZD09ACG | 4.6 | 4.6 |
| SUB6 | PCR | 13.3 | |
| SUB7 | PCR | ||
| Exoprotease genes | |||
| LAP1 | TrMZA03ACB | 10.5 | 11.1 |
| LAP2 | TrMZB03ACL | 33.7 | 60.4 |
| SCPA | PCR | −4.3 | |
| SCPB | PCR | 4.5 | |
| MCPA | TrMZG09AAA | 4.8 | 40.1 |
| DPPIV | PCR | ||
| DPPV | TrMZC07ABY | 2.1 | |
| PAP | PCR | −2.9 | |
| AMPP | PCR | ||
| NpII-1 | PCR | −14 | |
| NpII-2 | PCR | ||
For proteases represented by more than one cluster on the array, the one with the highest change in expression is indicated.
Genes which are represented by an added PCR product instead of EST clusters are marked with PCR.
Downregulation of a gene is indicated by a minus sign followed by the n-fold change in expression.
All of the probes in this table were significantly regulated with an FDR of <5%.
Transcriptional profiling in T. rubrum during growth on soy and keratin.
Growth media containing proteins as a sole source of carbon and nitrogen are known to induce stronger secretion of proteolytic activity of T. rubrum in the supernatant compared to Sabouraud medium (10, 11, 24). Therefore, T. rubrum cultures in Sabouraud medium were used as a control in our microarray experiments. To analyze the transcriptional response during the growth of T. rubrum on proteins, two substrates were used: liquid soy and keratin-soy media. The latter contained a small amount (0.01%) of soy protein to facilitate the initial growth of the fungus on the insoluble keratin granules. Fungal cells cultivated in soy medium were harvested for RNA extraction after 10 days, a time point when substantial proteolytic activity was recorded with concomitant clarification of the opaque medium. In contrast, the comparatively slower growing T. rubrum cells in keratin-soy medium were subjected to RNA isolation after 28 days, a time point when keratin granules were not completely dissolved by the fungus, yet substantial proteolytic activity was measured in the culture supernatant (data not shown). For microarray analysis, three independently prepared T. rubrum replicates grown in each of the three media, Sabouraud, soy, and keratin-soy media (designated SabA, -B, and -C, soyA, -B, and -C, and keratin-soyA, -B, and -C) were used. For the results of the pairwise transcriptional comparisons, i.e., soy medium versus Sabouraud medium (A) and keratin-soy medium versus Sabouraud medium (B), and the results from a linear model including all comparisons against Sabouraud medium plus three direct keratin-soy medium versus soy medium (C) comparisons, see Table S2A, B, and C in the supplemental material, which are accessible via Gene Expression Omnibus Series accession number GSE11711 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11711). An FDR of 5% and a twofold change in the gene expression level was set as the threshold.
During the growth of T. rubrum in soy medium, 179 and 110 genes were found to be up- and downregulated, respectively, compared to growth in Sabouraud medium. In keratin-soy medium, 258 genes were upregulated and 222 genes were downregulated. The expression profiles of soy medium versus Sabouraud medium and keratin-soy medium versus Sabouraud medium were very similar but not identical, as indicated by the higher number of differentially expressed T. rubrum genes in the latter profile (Fig. 1). Most of the genes specifically regulated in keratin-soy medium, however, were expressed at a comparatively low level, and only a minor set of genes was strongly activated (only 10 genes more than fivefold) or repressed (only 1 gene more than fivefold) (Table 4). Among the highly upregulated, keratin-specific genes, we identified sequences that encode a not-yet-described putative Lap1-like protease (TrMZE06ACL), a putative Rpn4 C2H2 transcription factor (TrMZA01AAW), and potentially dermatophyte-specific hypothetical protein-encoding genes.
FIG. 1.
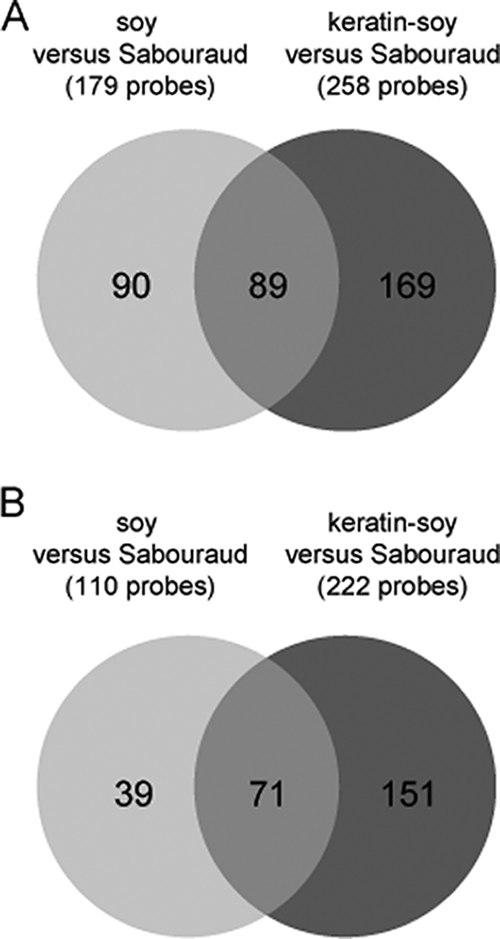
Venn diagrams showing the numbers of genes which are specifically upregulated (A) and downregulated (B) during the growth of T. rubrum in either soy or keratin-soy medium compared to Sabouraud medium, respectively.
TABLE 4.
T. rubrum genes only differentially regulated in keratin-soy medium versus Sabouraud medium and not in soy medium versus Sabouraud mediumd
| Cluster | Expression change (n-fold)a | Tentative annotation | KEGG orthology |
|---|---|---|---|
| TrMZE06ACL | 30.98 | Putative Lap1-like protease | |
| TrMZH10AAW | 9.34 | Hypothetical protein | |
| TrMZB09AAE | 8.65 | Hypothetical protein | |
| TrMZE06ACM | 5.83 | snoRNP (gar1), putative | |
| TrMZC05ACN | 5.72 | Oxalate decarboxylase | K01569c |
| TrMZA01AAW | 5.59 | C2H2 transcription factor Rpn4 | |
| TrMZC07ACO | 5.54 | Hypothetical protein | K01199 |
| Mep2b | 5.54 | Mep2 | |
| TrMZE07ACM | 5.24 | No significant hits found | |
| TrMZC07ACM | 5.22 | C2H2 transcription factor Rpn4 | |
| TrMZC07ABD | −7.61 | Hypothetical protein |
Only genes with an expression change of fivefold or greater are indicated.
This sequence was added as an additional PCR product on the microarray.
Entry belonging to the pathway ko00630, i.e., glyoxylate and dicarboxylate metabolism.
All of the probes in this table were significantly regulated with an FDR of <5%.
Identification of genes which suggest common adaptation mechanisms during the growth of T. rubrum on proteins.
As expected, many of the T. rubrum genes upregulated in both protein-containing media encode secreted proteases (see next paragraph). A pathway analysis with KEGG was systematically performed for all of the genes expressed more than fivefold in keratin-soy medium versus Sabouraud medium (Table 4) and both soy and keratin-soy media versus Sabouraud medium (Table 5), respectively. Of the annotated genes, three encode enzymes which are linked to the glyoxylate pathway, i.e., isocitrate lyase (TrMZA01AAT), malate synthase A (TrMZD06ACI), and oxalate decarboxylase (TrMZC05ACN); expression of the latter was only detected during the growth of T. rubrum in keratin-soy medium compared to Sabouraud medium. Other genes induced by both protein-containing media encode various enzymes, transcription factors such as a putative C6 Pro1 transcription factor (TrMZB02ABY), and a helix-loop-helix (HLH) transcription factor (TrMZA10AAT), as well as hypothetical proteins with unknown functions (Table 5). TrMZE08ACQ, the most upregulated T. rubrum sequence in keratin-soy medium and also among the most strongly activated genes in soy medium, shares homology with Aspergillus fumigatus and Neurospora crassa Hsp70 chaperones. Other putative heat shock-related genes, such as the HSP70-related sequence TrMZG01AAL or an HSP70 chaperone, HSP88 (TrMZH04ABZ), were only found to be activated in T. rubrum during growth on soy medium compared to Sabouraud medium, albeit at comparatively low levels.
Activation of multiple protease genes in T. rubrum during growth on proteins.
We examined in detail the expression of T. rubrum genes that encode secreted proteases because of their presumed role in both protein degradation and pathogenicity. Table 6 details the expression values of 22 distinct protease genes detected in the pairwise comparisons of T. rubrum grown in soy medium versus Sabouraud medium and keratin-soy medium versus Sabouraud medium, respectively. In both of the protein-containing media, a comparable set of protease genes was upregulated in the fungal cells. In particular, members of the two dermatophyte-specific endoprotease gene families of Subs and fungalysins were strongly induced, i.e., SUB3 and SUB4, as well as MEP3 and MEP4, respectively. SUB5 was found to be only moderately activated in both protein-containing media. In addition, genes that encode exoproteases were found to be strongly activated in both protein-containing media, for example, Lap genes LAP1 and LAP2, as well as the metallocarboxypeptidase gene MCPA.
Some of the commonly activated protease genes, such as SUB3, MEP1, MEP3, MEP4, LAP2, and MCPA, appeared to be even more strongly induced during growth in keratin-soy medium than in soy medium. Interestingly, induction of a small number of protease genes was detected only during growth on either soy or keratin-soy medium. For example, SUB1 was found to be activated only during growth in keratin-soy medium, albeit at a comparatively low level, as were MEP2 and DPPV. In contrast, the genes SUB6 and SCPB were shown to be upregulated only during growth in soy medium compared to keratin-soy medium. Other protease genes, such as SUB7 and DPPIV, were not found to be significantly activated during growth in either protein-containing medium. The direct comparison of T. rubrum cultures grown on keratin-soy and soy media confirmed that commonly activated protease genes such as MEP1, MEP4, SUB3, and MCPA were expressed at higher levels during growth in keratin-soy medium than during growth in soy medium (see Table S2C in the supplemental material). The gene with the highest n-fold expression change in this comparison encodes neutral protease II-1 (NpII-1), which was significantly downregulated in T. rubrum during growth on soy medium compared to growth on Sabouraud medium (Table 6).
Validation of microarray data by quantitative real-time RT-PCR.
Upregulation of the T. rubrum protease genes SUB3, SUB4, and MCPA in soy and keratin-soy media, as well as the differential activation of SUB6 in these two protein-containing media, was validated by quantitative real-time RT-PCR (Table 7). Differences in the expression levels among the three replicates were detected; i.e., upregulation of SUB3, SUB4, and MCPA in keratin-soy medium was stronger in samples keratin-soyB and keratin-soyC than in sample keratin-soyA, whereas SUB4 and SUB6 were expressed in soy medium at a higher level in samples soyA and soyC than in sample soyB. In summary, a large but comparable set of different proteolytic enzymes was activated in T. rubrum during growth in either soy or keratin-soy medium, supporting the view that multiple secreted proteases manage the efficient proteolysis of such substrates. Upregulation of the genes that encode isocitrate lyase (TrMZA01AAT) and malate synthase (TrMZD06ACI) of the glyoxylate cycle was also validated in soy and keratin-soy media (Table 7).
TABLE 7.
Expression of selected T. rubrum protease genes during growth in Sabouraud, soy, and keratin-soy media measured by quantitative RT-PCR
| Sample | Mean n-fold gene expression (SEM)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SUB3 | SUB4 | SUB6 | MCPA | ACUDa | ACUEb | ADRPRFc | CHSc,d | |
| SabA | 1.30 (0.21) | 13.42 (1.53) | 2.07 (0.15) | 1.47 (0.15) | 1.00 (0.11) | 1.08 (0.09) | 1.80 (0.06) | 1.09 (0.05) |
| SabB | 1.04 (0.13) | 1.37 (0.20) | 2.54 (0.16) | 1.03 (0.08) | 1.68 (0.06) | 1.00 (0.04) | 1.65 (0.16) | 1.19 (0.10) |
| SabC | 1.00 (0.14) | 1.00 (0.17) | 4.43 (0.20) | 1.00 (0.08) | 3.77 (0.14) | 1.79 (0.08) | 1.61 (0.11) | 1.21 (0.08) |
| SoyA | 140.83 (4.36) | 2,443.26 (197.38) | 69.98 (5.05) | 22.73 (0.31) | 16.01 (1.44) | 6.68 (0.33) | 1.96 (0.07) | 1.00 (0.05) |
| SoyB | 38.77 (3.05) | 93.20 (7.36) | 18.08 (2.13) | 5.56 (0.37) | 7.53 (0.59) | 4.73 (0.41) | 1.32 (0.14) | 1.49 (0.17) |
| SoyC | 41.15 (3.03) | 257.80 (24.10) | 92.58 (8.77) | 7.66 (0.42) | 23.00 (2.06) | 8.47 (0.58) | 1.68 (0.16) | 1.17 (0.12) |
| Keratin-soyA | 54.30 (2.85) | 122.03 (8.18) | 1.00 (0.10) | 23.82 (1.16) | 10.50 (0.32) | 8.10 (0.22) | 1.00 (0.05) | 1.96 (0.18) |
| Keratin-soyB | 2,527.09 (452.00) | 1,394.32 (181.39) | 1.28 (0.08) | 497.83 (84.68) | 7.20 (0.41) | 7.10 (0.48) | 1.17 (0.09) | 1.68 (0.16) |
| Keratin-soyC | 1,759.49 (292.60) | 1,464.24 (194.54) | 1.70 (0.13) | 296.72 (46.39) | 10.35 (0.68) | 8.19 (0.60) | 1.24 (0.10) | 1.58 (0.16) |
ACUD, isocitrate lyase AcuD.
ACUE, malate synthase A.
Control gene.
CHS, chitin synthase.
DISCUSSION
Dermatophytes are highly specialized pathogenic fungi which grow exclusively in keratinized host structures such as the stratum corneum, nails, or hair, utilizing them as a sole nitrogen and carbon source. A microarray was constructed with cDNA derived from transcripts of fungal cells grown in protein-containing medium to identify genes in T. rubrum which are involved in the basic adaptation mechanisms which support the growth of dermatophytes on proteins. Despite its limited total number of genes, covering approximately 20 to 25% of the estimated size of the T. rubrum genome, the present cDNA microarray appears useful for the specific analysis of protein utilization by dermatophytes. A previous comparison of our EST collection with the larger T. rubrum EST library of Wang et al. (33) already revealed that many of our ESTs were new, a finding which may be attributed to the different cultivation conditions preceding fungal mRNA isolation and cDNA preparation. Among these EST sequences were several genes that encode secreted proteases such as the major fungalysins Mep3 and Mep4 and metallocarboxypeptidase A, as well as many genes which do not share any homology with sequences deposited in public databases.
Two protein substrates were studied in our microarray analysis, soy and keratin-soy liquid media. Both media promote high secreted proteolytic activity by the fungus. Transcriptional analysis was performed at a time point of active fungal growth, when substantial proteolytic activity was recorded with a concomitant clarification of the media and dissolution of the water-insoluble keratin granules. In these conditions, a high degree of similarity was observed in the transcriptional response of T. rubrum during growth on each protein source, i.e., after 10 days of growth in soy medium and 28 days in keratin-soy medium, respectively. In particular, genes that encode major keratinases such as endoproteases Mep3 and Mep4, as well as Sub3 and Sub4, were commonly upregulated. In agreement with this finding, sequences that encode Sub3 and Mep3 were also among the most abundant transcripts in our EST collection. Among the exoproteases genes, a strong upregulation of the T. rubrum genes LAP1 and LAP2 was observed in both of the protein-containing media, and MCPA was particularly strongly upregulated in keratin-soy medium. In contrast to recent secretome analyses of T. rubrum soy medium cultures (8), we also identified the activation of protease genes MEP1, MEP2, and DPPV, notably, the latter two only during the growth of T. rubrum in keratin-containing medium. Expression of individual protease genes on keratin has also been detected by suppression-subtractive hybridization screens in T. mentagrophytes, i.e., dipeptidyl peptidase V (12) and in T. rubrum, i.e., Sub3, Sub5, Mep3, and Mep4 (19).
Individual protease genes were found to be activated at a significantly higher level in keratin-soy medium than in soy medium. However, these differences may not necessarily be substrate specific but could also be influenced by other parameters, e.g., different incubation times. In the filamentous fungus N. crassa, a coordinately regulated expression of genes with multiple cellular functions was demonstrated to be clock controlled (2; reviewed in reference 5), and a differential gene expression was detected in the concentric growth zones of Aspergillus niger on solid agar (15). In order to exclude the possibility that activation of distinct protease genes in keratin-soy medium is merely related to the age of the culture rather than substrate specific, an additional time course analysis by quantitative RT-PCR was conducted (data not shown). In this experiment, significant proteolytic activity was detected after 30 and 37 days of growth of T. rubrum in keratin-soy medium, along with keratinolysis and strong expression of protease genes SUB3, SUB4, and MCPA. In contrast, in poorly growing 23-day-old cultures, we detected neither keratin dissolution nor significant proteolytic activity and expression of the protease genes analyzed. This observation strengthens our present results and the hypothesis that proteolytic activity and protein degradation by T. rubrum are strongly correlated with the expression of distinct protease genes.
Transcriptional profiling in T. rubrum during growth on soy and keratin-soy protein-containing media revealed not only a common activation of proteases but also activation of other factors which could be involved in protein utilization. Notably, a sequence that encodes an Hsp70 protein (TrMZE08ACQ) was found to be strongly upregulated during growth in both protein-containing media, i.e., soy and keratin-soy media, compared to Sabouraud medium. Interestingly, the closely related Hsp70 protein Lhs1 (lumenal Hsp70) in Saccharomyces cerevisiae is located in the endoplasmic reticulum lumen and involved in protein precursor translocation and folding (3); S. cerevisiae LHS1 mutants were demonstrated to be defective in the translocation of several secretory preproteins. Since the induced T. rubrum HSP70 gene is concomitantly expressed with genes that encode major secreted proteases, the putative Hsp70 chaperone could be involved in the folding and/or secretion of these enzymes. Another heat shock gene, HSP70 (TrMZA12ACM), was the most abundant transcript in our EST collection. This gene was found to be expressed at higher levels in T. rubrum when it was exposed to elevated growth temperatures (27). However, our microarray analyses did not detect a differential expression of this HSP70 gene during the growth of T. rubrum in protein-containing media compared to Sabouraud medium at 30°C. Interestingly, a closely related, likely orthologous HSP70 sequence was recently identified as the most abundant transcript in a Pneumocystis carinii EST library derived from infected lung tissue (4). Seven other HSP70 sequences were detected in our EST collection. Heat shock proteins may not only be involved in environmental adaptation processes but are also known to be immunodominant antigens, e.g., in the related dermatophyte T. mentagrophytes and other fungal pathogens (1, 22).
Of major interest also appears to be the strong upregulation of key enzymes of the glyoxylate cycle in T. rubrum during growth on proteins, the putative malate synthase and isocitrate lyase. This observation suggests a particular function of this metabolic pathway in protein utilization by T. rubrum which could also be important in dermatophyte pathogenicity. In addition, the glyoxylate cycle is absent in mammals and therefore represents a potential drug target. Whereas this pathway appears to be dispensable for A. fumigatus-induced invasive aspergillosis (26, 28), it was found to be virulence associated in infections by other microbial pathogens such as Mycobacterium tuberculosis and the yeast Candida albicans (17, 21). In these pathogens, enzymes of the glyoxylate pathway were shown to contribute to the persistence of the microbes in phagocytic immune cells, a function which remains elusive for dermatophytes.
Exploring the adaptive response of dermatophytes during proteolysis, the identification of transcription factors also appears to be of interest. In both protein-containing media, the activation of a putative C6 Pro1 and an HLH transcription factor was detected in T. rubrum. The putative zinc finger transcription factor Pro1 shares homology with A. fumigatus RosA and NosA, which are known regulators of sexual development in A. nidulans. Interestingly, A. nidulans NosA is induced during late asexual development and also upon carbon starvation (32). In Sordaria macrospora, the C6 zinc finger transcription factor Pro1 was shown to be required for fruiting body development (20). Upregulation of a putative Rpn4 transcription factor was only found in keratin-soy medium. In S. cerevisiae, Rpn4 (also called Son1) is known to regulate genes that encode proteasomal subunits, the function of which is putatively linked to the unfolded protein response and endoplasmic reticulum-associated protein degradation processes (18, 25). Sequences that encode a putative HacA transcription factor were detected among the most abundant transcripts. HacA controls the unfolded protein response in eukaryotic cells, a regulatory pathway with multiple functions in the folding and secretion of proteins. A possible correlation of this pathway with the high secretory activity of dermatophytes appears to be of interest.
Since the particular ability of dermatophytes to grow on proteins such as keratin has long been discussed as the most important pathogenicity-related factor, our studies should contribute to a better understanding of the basic molecular mechanisms in the pathogenesis of these fungi. Molecular techniques in dermatophytes will probably further improve in the future, allowing us to functionally characterize candidate genes by genetic manipulation. Nevertheless, infection of the host is presumably much more complex, and transcriptional analysis in dermatophytes during infection is necessary to further decipher the relevant characteristics which make these fungi the most common agents of superficial mycoses.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Foundation for Scientific Research, grant 3100-105313/1. Peter Staib was part of the time a recipient of a postdoctoral fellowship from the Deutsche Akademie der Naturforscher Leopoldina (Förderkennzeichen BMBF-LPD 9901/8-146).
Footnotes
Published ahead of print on 19 December 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Burnie, J. P., T. L. Carter, S. J. Hodgetts, and R. C. Matthews. 2006. Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 3053-88. [DOI] [PubMed] [Google Scholar]
- 2.Correa, A., Z. A. Lewis, A. V. Greene, I. J. March, R. H. Gomer, and D. Bell-Pedersen. 2003. Multiple oscillators regulate circadian gene expression in Neurospora. Proc. Natl. Acad. Sci. USA 10013597-13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craven, R. A., M. Egerton, and C. J. Stirling. 1996. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 152640-2650. [PMC free article] [PubMed] [Google Scholar]
- 4.Cushion, M. T., A. G. Smulian, B. E. Slaven, T. Sesterhenn, J. Arnold, C. Staben, A. Porollo, R. Adamczak, and J. Meller. 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS ONE 2e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlap, J. C., and J. J. Loros. 2006. How fungi keep time: circadian system in Neurospora and other fungi. Curr. Opin. Microbiol. 9579-587. [DOI] [PubMed] [Google Scholar]
- 6.Fachin, A. L., M. S. Ferreira-Nozawa, W. Maccheroni, Jr., and N. M. Martinez-Rossi. 2006. Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J. Med. Microbiol. 551093-1099. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira-Nozawa, M. S., H. C. Silveira, C. J. Ono, A. L. Fachin, A. Rossi, and N. M. Martinez-Rossi. 2006. The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med. Mycol. 44641-645. [DOI] [PubMed] [Google Scholar]
- 8.Giddey, K., M. Monod, J. Barblan, A. Potts, P. Waridel, C. Zaugg, and M. Quadroni. 2007. Comprehensive analysis of proteins secreted by Trichophyton rubrum and Trichophyton violaceum under in vitro conditions. J. Proteome Res. 63081-3092. [DOI] [PubMed] [Google Scholar]
- 9.Hellemans, J., G. Mortier, A. De Paepe, F. Speleman, and J. Vandesompele. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jousson, O., B. Lechenne, O. Bontems, S. Capoccia, B. Mignon, J. Barblan, M. Quadroni, and M. Monod. 2004. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology 150301-310. [DOI] [PubMed] [Google Scholar]
- 11.Jousson, O., B. Lechenne, O. Bontems, B. Mignon, U. Reichard, J. Barblan, M. Quadroni, and M. Monod. 2004. Secreted subtilisin gene family in Trichophyton rubrum. Gene 33979-88. [DOI] [PubMed] [Google Scholar]
- 12.Kaufman, G., I. Berdicevsky, J. A. Woodfolk, and B. A. Horwitz. 2005. Markers for host-induced gene expression in Trichophyton dermatophytosis. Infect. Immun. 736584-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon-Chung, K. J., and J. E. Bennett (ed.). 1992. Medical mycology. Lea & Febiger, Philadelphia, PA.
- 14.Léchenne, B., U. Reichard, C. Zaugg, M. Fratti, J. Kunert, O. Boulat, and M. Monod. 2007. Sulphite efflux pumps in Aspergillus fumigatus and dermatophytes. Microbiology 153905-913. [DOI] [PubMed] [Google Scholar]
- 15.Levin, A. M., R. P. de Vries, A. Conesa, C. de Bekker, M. Talon, H. H. Menke, N. N. van Peij, and H. A. Wosten. 2007. Spatial differentiation in the vegetative mycelium of Aspergillus niger. Eukaryot. Cell 62311-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, T., Q. Zhang, L. Wang, L. Yu, W. Leng, J. Yang, L. Chen, J. Peng, L. Ma, J. Dong, X. Xu, Y. Xue, Y. Zhu, W. Zhang, L. Yang, W. Li, L. Sun, Z. Wan, G. Ding, F. Yu, K. Tu, Z. Qian, R. Li, Y. Shen, Y. Li, and Q. Jin. 2007. The use of global transcriptional analysis to reveal the biological and cellular events involved in distinct development phases of Trichophyton rubrum conidial germination. BMC Genomics 8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 41283-86. [DOI] [PubMed] [Google Scholar]
- 18.Mannhaupt, G., R. Schnall, V. Karpov, I. Vetter, and H. Feldmann. 1999. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 45027-34. [DOI] [PubMed] [Google Scholar]
- 19.Maranhão, F. C., F. G. Paiao, and N. M. Martinez-Rossi. 2007. Isolation of transcripts over-expressed in human pathogen Trichophyton rubrum during growth in keratin. Microb. Pathog. 43166-172. [DOI] [PubMed] [Google Scholar]
- 20.Masloff, S., S. Poggeler, and U. Kuck. 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406735-738. [DOI] [PubMed] [Google Scholar]
- 22.Milan, R., R. Alois, C. Josef, B. Jana, and W. Evzen. 2004. Recombinant protein and DNA vaccines derived from hsp60 Trichophyton mentagrophytes control the clinical course of trichophytosis in bovine species and guinea-pigs. Mycoses 47407-417. [DOI] [PubMed] [Google Scholar]
- 23.Monod, M. 2008. Secreted proteases from dermatophytes. Mycopathologia 166285-294. [DOI] [PubMed] [Google Scholar]
- 24.Monod, M., B. Lechenne, O. Jousson, D. Grand, C. Zaugg, R. Stocklin, and E. Grouzmann. 2005. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology 151145-155. [DOI] [PubMed] [Google Scholar]
- 25.Ng, D. T., E. D. Spear, and P. Walter. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 15077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivas, I., M. Royuela, B. Romero, M. C. Monteiro, J. M. Minguez, F. Laborda, and J. R. De Lucas. 2008. Ability to grow on lipids accounts for the fully virulent phenotype in neutropenic mice of Aspergillus fumigatus null mutants in the key glyoxylate cycle enzymes. Fungal Genet. Biol. 4545-60. [DOI] [PubMed] [Google Scholar]
- 27.Rezaie, S., J. Ban, M. Mildner, C. Poitschek, C. Brna, and E. Tschachler. 2000. Characterization of a cDNA clone, encoding a 70 kDa heat shock protein from the dermatophyte pathogen Trichophyton rubrum. Gene 24127-33. [DOI] [PubMed] [Google Scholar]
- 28.Schöbel, F., O. Ibrahim-Granet, P. Ave, J. P. Latge, A. A. Brakhage, and M. Brock. 2007. Aspergillus fumigatus does not require fatty acid metabolism via isocitrate lyase for development of invasive aspergillosis. Infect. Immun. 751237-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3Article3. [DOI] [PubMed] [Google Scholar]
- 30.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31265-273. [DOI] [PubMed] [Google Scholar]
- 31.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 18 June 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3research0034.1-0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed]
- 32.Vienken, K., and R. Fischer. 2006. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol. Microbiol. 61544-554. [DOI] [PubMed] [Google Scholar]
- 33.Wang, L., L. Ma, W. Leng, T. Liu, L. Yu, J. Yang, L. Yang, W. Zhang, Q. Zhang, J. Dong, Y. Xue, Y. Zhu, X. Xu, Z. Wan, G. Ding, F. Yu, K. Tu, Y. Li, R. Li, Y. Shen, and Q. Jin. 2006. Analysis of the dermatophyte Trichophyton rubrum expressed sequence tags. BMC Genomics 7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada, T., K. Makimura, and S. Abe. 2006. Isolation, characterization, and disruption of dnr1, the areA/nit-2-like nitrogen regulatory gene of the zoophilic dermatophyte, Microsporum canis. Med. Mycol. 44243-252. [DOI] [PubMed] [Google Scholar]
- 36.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaugg, C., O. Jousson, B. Lechenne, P. Staib, and M. Monod. 2008. Trichophyton rubrum secreted and membrane-associated carboxypeptidases. Int. J. Med. Microbiol. 298669-682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


