Abstract
The inositol 1,3,4,5,6-pentakisphosphate (IP5) 2-kinase (Ipk1) catalyzes the production of inositol hexakisphosphate (IP6) in eukaryotic cells. Previous studies have shown that IP6 is required for efficient nuclear mRNA export in the budding yeast Saccharomyces cerevisiae. Here, we report the first functional analysis of ipk1+ in Schizosaccharomyces pombe. S. pombe Ipk1 (SpIpk1) is unique among Ipk1 orthologues in that it harbors a novel amino (N)-terminal domain with coiled-coil structural motifs similar to those of BAR (Bin-amphiphysin-Rvs) domain proteins. Mutants with ipk1+ deleted (ipk1Δ) had mRNA export defects as well as pleiotropic defects in polarized growth, cell morphology, endocytosis, and cell separation. The SpIpk1 catalytic carboxy-terminal domain was required to rescue these defects, and the mRNA export block was genetically linked to SpDbp5 function and, likely, IP6 production. However, the overexpression of the N-terminal domain alone also inhibited these functions in wild-type cells. This revealed a distinct noncatalytic function for the N-terminal domain. To test for connections with other inositol polyphosphates, we also analyzed whether the loss of asp1+ function, encoding an IP6 kinase downstream of Ipk1, had an effect on ipk1Δ cells. The asp1Δ mutant alone did not block mRNA export, and its cell morphology, polarized growth, and endocytosis defects were less severe than those of ipk1Δ cells. Moreover, ipk1Δ asp1Δ double mutants had altered inositol polyphosphate levels distinct from those of the ipk1Δ mutant. This suggested novel roles for asp1+ upstream of ipk1+. We propose that IP6 production is a key signaling linchpin for regulating multiple essential cellular processes.
Inositol polyphosphates (IPs) constitute an emerging class of signaling molecules that regulate multiple cellular activities including chromatin remodeling and transcription, mRNA export, telomere length regulation, RNA editing, exocytosis, ciliary beating and length maintenance, and translation (8, 21, 32, 48, 51, 56, 59, 70-72). IP production is initiated with the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by phospholipase C (PLC), producing diacylglycerol and soluble inositol 1,4,5-trisphosphate (IP3). IP3 is sequentially phosphorylated by the coordinated actions of specific kinases to produce more highly phosphorylated IP molecules, including inositol 1,3,4,5-tetrakisphosphate (IP4), inositol 1,3,4,5,6-pentakisphosphate (IP5), inositol hexakisphosphate (IP6), and inositol pyrophosphate isomers (e.g., PP-IP4 and IP7) (2, 24, 31, 41, 43, 55, 71). The perturbation of IP synthesis pathways is linked to defects in nutrient homeostasis in fungi (30, 41, 43) and developmental defects in vertebrates (16, 50, 51, 63). In mammalian tissue culture cells, the total cellular IP pool undergoes complex changes during transit through the cell cycle, with IP5, IP6, and IP7 being most abundant in G1 phase, decreasing during S phase, and rising again during G2/M phase (4). However, a functional link between IP flux and cell cycle progression has not been defined.
In Saccharomyces cerevisiae, IP metabolic flux is apparently regulated at the level of the lone Plc1 enzyme, which is most closely related to the vertebrate PLC-δ isoform (71). The multiple PLC isoforms in vertebrates (23) all apparently converge on a single IP5 2-kinase, Ipk1 (64). Ipk1 enzymes have highly conserved putative catalytic site motifs and display functional cross-species complementation (25, 50, 64) (see Fig. S1 in the supplemental material). However, conservation at the overall protein sequence level is relatively low (∼11%), suggesting potential functional and/or regulatory diversification in different organisms. The most striking difference in structural regions is observed in Schizosaccharomyces pombe Ipk1 (SpIpk1), wherein a distinctive N-terminal domain exists with coiled-coil structural motifs similar to those of BAR (Bin-amphiphysin-Rvs) domain proteins (25). In the mammalian amphiphysin and S. cerevisiae Rvs161/167 proteins, such BAR domains are dimerization, membrane-binding, and membrane curvature-sensing modules (45). Functional analysis of the SpIpk1 N-terminal domain has not been reported, and this domain might mediate specialized cellular roles of the protein.
Several recent studies have made key insights into defining the cellular targets for IP6. The human RNA-editing enzyme ADAR2 and the S. cerevisiae tRNA-editing enzyme ADAT1 both require IP6 binding for protein function (32). The efficient nuclear export of mRNA also specifically requires the Ipk1-catalyzed production of IP6 (71). mRNAs are exported as large ribonucleoprotein (mRNP) complexes in a unidirectional manner through nuclear pore complexes (NPCs), embedded in the nuclear envelope (28). The targeting of export-competent mRNPs to NPCs is dependent on the essential mRNA export receptor dimer Mex67-Mtr2 in the budding yeast S. cerevisiae and TAP/NXF1-p15/NXT1 in metazoan cells (18, 26, 54); however, Mex67 is not essential in S. pombe (69). Two factors that are essential for mRNA export in S. cerevisiae are the DEAD box protein Dbp5 and its IP6-bound activator Gle1 (1, 19, 52, 58, 62, 67). Gle1/IP6 activation of Dbp5 at the NPC cytoplasmic face results in a nucleotide-dependent switch in Dbp5 and triggers changes in mRNP protein composition, thus providing directionality to the export process (61). Interestingly, the IPK1 gene was first discovered in an S. cerevisiae genetic screen aimed at studying Gle1 function (71). Global IP production is also required for efficient mRNA export in mammalian cells (13). To date, S. cerevisiae has been the primary model system used to study the mechanism of IP6 function in mRNA export. Whether metazoans or other fungi also specifically require IP6 production for mRNA export has not been directly tested.
In addition to direct protein binding targets for IP6 function, IP6 is also the substrate for downstream IP6 kinases and is inherently required for the production of IP7 pyrophosphates (1). SpAsp1 and its S. cerevisiae orthologue, Vip1, have recently been defined as IP6 and IP7 kinases, with Vip1 acting as a 1/3-kinase contributing to the synthesis of 1/3-PP-IP5 and 1/3,5-(PP)2-IP4 (31, 41). One S. cerevisiae IP7 target has been defined, the Pho80-Pho85-Pho81 cyclin-CDK-CKI system required for nutrient homeostasis (29, 30). However, even if this regulation is conserved in S. pombe, it does not account for the phenotypes observed in S. pombe asp1Δ cells. Notably, S. pombe asp1Δ cells are defective in cell morphology, polarized growth, and endocytosis, and asp1Δ cells are synthetically lethal with mutations in genes encoding components of the Arp2/3 complex and actin (14).
S. pombe cells grow in a polarized fashion. Immediately after cell division, the daughter cells initially grow in a monopolar manner from the cell end that existed before division. Subsequently, cells initiate growth from the new end and resume bipolar growth until mitosis (38). The actin cytoskeleton is critical for such polarized growth, and cytoskeleton perturbations result in round, swollen cells. Actin is organized at the growing surfaces of the cell as cortical patches, which function in membrane growth and endocytosis, and along the long axis of the cell as actin cables, which function as tracks for the delivery of secretory vesicles to growing cell ends (9, 17, 42). Actin patches are delocalized during mitosis and concentrated around the medial septum during cytokinesis (33). Dynamic actin assembly (and disassembly) is essential for the assembly, maintenance, and closure of the contractile actomyosin ring and cytokinesis (44). A functional actin cytoskeleton is also required for the proper trafficking of secretory cargoes during cytokinesis (17). For example, secretory vesicles containing Eng1 and Agn1 endoglucanases are delivered to the septum region by the exocyst complex, allowing the digestion of the division septum and the surrounding cell wall and the final physical separation of the daughter cells (10, 34, 66).
We speculated that an interspecies comparison of Ipk1 between S. cerevisiae and S. pombe, two phylogenetically distant yeasts, would allow the dissection of functional conservation and divergence in the soluble IP pathway. To test this, we used a combined genetic and cell biological approach to investigate SpIpk1 function. In addition to conserved defects in mRNA export, a loss of ipk1+ function resulted in pleiotropic defects in cell morphology, polarized growth, endocytosis, and cell separation. By analyzing ipk1Δ asp1Δ double mutants, we gained unique insights into the independent roles of the respective kinases in IP production and cell physiology.
MATERIALS AND METHODS
S. pombe strain construction, media, and genetic methods.
S. pombe strains were grown in YE medium or Edinburgh minimal medium (EMM) with the appropriate supplements as previously described (40). Strains were constructed by a PCR-based gene disruption strategy and tetrad dissection (see below). Crosses were performed on glutamate medium (EMM lacking ammonium chloride and containing 10 mM glutamate). DNA transformations were done by the lithium acetate method (27). For the regulated expression of genes by the nmt1 promoter, cells were grown in EMM either lacking thiamine to allow expression or with the addition of 10 μM thiamine to repress expression (36).
For the generation of the ipk1Δ strain, the ipk1+ open reading frame (ORF) was completely deleted by PCR-based one-step homologous recombination as previously described (3), using ura4+ as a selectable marker. ura4+ was amplified by PCR from plasmid pKG358 using a forward oligonucleotide primer (spipk1D-F) (see Table S1 in the supplemental material) corresponding to 80 bp upstream of the ATG start codon and a reverse primer (spipk1D-R) (see Table S1 in the supplemental material) corresponding to 80 bp downstream of the TAA stop codon of ipk1+ and transformed into the h−/h+ ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 diploid strain, and stable integrants were selected. The deletion of one copy of ipk1+ in strain SWY2558 was confirmed by PCR. For generating ipk1Δ haploid strains SWY2559 and SWY2560, SWY2558 was sporulated, and tetrads were dissected. Strain SWY2559 (h− ipk1::ura4+ leu1-32 ura4-D18 ade6-M210) was used in this study.
For the generation of the ipk1Δ asp1Δ strain, SWY2559 was crossed with KGY956 (h+ asp1::ura4+ leu1-32 ura4-D18 ade6-M210) and sporulated, and double mutants were identified by tetrad analysis and confirmed by PCR.
Gene cloning and deletion constructs.
To clone ipk1+, specific cDNA was amplified by PCR with oligonucleotide primers spipk1-NdeI-F and spipk1-SmaI-R from a Superscript-II-RT- and oligo(dT) primer (Invitrogen)-derived total cDNA preparation from S. pombe wild-type cells. The PCR product was cloned into the EcoRV site of pBluescript SK (Stratagene), resulting in pSW3019. The NdeI-SmaI fragment of pSW3019 was cloned into vectors pREP1 and pREP81GFP (5, 11, 36), placing ipk1+ under the control of the nmt promoters, giving pSW3021 and pSW3360, respectively. For generating the ipk1+ genomic DNA (gDNA) clone, S. pombe gDNA was PCR amplified using primers spipk1-PstI-F and spipk1-BamHI-R and cloned in pREP1, replacing the nmt1 promoter with the ipk1+ ORF and the ∼1-kb ipk1+ sequence upstream of the ATG start codon to give pSW2023. For cloning of the ipk1 C terminus (ipk1 C-term) under the nmt1 promoter, a specific ipk1+ sequence was PCR amplified from pSW3019 using primers spipk1-NdeI-F1 and spipk1-SmaI-R, cut with NdeI and SmaI, and cloned into pREP1, resulting in pSW3357. ipk1 N-terminal (N-term), ipk1 ΔRVS (RVS homology), and ipk1 ΔCC (coiled coil) clones were derived from pSW3021 using an inverse-PCR-based strategy using Pfu DNA polymerase (Stratagene) and primer pairs spipk1-F2 and spipk1-R2, spipk1-F763 and spipk1-R321, and spipk1-F571 and spipk1-R480, respectively. The PCR product was digested with DpnI (New England Biolabs) to remove template DNA, resolved in agarose gel, purified, and 5′-end phosphorylated. The DNA was then self-ligated and transformed into Escherichia coli DH5α competent cells (Invitrogen). For cloning of S. cerevisiae IPK1 (ScIPK1) under the control of the nmt1 promoter, the coding sequence was amplified from pSW1273 (37) using primers sc-IPK1-NdeI-F and sc-IPK1-R and cloned into the NdeI-SmaI sites of pREP1, giving pSW3233. Similarly, zebrafish ipk1 (Zfipk1)was amplified from pSW3007 (50) using primers zf-ipk1-NdeI-F and zf-ipk1-3′R1 and cloned into the NdeI-SmaI sites of pREP1, resulting in pSW3232. For cloning of S. pombe dbp5+ under the control of the nmt1 promoter, the dbp5+coding sequence was amplified from a total cDNA preparation using primers spdbp5-NdeI-F and spdbp5-BamHI-R and cloned into the NdeI-SmaI sites of pREP1, giving pSW3363. All the clones that involved PCR-based cloning strategies were confirmed by sequencing. Sequences of oligonucleotide primers used in this study are presented in Table S1 in the supplemental material.
Analysis of levels of cellular [3H]inositol-labeled IP.
The soluble-IP profiles of S. pombe cells were determined according to a protocol previously described for S. cerevisiae cells (71). Briefly, S. pombe cells were grown in EMM containing 20 μCi/ml [3H]inositol (Perkin-Elmer) to mid-logarithmic phase. Cells (1 ml) were harvested, washed in H2O, and resuspended in 100 μl of 0.5 N HCl. Soluble IPs were extracted by adding 372 μl of chloroform-methanol (1:2, vol/vol) and 100 μl of glass beads. The mixture was vortexed at maximum speed for 2 min, followed by the addition of 125 μl of chloroform and 125 μl of 2 M potassium chloride and another 2 min of vortexing. The lysates were clarified by a spin at 13,000 × g for 5 min, and the supernatant was recovered. Samples were analyzed by high-performance liquid chromatography (HPLC), with the IPs resolved by use of a Whatman Partisphere strong-anion-exchange column (4.6 by 125 mm) and a linear gradient from 10 mM to 1.7 M ammonium phosphate (pH 3.5) over 25 min, followed by elution with 1.7 M ammonium phosphate for 20 min.
Transmission electron microscopy.
S. pombe cells were grown in YE medium to early log phase, fixed in 4% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), postfixed in 1% OsO4 in cacodylate buffer, dehydrated through an ethanol series, equilibrated in propylene oxide, and then embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and examined using a Hitachi H-800 electron microscope.
In situ hybridization and indirect immunofluorescence.
S. pombe cells were grown in YE medium or EMM to early log phase at 30°C. Additionally, the EMM-grown cells were shifted to cell growth at 36°C for 90 min. Cells were fixed for 10 min and processed as previously described (22, 68). Cells were incubated overnight with a digoxigenin-dUTP-labeled oligo(dT) probe and were detected with fluorescein-labeled antidigoxigenin Fabs (1:25; Boehringer). DNA was stained with 0.1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole), and samples were mounted for imaging in 90% glycerol and 1 mg/ml p-phenylenediamine (pH 8.0; Sigma-Aldrich). Images were acquired using a microscope (BX50; Olympus) with a UPlanF1 100× 1.30-numerical-aperture oil immersion objective (Olympus) and a camera (CoolSNAP HQ; Photometrics). Within each experiment, all images were collected and scaled identically using MetaVue, version 4.6 (Molecular Devices), or Image-Pro Express (Media Cybernetics) and processed with Photoshop 9.0 software (Adobe).
Cytology and microscopy.
Septa were visualized by staining ethanol-fixed cells with 1 mg/ml methyl blue solution (Sigma-Aldrich). To visualize DNA, cells were stained with 0.1 μg/ml DAPI. Strains expressing green fluorescent protein-tagged proteins were grown in liquid YE medium and visualized and photographed live. All images were acquired and processed as described above.
F-actin and FM4-64 staining.
For F-actin staining, cells were grown to early log phase, fixed by adding formaldehyde to a final concentration of 3.7% for 10 min, and suspended in 0.1 M potassium phosphate buffer (pH 6.5). Cells were fixed in 3.7% formaldehyde again for 45 min, washed twice in phosphate-buffered saline, and stained with rhodamine-phalloidin (Molecular Probes) on ice for 30 min.
FM4-64 staining was performed as described previously by Feoktistova et al. (14). Briefly, cells were grown in YE medium at 30°C to mid-log phase, concentrated 100-fold by centrifugation, and suspended in fresh YE medium. FM4-64 (Molecular Probes) was added to a final concentration of 16 μM, and cells were incubated on ice for 15 min. Cells were then washed and suspended in YE medium and incubated at 30°C. Aliquots of cells were collected at 15-min intervals, mounted onto slides, and visualized immediately. Images were collected using Image-Pro Express (Media Cybernetics) and processed with Photoshop 9.0 software (Adobe).
RESULTS
ipk1Δ S. pombe cells are defective in nuclear mRNA export, cell morphology, polarized growth, and cell separation.
We cloned full-length S. pombe ipk1+ based on the previously published sequence information (25); additionally, we generated an ipk1Δ strain. The ipk1Δ cells were viable at 30°C, temperature sensitive at 36°C, and cold sensitive at 18°C (Fig. 1B and see Fig. 7A and 8A). To test the metabolic effect of the ipk1+ deletion on IP production, we compared the levels of soluble IPs in extracts isolated from ipk1Δ cells to those of wild-type cells. Cells were labeled to steady state with [3H]inositol, and extracted IPs were separated and analyzed by HPLC. In wild-type cells, IP6 was most abundant, with markedly lower levels of IP5 and minimal detection of other IPs (see Fig. 7C). In contrast, IP6 was absent in extracts from ipk1Δ cells, and IP3, IP4, IP5, and PP-IP4 levels were distinctly elevated (see Fig. 7C). Others have shown that increased upstream IP levels are an established indicator of inhibited IP6 production in S. cerevisiae, Drosophila melanogaster S2 cells, and zebrafish embryos (25, 50, 53, 71). Thus, our results indicate that the deletion of ipk1+ results in a loss of IP6 production and a general perturbation of the IP synthesis pathway.
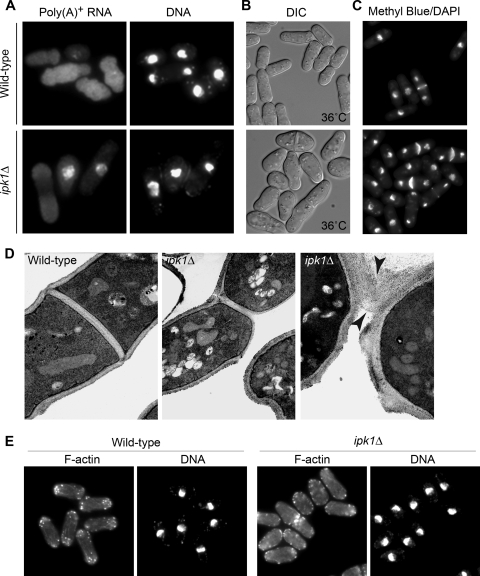
FIG. 1.
ipk1+ is required for nuclear mRNA export, polarized growth, and morphology. (A) mRNA export is inhibited in ipk1Δ cells. In situ hybridization with a digoxigenin-coupled oligo(dT) probe was conducted in wild-type and ipk1Δ cells grown in YE medium at 30°C. Poly(A)+ RNA localization was visualized by indirect immunofluorescence microscopy with a fluorescein isothiocyanate-coupled anti-digoxigenin antibody (left), and nuclei were visualized with coincident DAPI staining (right). (B) ipk1Δ cells developed a swollen, rounded cell shape at 36°C. Presented are the differential interference contrast (DIC) images of wild-type and ipk1Δ cells grown in YE medium at 36°C for 6 h. (C) ipk1Δ cells are defective in cell separation. Shown are wild-type and ipk1Δ cells grown in YE medium at 30°C, ethanol fixed, and then stained with methyl blue and DAPI to reveal division septum and DNA, respectively. (D) ipk1Δ cells formed normal septa but exhibited defects in cell wall digestion and cleavage. Wild-type and ipk1Δ cells were grown at 30°C, fixed, and processed for transmission electron microscopic analysis as described in Materials and Methods. Presented are representative images of wild-type cells with a division septum (left) and ipk1Δ cells in the early (middle) and late (right) stages of septum digestion. Black arrowheads (right) point to the undigested cell wall materials. (E) ipk1Δ cells are defective in polarized growth. After growth at 30°C, wild-type (left) and ipk1Δ (right) cells were stained with rhodamine-phalloidin and DAPI for actin cytoskeleton organization and DNA, respectively.
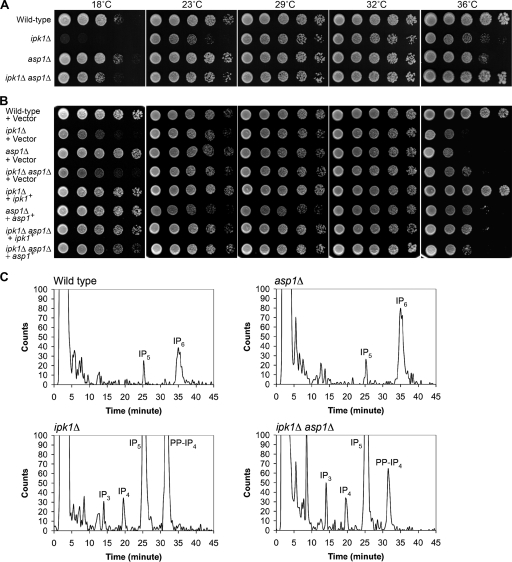
FIG. 7.
ipk1Δ cells exhibit both distinct and shared growth phenotypes compared with those of asp1Δ cells. (A) ipk1Δ cells have both cold-sensitive and temperature-sensitive growth defects. Wild-type, ipk1Δ, asp1Δ, and ipk1Δ asp1Δ cells were spotted onto YE medium in fivefold serial dilutions and grown at the temperatures shown. (B) ipk1+ overexpression rescues the growth defects of ipk1Δ cells. Wild-type, ipk1Δ, asp1Δ, and ipk1Δ asp1Δ cells containing the indicated plasmids were grown in EMM lacking Leu and with 10 μM thiamine (to repress expression) and spotted onto EMM lacking Leu. (C) Deletion of the ipk1+ and asp1+ genes perturbs soluble IP levels. The strains in A were grown in minimal medium containing [3H]inositol. Soluble IPs were extracted and separated by Partisphere strong-anion-exchange HPLC. Presented are the IP profiles obtained from the same total cell number from each strain. Labels indicate IP elution positions.
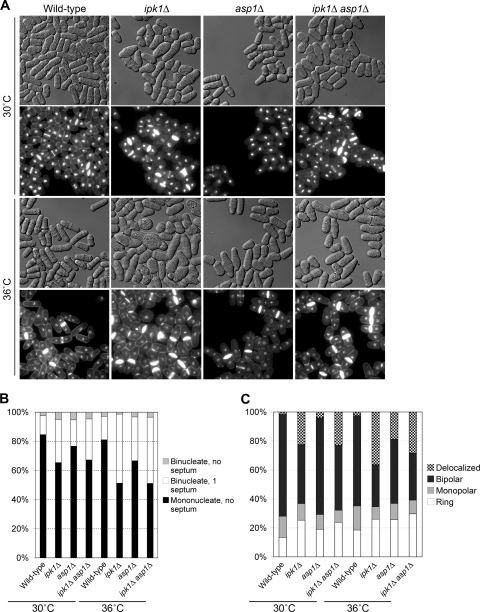
FIG. 8.
Cells without ipk1+ and/or asp1+ function(s) are defective in cell separation and polarized growth. Wild-type, ipk1Δ, asp1Δ, and ipk1Δ asp1Δ cells were grown in YE medium at 30°C, and aliquots were shifted to 36°C for 6 h. (A) To analyze cell separation, cells were fixed in ethanol and costained with methyl blue and DAPI. Representative images (rows 2 and 4) along with their respective DIC images (rows 1 and 3) are presented. (B) Quantification of the number of septa and nuclei in cells from A (n > 300 cells). (C) Bar graph showing F-actin distribution in cells from A (n > 300 cells). Ring, actin localized to medial septum; monopolar, actin only at one end; bipolar, actin at both ends; delocalized, actin patches throughout the cell.
We also examined if the expression of ScIPK1 or Zfipk1 could restore IP6 production in ipk1Δ cells. Both ScIPK1 and Zfipk1 expressions rescued IP6 production in ipk1Δ cells (see Fig. S3 in the supplemental material), suggesting enzymatic complementation across species by the IP5 2-kinase enzymes. Next, we analyzed the ipk1Δ cells for mRNA export defects using oligo(dT) in situ hybridization for the subcellular distribution of poly(A)+ RNA. The ipk1Δ cells showed an accumulation of poly(A)+ RNA in the nucleus at a growth temperature of 30°C (Fig. 1A). Thus, we conclude that IP6 production is required for efficient mRNA export in S. pombe.
Although ipk1+ is not essential, ipk1Δ cells exhibited morphological and cell separation defects. The ipk1Δ cells were rounder than wild-type cells, and the cell morphology defects were exacerbated by growth at an elevated temperature (36°C) (Fig. 1B and see Fig. 8A). We also observed an increased number of binucleate-septated cells in a nonsynchronous exponentially growing cell population (Fig. 1C and see Fig. 8A and B). To further assess the cell separation defect in ipk1Δ cells, the morphology of the septum region was examined by thin-section transmission electron microscopy. The formation and organization of septa in wild-type and ipk1Δ cells were normal (Fig. 1D), with the three-layer septum structure (a clear primary septum surrounded by two darker secondary septa) apparent in both cell types. However, there were distinctions in the apparent degradations of the primary septum. Compared to wild-type cells, where the primary septum is degraded centripetally from the cortex to the septum midpoint, in ipk1Δ cells, degradation of the primary septum material appeared asymmetric, with the daughter cells remaining attached by remnants of the cell wall at one end of the division plane (Fig. 1D). This finding indicates that ipk1Δ cells are defective in the dissolution of both the septum and the cell wall that surrounds the septum. We speculate that ipk1Δ cells fail to disassemble the division septa, leading to an accumulation of binucleate cells with a medial division septum. Such cell shape and separation defects have not been reported for ipk1Δ S. cerevisiae cells, suggesting that roles for Ipk1 in these cellular processes are specific to S. pombe.
Defects in morphology and cell separation in ipk1Δ cells might reflect a perturbation of the polarized growth. We first examined the organization of the F-actin cytoskeleton by staining exponentially growing cells with rhodamine-conjugated phalloidin. In 70% of wild-type cells at 30°C, actin was organized in cortical patches at both cell ends and in thin cables running along the long axis of the cell (Fig. 1E and see Fig. 8C). In contrast, only 41% of ipk1Δ cells at 30°C showed bipolar cortical patches, whereas 22% displayed disorganized actin structures and actin patches delocalized throughout the cell body (Fig. 1E and see Fig. 8C). After shifting to 36°C for 6 h, ipk1Δ cells were swollen and round, with 36% of the cells exhibiting delocalized cortical actin patches (see Fig. 8C). At 36°C, only 2% of the wild-type cells showed an altered actin distribution. This delocalization of F-actin patches in the ipk1Δ cells was at the expense of its normal bipolar and monopolar distributions (see Fig. 8C). Compared to the wild type, we also observed an increase in the medial ring localization of F-actin in ipk1Δ cells at both growth temperatures (see Fig. 8C). These results indicate that ipk1+ plays a critical role in the organization of cortical actin patches, whereas it is dispensable for the formation of medial actin rings.
The SpIpk1 C-terminal domain is sufficient to rescue mRNA export, cell separation, and polarized growth defects in ipk1Δ S. pombe cells.
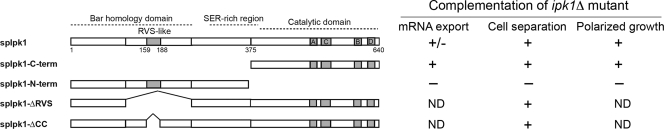
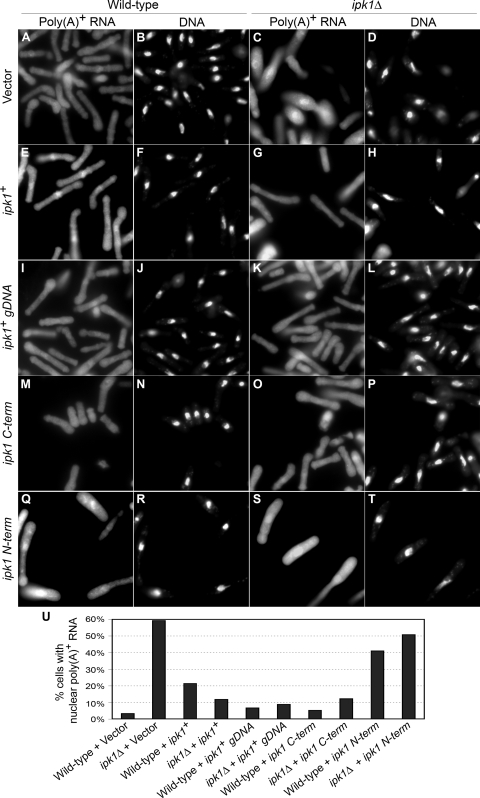
We previously reported that the purified recombinant C-terminal domain of SpIpk1 has substrate selectivity and catalytic efficiency similar to those of ScIpk1 despite sharing only 24% sequence identity (25). The most striking difference between the ScIpk1 and SpIpk1 proteins is the unique N-terminal region of the S. pombe protein (see Fig. S1 in the supplemental material). To identify the relevant protein activity responsible for the defects exhibited by ipk1Δ S. pombe cells, we constructed a series of plasmids expressing ipk1+ deletion mutants under the control of the nmt1 promoter (Fig. 2). As complementation controls, plasmids harboring either the full-length ipk1+ under the control of the nmt1 promoter or a genomic fragment of ipk1+ with the endogenous promoter (ipk1+ gDNA) were used. The mRNA export, cell separation, and polarized growth phenotypes were assayed after induction for overexpression by the nmt1 promoter. The results from these analyses are summarized in Fig. 2. The ipk1Δ cells exhibited nuclear poly(A)+ RNA accumulation in 59% of the cells (Fig. 3C, D, and U). The overexpression of ipk1+, ipk1+ gDNA, and ipk1 C-term suppressed the mRNA export defect of ipk1Δ cells (Fig. 2 and 3) and restored IP6 production (see Fig. S2 in the supplemental material). However, the level of rescue of the mRNA export defect by full-length ipk1+ was partial. Of note, in wild-type cells, the overexpression of ipk1+ resulted in a weak mRNA export defect, with nuclear poly(A)+ accumulation in 21% of the cells (Fig. 3E, F, and U). Most clearly, the overexpression of ipk1 N-term did not rescue the ipk1Δ mRNA export defect (Fig. 3S, T, and U), and IP6 production was not restored (see Fig. S2 in the supplemental material). In addition, 41% of wild-type cells overexpressing ipk1 N-term showed nuclear poly(A)+ accumulation (Fig. 3Q, R, and U). Together, we conclude that the loss of the Ipk1 catalytic domain is the critical defect linked to the mRNA export perturbation in ipk1Δ cells. For wild-type cells, there are also dominant negative effects on mRNA export from overexpressing either ipk1+ or ipk1 N-term.
FIG. 2.
Ipk1 kinase activity is critical for mRNA export, cell separation, and polarized growth. Shown are schematic diagrams of the ipk1+ ORF and deletion constructs used in this study. Putative domains and conserved motifs (see Fig. S1 in the supplemental material) are marked in the ORF schematic. Presented is a summary of the results obtained from the overexpression of the constructs in wild-type and ipk1Δ cells shown in Fig. 3 [subcellular distribution of poly(A)+ RNA], Fig. 4 (cell separation), and Fig. 5 (polarized growth). ND, not determined.
FIG. 3.
Overexpression of ipk1 C-term is sufficient to restore mRNA export in ipk1Δ cells, whereas ipk1 N-term has an inhibitory effect in both wild-type and ipk1Δ cells. Wild-type (A, B, E, F, I, J, M, N, Q, and R) and ipk1Δ (C, D, G, H, K, L, O, P, S, and T) cells were transformed with the empty vector (A to D) or the plasmid constructs carrying ipk1+ (E to H), ipk1 gDNA with ipk1+ under the transcriptional control of its own promoter (I to L), ipk1 C-term (M to P), and ipk1 N-term (Q to T); grown in EMM at 30°C for 18 h; and shifted to cell growth at 36°C for 90 min. The subcellular distribution of poly(A)+ RNA was visualized by in situ hybridization with oligo(dT) (columns 1 and 3). DNA was visualized by subsequent DAPI staining (columns 2 and 4). (U) Bar graph quantifying nuclear poly(A)+ RNA distribution in wild-type and ipk1Δ cells harboring different plasmids (n > 200 cells).
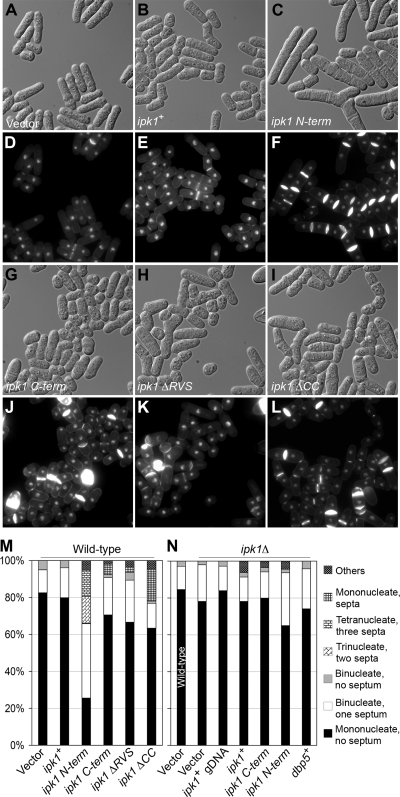
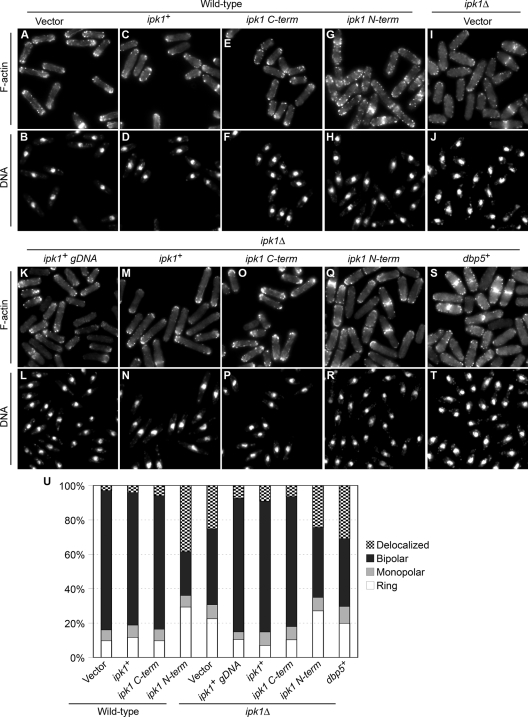
We also examined the effects of overexpressing the ipk1+ deletions on cell separation and polarized growth in both wild-type and ipk1Δ cells. The overexpression of the ipk1+, ipk1 C-term, ipk1 ΔRVS (Rvs homology), or ipk1 ΔCC (coiled-coil) constructs in wild-type cells did not significantly alter the ratios between binucleate-septated and mononucleate-unseptated cells (Fig. 4A to M). Similarly, wild-type vector-only cells and those overexpressing ipk1+ or ipk1 C-term did not display any significant difference in F-actin distribution (Fig. 5A to F and U). Strikingly, the overexpression of ipk1+ gDNA, ipk1+, or ipk1 C-term suppressed both cell separation and polarized growth defects exhibited by ipk1Δ cells (Fig. 4N and 5I to P and U). There were differences in the cell separation defect levels between ipk1Δ cells grown in minimal medium and those grown in rich medium (see Fig. S4 in the supplemental material). The ipk1Δ cells grown in rich medium had a significant defect, with 30% and 47% of the cells being binucleate-septated at 30°C and 36°C, respectively. However, the ipk1Δ cells grown in minimal medium had a more modest defect, with 16% and 19% of the cells being binucleate-septated at 30°C and 36°C, respectively. Overall, we conclude that the SpIpk1 C-terminal domain, and potentially IP6 production as well as the proper maintenance of other IP levels, is required for correct cell separation and polarized growth.
FIG. 4.
Overexpression of ipk1 C-term and ipk1 N-term differentially impacts cell separation. (A to L) Wild-type cells were transformed with the empty vector (pREP1) (A and D) and plasmids expressing ipk1+ (B and E), ipk1 N-term (C and F), ipk1 C-term (G and J), ipk1 ΔRVS (H and K), and ipk1 ΔCC (I and L). Cells harboring different plasmid constructs were grown at 30°C for 24 h in EMM lacking thiamine, fixed in ethanol, and costained with methyl blue and DAPI to visualize septa and DNA (D, E, F, J, K, and L; the respective DIC images are presented in A, B, C, G, H, and I, respectively). (M) Quantification of the number of septa vis-à-vis nuclei in cells from D, E, F, J, K, and L (n > 300 cells). (N) ipk1Δ cells were transformed with the empty vector, and plasmids expressing ipk1+ from its own promoter (ipk1+ gDNA), ipk1+, ipk1 C-term, ipk1 N-term, and dbp5+ and effects of gene overexpression were analyzed as described above. Presented is a bar graph quantifying the number of septa and nuclei in ipk1Δ cells harboring different plasmids (n > 300 cells); wild-type cells transformed with empty vector served as a control. Others represent percent multinucleate (at least three nuclei), multiseptum (at least one septum) cells (ipk1 N-term overexpression) and multinucleate (at least one nucleus), multiseptum (more than one septum) cells (ipk1+, ipk1 C-term, ipk1 ΔRVS, or ipk1 ΔCC overexpression).
FIG. 5.
Overexpression of ipk1 C-term and ipk1 N-term have opposite effects on polarized growth. Wild-type (A to H) and ipk1Δ (I to R) cells were transformed with the empty vector (A, B, I, and J) or plasmids carrying ipk1+ (C, D, M, and N), ipk1 gDNA with ipk1+ under the control of its own promoter (K and L), ipk1 C-term (E, F, O, and P), and ipk1 N-term (G, H, Q, and R). (S and T) ipk1Δ cells were also transformed with plasmid pREP1 harboring dbp5+. Cells harboring different plasmids were grown in EMM lacking thiamine at 30°C for 18 h and stained with rhodamine-phalloidin (rows 1 and 3) and DAPI (rows 2 and 4) to visualize F-actin distribution and DNA, respectively. (U) Bar graph quantifying F-actin distribution in wild-type and ipk1Δ cells harboring different plasmids (n > 300 cells). Ring, actin localized to medial septum; monopolar, actin only at one end; bipolar, actin at both ends; delocalized, actin patches throughout the cell.
Next, we analyzed the effects of the Ipk1 N-terminal domain. A high percentage of wild-type cells overexpressing ipk1 N-term were multinucleated and multiseptated (Fig. 4C, F, and M). In contrast, the ipk1Δ cells overexpressing ipk1 N-term exhibited an increase in the number of binucleate-septated cells only (Fig. 4N). Additionally, we observed that ipk1 N-term overexpression perturbed polarized growth. Thirty-nine percent of wild-type cells overexpressing ipk1 N-term had delocalized cortical actin patches, whereas only 3% of wild-type vector-only cells showed an altered F-actin distribution (Fig. 5A, B, G, and H). Together, these data suggest that ipk1 N-term overexpression specifically perturbs septum cleavage, cell separation, and polarized growth.
Specific rescue of the mRNA export defect in ipk1Δ cells by overexpressing dbp5+.
Given that the SpIpk1 C-terminal domain is sufficient to rescue both the mRNA export and cell separation defects in ipk1Δ cells, we speculated that the cell separation defects might be an indirect effect due to a lack of an IP6-dependent export of an mRNA(s) encoding proteins involved in cell separation. In S. cerevisiae, the IP6 target in the mRNA export pathway has been pinpointed to Gle1, a cofactor for Dbp5, and DBP5 overexpression specifically suppresses the mRNA export defect of an S. cerevisiae ipk1Δ nup42Δ mutant (1). Thus, we tested whether S. pombe dbp5+ overexpression suppresses the mRNA export defect of ipk1Δ S. pombe cells. Strikingly, the overexpression of dbp5+ fully rescued the mRNA export defect in ipk1Δ cells, whereas its overexpression in wild-type cells had no effect (Fig. 6). This suggests that the mRNA export defect in the ipk1Δ S. pombe cells is specifically linked to SpDbp5 function, similar to what is known for S. cerevisiae (1). In sharp contrast, the cell separation and polarized growth defects in ipk1Δ cells were not rescued by dbp5+ overexpression (Fig. 4N and 5S, T, and U). We conclude that the role of SpIpk1/IP6 in cell separation and polarized growth is independent of its role in mRNA export.
FIG. 6.
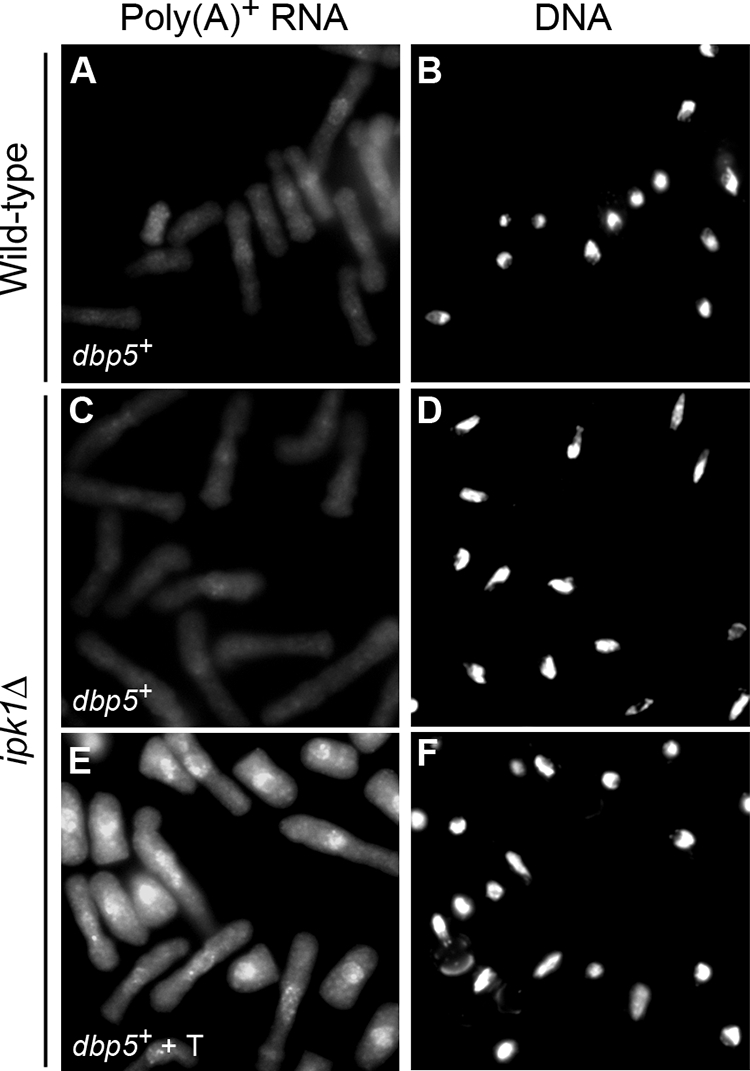
Overexpression of dbp5+ suppresses the mRNA export defect in ipk1Δ cells. Wild-type and ipk1Δ cells were transformed with plasmid pREP1 harboring dbp5+. Cells were grown in minimal medium (EMM lacking Leu) (A to D) or in EMM lacking Leu and with 10 μM thiamine (T) (to repress dbp5+ expression) (E and F) at 30°C for 18 h and shifted to 36°C for 90 min, and in situ hybridization with an oligo(dT) probe was conducted. The signal for the oligo(dT) probe indicates the subcellular distribution of poly(A)+ RNA (A, C, and E), in comparison to the nuclear signal revealed by coincident DAPI staining (B, D, and F).
Elevated levels of PP-IP4 production in ipk1Δ cells are linked to Asp1 function.
A loss of the IP6 kinase activity in asp1Δ cells results in cell morphology, cell separation, and polarized growth defects that are similar to those observed here in ipk1Δ cells (14, 41). Thus, we reasoned that these ipk1Δ phenotypes might be due to the indirect effect of a loss of IP7. To test this, we directly compared the ipk1Δ mutant, the asp1Δ mutant, and an ipk1Δ asp1Δ double mutant for growth in rich medium. As shown in Fig. 7A, wild-type and ipk1Δ asp1Δ cells showed similar levels of growth at 23°C, 29°C, 32°C, and 36°C. The growth of ipk1Δ asp1Δ cells was slightly compromised at 18°C. In comparison, the asp1Δ cells displayed modest temperature sensitivity at 36°C. The ipk1Δ cells showed a similar level of temperature sensitivity at 36°C; however, it was severely cold sensitive at 18°C (Fig. 7A). The expression of wild-type ipk1+ or asp1+ in the respective mutants resulted in a partial to complete rescue of the temperature- and/or cold-sensitive growth defects (Fig. 7B). However, there were relative differences in the levels of growth defects between rich and minimal media, potentially reflecting differential effects of culture media on growth. Overall, the ipk1Δ cells had the most severe growth perturbations. This is not unanticipated, because ipk1Δ cells fail to produce both IP6 and IP7 isomers. However, the difference in the growth characteristics between the ipk1Δ asp1Δ double mutant and the ipk1Δ single mutant is surprising.
To directly examine the effects on the IP metabolic pathways, we compared the IP profiles of the mutant strains. Following steady-state radiolabeling with [3H]inositol, lysates from equivalent total cell numbers were prepared, and total soluble IPs were resolved by HPLC. As shown in Fig. 7C, IP5 and IP6 peaks were detected in wild-type cells. The ipk1Δ cells had elevated levels of all upstream IPs (e.g., IP3, IP4, IP5, and PP-IP4) and did not have the IP6 peak. In contrast, only the level of IP6 was elevated in the asp1Δ cells compared to those of the wild type. Interestingly, although the ipk1Δ asp1Δ cells had elevated levels of IP3, IP4, and IP5, the relative level of the ratio of PP-IP4 to IP5 was significantly lower than that in ipk1Δ samples. In ipk1Δ asp1Δ cells, the PP-IP4-to-IP5 ratio was ∼0.09, whereas in ipk1Δ cells, it was ∼0.43. This indicates that Asp1 has an IP5 kinase activity that contributes to PP-IP4 synthesis in the ipk1Δ cells. Others reported previously that ScVip1, the Asp1 orthologue, can produce PP-IP4 in vitro (41). As some PP-IP4 is still present in the ipk1Δ asp1Δ strain, there must be an additional kinase(s) responsible for this synthesis. Taken together, the elevated PP-IP4 level in the ipk1Δ strain might be responsible for the mutant's more severe cold-sensitive growth defect, with the more modest level of PP-IP4 accumulation in the ipk1Δ asp1Δ mutant having a lesser effect.
Comparison of roles of SpIpk1 and Asp1 in mRNA export, cell morphology, cell separation, polarized growth, and endocytosis.
To further dissect how the cellular defects were linked to specific perturbations in IP production for the ipk1Δ, asp1Δ, and ipk1Δ asp1Δ mutants, we compared the relative defects in mRNA export, polarized growth, and cell separation. In situ hybridization for poly(A)+ RNA showed that the asp1Δ cells did not accumulate poly(A)+ RNA in the nucleus (see Fig. S5 in the supplemental material). In contrast, the ipk1Δ asp1Δ cells accumulated poly(A)+ RNA in the nucleus at a level similar to that for the ipk1Δ cells (see Fig. S5 in the supplemental material). Because the common IP perturbation between the ipk1Δ and ipk1Δ asp1Δ mutants (Fig. 7C) is the loss of IP6, we conclude that proper IP6 production is the most critical effector of mRNA export in S. pombe.
Interestingly, the ipk1Δ and ipk1Δ asp1Δ mutants had comparable perturbations in the levels of binucleate-septated cells at both 30°C and 36°C (Fig. 8A and B). The cell separation defect in each mutant was exacerbated to a similar extent at 36°C, with ∼50% of the cells being binucleate-septated (compared to less than 20% in the wild-type cell population). Again, the asp1Δ mutant exhibited a more modest cell separation defect, with ∼30% of the cells being binucleate-septated at 36°C (Fig. 8A and B). We also observed that both the ipk1Δ and ipk1Δ asp1Δ cells developed a rounded shape, which was more pronounced at 36°C (Fig. 8A). The asp1Δ mutant had only a subtle cell shape defect (Fig. 8A).
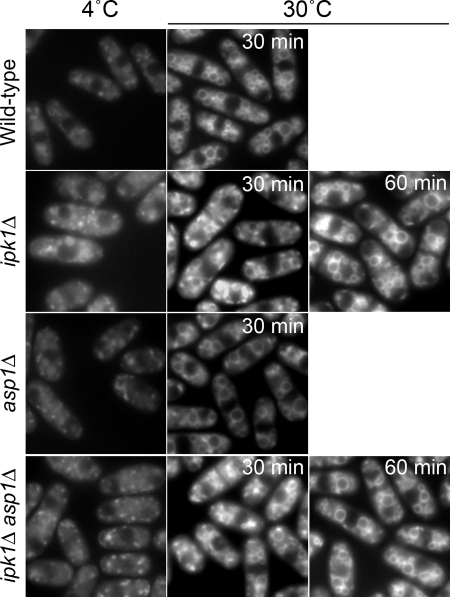
The ipk1Δ and ipk1Δ asp1Δ mutants also had comparable perturbations in polarized growth at both 30°C and 36°C (Fig. 8C). In contrast, asp1Δ cells exhibited a modest polarized growth defect, with only 19% of the cells displaying delocalized F-actin patches (Fig. 8C). Because defects in the actin cytoskeleton result in perturbations of endocytosis (17, 39), we measured endocytosis with a qualitative assay. The amphiphilic fluorescent dye FM4-64 enters cells through endocytosis and is transported to the vacuolar membrane (17, 65). As shown in Fig. 9, at 4°C in wild-type, ipk1Δ, asp1Δ, and ipk1Δ asp1Δ cells, FM4-64 localized in a speckled pattern, indicating that dye uptake was normal. Within 30 min of incubation at 30°C, vacuolar membranes were intensely stained in wild-type and asp1Δ cells (Fig. 9). In contrast, a similar level of staining in the ipk1Δ and ipk1Δ asp1Δ cells was not observed until after 1 h (Fig. 9). These results suggest that endocytosis is more strongly inhibited in the ipk1Δ and ipk1Δ asp1Δ cells than in the asp1Δ cells. We conclude that the more severe defects in cell separation, cell morphology, polarized growth, and endocytosis found in the ipk1Δ mutant are due to the loss of both IP6 and IP7.
FIG. 9.
Endocytosis is inhibited in ipk1Δ cells. Wild-type, ipk1Δ, asp1Δ, and ipk1Δ asp1Δ cells were treated with the fluorescent dye FM4-64 on ice for 15 min (left), washed and suspended in YE medium, and incubated at 30°C. Images of FM4-64 distribution within cells were digitally acquired at 15-min intervals, and the time required for the vacuolar membranes to become fully fluorescent was noted for each strain (right).
DISCUSSION
Here, we report the first analysis of cellular functions for ipk1+ in S. pombe. This work directly complements and extends prior analyses of S. cerevisiae in several important ways. We find that SpIpk1 is required for mRNA export and is genetically linked to SpDbp5 function. More strikingly, the ipk1Δ mutants have pleiotropic defects in cell morphology, polarized growth, endocytosis, and cell separation. These defects are potentially due to the loss of production of both IP6 and IP7. In addition, increased PP-IP4 levels from Asp1 kinase activity are correlated with cold-sensitive ipk1Δ cell growth. However, the noncatalytic unique N-terminal SpIpk1 domain is also required. Taken together, the phenotypes of the ipk1Δ and asp1Δ mutants delineate multiple roles for SpIpk1 function and IP6 production and highlight the cellular consequences of perturbing IP flux.
This work provides direct evidence implicating IP6 production as being required for mRNA export in an organism other than S. cerevisiae. Our data suggest that the steps and factors mediating the highly intricate export process are conserved between S. cerevisiae and S. pombe and potentially across all eukaryotes. Based on the complementation of ipk1Δ S. pombe cells by dbp5+ overexpression (Fig. 6), we predict that SpGle1 will be an S. pombe IP6 target for the activation of SpDbp5, similar to the mechanism in S. cerevisiae (1, 67). Of note, as with Kcs1 in S. cerevisiae (46), mRNA export in S. pombe is not dependent on asp1+ function or, presumably, IP7 production. It is intriguing that the overexpression of ipk1+ results in a modest mRNA export defect in wild-type cells. Thus, the SpIpk1 protein might compete for an essential mRNA export factor(s). These findings are similar to the reported effects of an overproduction of SpMex67 (69). We predict that the noncatalytic, unique N-terminal domain of SpIpk1 is mediating specialized cellular functions (see below).
Previous studies of S. pombe have revealed roles for asp1+ function in endocytosis and the actin cytoskeleton (14). As SpIpk1 activity is upstream of Asp1 and inherently required for all IP7 production, it is not unexpected that ipk1Δ cells show similar defects. IP7 might regulate Arp2/3 complexes that participate in actin cytoskeleton and cellular morphology (14, 41). The fusion of the exocyst complex to the plasma membranes for the release of secretory vesicles requires Arp2/3 complex-mediated actin assembly (6). Thus, a functional actin cytoskeleton is critical for polarized membrane growth, protein secretion, and endocytosis (15, 17, 39), and the endocytic and morphological defects in the ipk1Δ and asp1Δ cells are potentially indirect effects of perturbations in the actin cytoskeleton.
A direct role for IP6 and/or IP7 in vesicular trafficking is also possible. It is known that IP6 and IP7 both can modulate vesicular trafficking in fungi and mammalian cells (12, 20, 21, 49), and IP6 promotes dynamin-mediated endocytosis in pancreatic β cells (20). Additionally, a recent report suggested that IP7 is required for full exocytic capacity in insulin-secreting pancreatic β cells (21). Interestingly, in ipk1Δ cells, the septum assembles normally, but the septum and its surrounding cell wall are not cleaved completely, resulting in an accumulation of septated cells. The overexpression of dbp5+ does not suppress the cell separation defect in ipk1Δ cells (Fig. 4N), suggesting that the phenotype is not related to mRNA export or SpDbp5 function. ipk1Δ cells might be defective in the trafficking and secretion of hydrolytic enzymes or their release at the medial region.
The ipk1Δ mutant phenotypes presumably result from the combined effects of the loss of IP6 and IP7 production and the accumulation of upstream IPs. As such, IP6 and IP7 could have nonoverlapping independent functions that mediate distinct events during cell separation. For example, the membrane fusion step of the vesicles might be perturbed in the absence of IP6, whereas the Arp2/3 complex mediating actin organization is defective in the absence of IP7. Consistent with this hypothesis, the ipk1Δ mutant phenotypes are consistently more severe than the asp1Δ mutant phenotypes. Alternatively, the more severe ipk1Δ phenotypes could reflect a role for an additional IP6 kinase that partially compensates for the absence of Asp1 and allows some IP7 production in the asp1Δ mutant. In S. cerevisiae, Kcs1 also acts as an IP6 kinase (47). A Kcs1 orthologue in S. pombe has not been fully characterized. With regard to the IP synthesis pathway, it is intriguing that ipk1Δ mutants are more severely cold sensitive than the ipk1Δ asp1Δ double mutant. Our results suggest that Asp1-dependent elevated PP-IP4 levels in ipk1Δ cells might be responsible. However, physiological targets and functions for such a PP-IP4 molecule are unknown. Future analysis of the ipk1Δ and asp1Δ mutants might reveal such targets.
Several pieces of evidence implicate a role for the noncatalytic N-terminal domain of SpIpk1 in cell function. The mRNA export, temperature-sensitive growth, and morphological defects exhibited by ipk1Δ cells are rescued by overexpressing the ipk1+ catalytic C-terminal domain but not the N-terminal domain. Moreover, the overexpression of the SpIpk1 N-terminal domain in wild-type cells perturbs mRNA export, cell separation, and polarized growth. A high percentage of the ipk1 N-term-overexpressing wild-type cells are multinucleate-multiseptate (Fig. 4). This ipk1 N-term phenotype is similar to those of mutants with a loss or reduction of glucanases (e.g., ace2, septin genes, and mid2) (7, 34, 57, 60). In these mutants, septa also form normally; however, cells are defective in cell separation, with a chain of cells connected by septa upon subsequent rounds of nuclear division. We speculate that the N-terminal domain serves as a protein-protein-interacting module, and when overexpressed, it sequesters a critical cellular factor that mediates IP synthesis pathway functions. One potential candidate for such interactions could be SpIpk1 itself, with the truncated N-terminal domain alone heterodimerizing to inhibit IP production and possibly block effective substrate exchange among different IP kinases. Additionally, the coiled-coil BAR homology domain might act as a membrane-binding and curvature-sensing module to localize SpIpk1 in distinct cellular microenvironments. Interestingly, SpIpk1 is localized predominantly in the cytoplasm (http://cgl.riken.go.jp) (35; our unpublished data for an ectopically expressed, N-terminal green fluorescent protein-tagged fusion protein). The nuclear envelope, the medial cortex region where actomyosin ring assembles, and the cytosolic membrane components involved in secretory cargo formation represent potential membrane microenvironments for SpIpk1 function. We propose that the unique N-terminal domain of SpIpk1 allows the spatially restricted production of IP6 for roles in mRNA export, endocytosis, polarized cell growth, and cell separation.
In summary, our studies show that alterations in SpIpk1 function lead to defects in mRNA export and polarized growth and morphology and demonstrate that IP6 acts as a critical regulator of these cellular processes. This places proper IP6 production as a key mediator for a general cellular signaling mechanism that regulates vital cellular processes in all eukaryotes.
Supplementary Material
Acknowledgments
We are indebted to Kathy Gould for critical input and guidance throughout the project and to Anna Feoktistova for generous assistance with strain generation. We thank Gary Olson and Virginia Winfrey for assistance and expertise with the electron microscopy experiments; Elizabeth Tran, Laura Terry, Timothy Bolger, and Li-En Jao for comments on the manuscript; and Srinivas Venkatram and members of the Wente and Gould laboratories for discussions.
This work was supported by a grant from the National Institutes of Health (R01 GM51912 to S.R.W.).
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alcazar-Roman, A. R., E. J. Tran, S. Guo, and S. R. Wente. 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8711-716. [DOI] [PubMed] [Google Scholar]
- 2.Alcazar-Roman, A. R., and S. R. Wente. 2008. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma 1171-13. [DOI] [PubMed] [Google Scholar]
- 3.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14943-951. [DOI] [PubMed] [Google Scholar]
- 4.Barker, C. J., J. Wright, P. J. Hughes, C. J. Kirk, and R. H. Michell. 2004. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem. J. 380465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123131-136. [DOI] [PubMed] [Google Scholar]
- 6.Basu, R., and F. Chang. 2007. Shaping the actin cytoskeleton using microtubule tips. Curr. Opin. Cell Biol. 1988-94. [DOI] [PubMed] [Google Scholar]
- 7.Berlin, A., A. Paoletti, and F. Chang. 2003. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J. Cell Biol. 1601083-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolger, T. A., A. W. Folkmann, E. J. Tran, and S. R. Wente. 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134624-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, F., and M. Peter. 2003. Yeasts make their mark. Nat. Cell Biol. 5294-299. [DOI] [PubMed] [Google Scholar]
- 10.Dekker, N., D. Speijer, C. H. Grun, M. van den Berg, A. de Haan, and F. Hochstenbach. 2004. Role of the alpha-glucanase Agn1p in fission-yeast cell separation. Mol. Biol. Cell 153903-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond, D. R., and I. M. Hagan. 1998. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J. Cell Sci. 111(Pt. 7)853-865. [DOI] [PubMed] [Google Scholar]
- 12.Efanov, A. M., S. V. Zaitsev, and P. O. Berggren. 1997. Inositol hexakisphosphate stimulates non-Ca2+-mediated and primes Ca2+-mediated exocytosis of insulin by activation of protein kinase C. Proc. Natl. Acad. Sci. USA 944435-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, Y., S. R. Wente, and P. W. Majerus. 2001. Overexpression of the inositol phosphatase SopB in human 293 cells stimulates cellular chloride influx and inhibits nuclear mRNA export. Proc. Natl. Acad. Sci. USA 98875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feoktistova, A., D. McCollum, R. Ohi, and K. L. Gould. 1999. Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 152895-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finger, F. P., and P. Novick. 1998. Spatial regulation of exocytosis: lessons from yeast. J. Cell Biol. 142609-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frederick, J. P., D. Mattiske, J. A. Wofford, L. C. Megosh, L. Y. Drake, S. T. Chiou, B. L. Hogan, and J. D. York. 2005. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. USA 1028454-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gachet, Y., and J. S. Hyams. 2005. Endocytosis in fission yeast is spatially associated with the actin cytoskeleton during polarised cell growth and cytokinesis. J. Cell Sci. 1184231-4242. [DOI] [PubMed] [Google Scholar]
- 18.Gruter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1649-659. [DOI] [PubMed] [Google Scholar]
- 19.Hodge, C. A., H. V. Colot, P. Stafford, and C. N. Cole. 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 185778-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoy, M., A. M. Efanov, A. M. Bertorello, S. V. Zaitsev, H. L. Olsen, K. Bokvist, B. Leibiger, I. B. Leibiger, J. Zwiller, P. O. Berggren, and J. Gromada. 2002. Inositol hexakisphosphate promotes dynamin I-mediated endocytosis. Proc. Natl. Acad. Sci. USA 996773-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illies, C., J. Gromada, R. Fiume, B. Leibiger, J. Yu, K. Juhl, S. N. Yang, D. K. Barma, J. R. Falck, A. Saiardi, C. J. Barker, and P. O. Berggren. 2007. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science 3181299-1302. [DOI] [PubMed] [Google Scholar]
- 22.Iovine, M. K., J. L. Watkins, and S. R. Wente. 1995. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 1311699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irvine, R. F. 2003. 20 years of Ins(1,4,5)P3, and 40 years before. Nat. Rev. Mol. Cell Biol. 4586-590. [DOI] [PubMed] [Google Scholar]
- 24.Irvine, R. F., and M. J. Schell. 2001. Back in the water: the return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2327-338. [DOI] [PubMed] [Google Scholar]
- 25.Ives, E. B., J. Nichols, S. R. Wente, and J. D. York. 2000. Biochemical and functional characterization of inositol 1,3,4,5,6-pentakisphosphate 2-kinases. J. Biol. Chem. 27536575-36583. [DOI] [PubMed] [Google Scholar]
- 26.Katahira, J., K. Strasser, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 182593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keeney, J. B., and J. D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler, A., and E. Hurt. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8761-773. [DOI] [PubMed] [Google Scholar]
- 29.Lee, Y. S., K. Huang, F. A. Quiocho, and E. K. O'Shea. 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 425-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y. S., S. Mulugu, J. D. York, and E. K. O'Shea. 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316109-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, H., P. C. Fridy, A. A. Ribeiro, J. H. Chol, D. K. Barma, G. Vogel, J. R. Falck, S. B. Shears, J. D. York, and G. W. Mayr. 3 November 2008. Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3 kinases. J. Biol. Chem. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed]
- 32.Macbeth, M. R., H. L. Schubert, A. P. Vandemark, A. T. Lingam, C. P. Hill, and B. L. Bass. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 3091534-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marks, J., I. M. Hagan, and J. S. Hyams. 1986. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. Suppl. 5229-241. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Cuadrado, A. B., E. Duenas, M. Sipiczki, C. R. Vazquez de Aldana, and F. del Rey. 2003. The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 1161689-1698. [DOI] [PubMed] [Google Scholar]
- 35.Matsuyama, A., R. Arai, Y. Yashiroda, A. Shirai, A. Kamata, S. Sekido, Y. Kobayashi, A. Hashimoto, M. Hamamoto, Y. Hiraoka, S. Horinouchi, and M. Yoshida. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24841-847. [DOI] [PubMed] [Google Scholar]
- 36.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123127-130. [DOI] [PubMed] [Google Scholar]
- 37.Miller, A. L., M. Suntharalingam, S. L. Johnson, A. Audhya, S. D. Emr, and S. R. Wente. 2004. Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J. Biol. Chem. 27951022-51032. [DOI] [PubMed] [Google Scholar]
- 38.Mitchison, J. M., and P. Nurse. 1985. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75357-376. [DOI] [PubMed] [Google Scholar]
- 39.Moreau, V., J. M. Galan, G. Devilliers, R. Haguenauer-Tsapis, and B. Winsor. 1997. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol. Biol. Cell 81361-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 41.Mulugu, S., W. Bai, P. C. Fridy, R. J. Bastidas, J. C. Otto, D. E. Dollins, T. A. Haystead, A. A. Ribeiro, and J. D. York. 2007. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316106-109. [DOI] [PubMed] [Google Scholar]
- 42.Mulvihill, D. P., S. R. Edwards, and J. S. Hyams. 2006. A critical role for the type V myosin, Myo52, in septum deposition and cell fission during cytokinesis in Schizosaccharomyces pombe. Cell Motil. Cytoskelet. 63149-161. [DOI] [PubMed] [Google Scholar]
- 43.Odom, A. R., A. Stahlberg, S. R. Wente, and J. D. York. 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 2872026-2029. [DOI] [PubMed] [Google Scholar]
- 44.Pelham, R. J., and F. Chang. 2002. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature 41982-86. [DOI] [PubMed] [Google Scholar]
- 45.Peter, B. J., H. M. Kent, I. G. Mills, Y. Vallis, P. J. Butler, P. R. Evans, and H. T. McMahon. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303495-499. [DOI] [PubMed] [Google Scholar]
- 46.Saiardi, A., J. J. Caffrey, S. H. Snyder, and S. B. Shears. 2000. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 27524686-24692. [DOI] [PubMed] [Google Scholar]
- 47.Saiardi, A., H. Erdjument-Bromage, A. M. Snowman, P. Tempst, and S. H. Snyder. 1999. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 91323-1326. [DOI] [PubMed] [Google Scholar]
- 48.Saiardi, A., A. C. Resnick, A. M. Snowman, B. Wendland, and S. H. Snyder. 2005. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. USA 1021911-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saiardi, A., C. Sciambi, J. M. McCaffery, B. Wendland, and S. H. Snyder. 2002. Inositol pyrophosphates regulate endocytic trafficking. Proc. Natl. Acad. Sci. USA 9914206-14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarmah, B., A. J. Latimer, B. Appel, and S. R. Wente. 2005. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev. Cell 9133-145. [DOI] [PubMed] [Google Scholar]
- 51.Sarmah, B., V. P. Winfrey, G. E. Olson, B. Appel, and S. R. Wente. 2007. A role for the inositol kinase Ipk1 in ciliary beating and length maintenance. Proc. Natl. Acad. Sci. USA 10419843-19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitt, C., C. von Kobbe, A. Bachi, N. Pante, J. P. Rodrigues, C. Boscheron, G. Rigaut, M. Wilm, B. Seraphin, M. Carmo-Fonseca, and E. Izaurralde. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 184332-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seeds, A. M., J. C. Sandquist, E. P. Spana, and J. D. York. 2004. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J. Biol. Chem. 27947222-47232. [DOI] [PubMed] [Google Scholar]
- 54.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 163256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shears, S. B. 2004. How versatile are inositol phosphate kinases? Biochem. J. 377265-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen, X., H. Xiao, R. Ranallo, W. H. Wu, and C. Wu. 2003. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299112-114. [DOI] [PubMed] [Google Scholar]
- 57.Sipiczki, M., B. Grallert, and I. Miklos. 1993. Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J. Cell Sci. 104(Pt. 2)485-493. [DOI] [PubMed] [Google Scholar]
- 58.Snay-Hodge, C. A., H. V. Colot, A. L. Goldstein, and C. N. Cole. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 172663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steger, D. J., E. S. Haswell, A. L. Miller, S. R. Wente, and E. K. O'Shea. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tasto, J. J., J. L. Morrell, and K. L. Gould. 2003. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 1601093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tran, E. J., Y. Zhou, A. H. Corbett, and S. R. Wente. 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28850-859. [DOI] [PubMed] [Google Scholar]
- 62.Tseng, S. S., P. L. Weaver, Y. Liu, M. Hitomi, A. M. Tartakoff, and T. H. Chang. 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 172651-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verbsky, J., K. Lavine, and P. W. Majerus. 2005. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc. Natl. Acad. Sci. USA 1028448-8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verbsky, J. W., M. P. Wilson, M. V. Kisseleva, P. W. Majerus, and S. R. Wente. 2002. The synthesis of inositol hexakisphosphate. Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J. Biol. Chem. 27731857-31862. [DOI] [PubMed] [Google Scholar]
- 65.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, H., X. Tang, J. Liu, S. Trautmann, D. Balasundaram, D. McCollum, and M. K. Balasubramanian. 2002. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 13515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weirich, C. S., J. P. Erzberger, J. S. Flick, J. M. Berger, J. Thorner, and K. Weis. 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8668-676. [DOI] [PubMed] [Google Scholar]
- 68.Wente, S. R., M. P. Rout, and G. Blobel. 1992. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119705-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoon, J. H., D. C. Love, A. Guhathakurta, J. A. Hanover, and R. Dhar. 2000. Mex67p of Schizosaccharomyces pombe interacts with Rae1p in mediating mRNA export. Mol. Cell. Biol. 208767-8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.York, J. D. 2006. Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761552-559. [DOI] [PubMed] [Google Scholar]
- 71.York, J. D., A. R. Odom, R. Murphy, E. B. Ives, and S. R. Wente. 1999. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 28596-100. [DOI] [PubMed] [Google Scholar]
- 72.York, S. J., B. N. Armbruster, P. Greenwell, T. D. Petes, and J. D. York. 2005. Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 2804264-4269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.