Abstract
In Saccharomyces cerevisiae, TBF1, an essential gene, influences telomere function but also has other roles in the global regulation of transcription. We have identified a new member of the tbf1 gene family in the mammalian pathogen Pneumocystis carinii. We demonstrate by transspecies complementation that its ectopic expression can provide the essential functions of Schizosaccharomyces pombe tbf1 but that there is no rescue between fission and budding yeast orthologues. Our findings indicate that an essential function of this family of proteins has diverged in the budding and fission yeasts and suggest that effects on telomere length or structure are not the primary cause of inviability in S. pombe tbf1 null strains.
The phylogenetic pedigree of Pneumocystis species implies they are likely to employ conserved fungus-specific proteins in pathways that are essential for viability. Identification of such modules in the genome of the rat-specific pathogen P. carinii may help to identify novel targets for directed pharmaceutical intervention to treat opportunistic infection of immunocompromised human hosts by P. jirovecii. To identify conserved functional modules, we are assessing the ability of genes from P. carinii to function in the budding yeast Saccharomyces cerevisiae as well as in the fission yeast Schizosaccharomyces pombe, the closest evolutionary relationship for which molecular genetic analysis is well developed (25, 36).
In the course of this study, we identified a potential homologue of the S. pombe tbf1 (Sptbf1) and S. cerevisiae TBF1 (ScTBF1) genes. Sequence comparisons indicate that both SpTbf1p and ScTbf1p are members of a conserved fungus-specific family (50, 51). All the Tbf1 family members contain a C-terminal “telobox” DNA binding domain (8, 9) but bear additional significant homology throughout their coding sequences. The telobox, a particular variant of the Myb family motif, is also found in the mammalian telomere factors TRF1 and TRF2 as well as in the fission yeast telomeric protein Taz1p (13). The telobox recognizes sequences similar to the mammalian telomeric repeat TTAGGG. High-affinity binding sites for ScTbf1p have also been identified in the STARs (subtelomeric antisilencing regions) of the subtelomeric X and Y′ elements. Several studies have proposed a regulatory role for ScTbf1p at telomeres (2, 6, 19, 33). The ScTBF1 gene is essential, but this is commonly assumed to be due to its function as a global transcriptional regulator that binds to many sites throughout chromatin rather than to direct effects on telomeres (10, 33).
SpTbf1p was identified in S. pombe whole-cell extracts as one of several activities that exhibited differential affinity in vitro for tandem copies of the human and S. pombe telomere repeat sequences (55, 62). The Sptbf1 gene is essential, although its full range of cellular functions is unknown. Overexpression of Sptbf1 has been shown to slightly increase the mean length of telomeres in vivo (51).
We have examined whether the P. carinii tbf1 homologue (Pctbf1) can rescue deletions of ScTBF1 and Sptbf1. Our data lead us to infer that SpTbf1p and PcTbf1p are also likely to be global regulators of chromatin structure in their respective organisms.
MATERIALS AND METHODS
Identification and cloning of the full-length P. carinii tbf1 gene.
A partial sequence corresponding to the Pctbf1 open reading frame (ORF) was identified in the unigene set of 1,042 expressed sequence tags (ESTs) isolated from a P. carinii cDNA library (the Pneumocystis Genome Project, http://pgp.cchmc.org) (14) by its homology to the Sptbf1 and ScTBF1 genes. Longer fragments of the Pctbf1 gene were obtained from the randomly amplified cDNA library by PCR (GenomiPhi DNA amplification kit; GE Healthcare, Otelfingen, Switzerland) with primers corresponding to adjacent regions of the genomic sequence in conjunction with the T3 primer located upstream of the multiple-cloning site in the Uni-ZAP XR vector. The longest Pctbf1 fragment that could be amplified from the cDNA library was obtained using the T3 and the PcONTIG609_5ter primers and was considered to contain the full-length ORF since the sequences between all potential further upstream start sites were punctuated by stop codons. The locus was also identified within the genomic databases of the Pneumocystis Genome Project. The Pctbf1 gene contains six introns with the canonical donor and acceptor sites found in P. carinii.
All multiple-sequence alignments were constructed using MAFFT (G-INS-i mode) (28). HHalign was used with standard parameters (http://toolkit.tuebingen.mpg.de). The sequences used as input for HHalign are as follows: for the TRF domain, the full alignment of TRF domain provided by Pfam (http://pfam.sanger.ac.uk) or a multiple alignment of full-length vertebrata sequences containing both TRF and Myb DNA-binding domains (Q4QRH9_DANRE, Q4FZZ9_DANRE, Q8JGS4_DANRE, Q1WM12_XENLA, Q71E47_XENLA, Q2LK75_XENLA, TERF2_HUMAN, TERF1_HU MAN, Q8NHT6_HUMAN, Q5R6X2_PONPY, Q8CH10_MUSSP, Q5EB98_RAT, TERF1_CRIGR, Q3MHY0_BOVIN, Q539Y9_MUNMU, Q539Y6_MUNMU, Q539Z0_MUNMU, Q539Y8_MUNRE, Q539Y5_MUNRE, Q539Y7_MUNRE, Q71M47_CHICK, Q7T1R9_CHICK, TERF2_CHICK, Q5F3M6_CHICK, and Q802C2_CHICK); for fungal tbf sequences, the full alignment of 11 fungal tbf1 sequences as shown in Fig. S1 in the supplemental material (pc-tbf1/1-566, sp Q6E434 TRF1_SCHPO/1-485, sp Q02457TBF1_YEAST/1-562, tr Q6FJX7 Q6FJX7_CANGA/1-525, tr Q5AHX1 Q5AHX1_CANAL/1-849, tr Q6BTS1 Q6BTS1_DEBHA/1-815, tr Q75C21 Q75C21_ASHGO/1-505, and tr Q6CE73 Q6CE73_YARLI/1-710 [all from Uniprot], gi 85082158ref XP_956863.1 1-1146 [from NCBI], and Cd36_18830/1-817 [from GeneDB website version 2.1]).
The full-length cDNA Pctbf1 ORF was amplified from the cDNA library using primers MC01 and MC02 for subcloning into the S. pombe expression vector pREP41 (5). Primers P609StartEcoRI and P609EndSalI were also used to amplify the full-length Pctbf1 ORF (1,701 bp) from the cDNA library, and the product was cloned into an S. cerevisiae centromeric expression vector, p416scTBF1pro. The vector was derived from p416GPD (47) by replacing GPD sequences with a 1,432-bp fragment immediately upstream of the ScTBF1 coding region. All cloned PCR fragments were verified by sequencing in the vectors.
Cloning of Sptbf1, ScTBF1 and CgTBF1.
The Sptbf1 ORF (1,458 bp) was amplified from S. pombe 972 h− genomic DNA, using primers MC04 and MC05, and cloned into pREP41 (5). The ScTBF1 ORF (1,689 bp) was amplified from S. cerevisiae BY4741 genomic DNA, using primers pQ02457StartBamHI and pQ02457EndSalI, and cloned into p416scTBF1pro. The Candida glabrata TBF1 (CgTBF1) ORF (1,578 bp) was amplified from Candida glabrata DSY739 genomic DNA, using primers cgTBF1StartBamHI and cgTBF1EndSalI, and subcloned into the vector p416scTBF1pro. The ScTBF1 ORF was reamplified from the budding yeast expression vectors using primers MC40 and MC41 and subcloned into the pREP41 fission yeast expression vector.
Strains and media.
S. pombe was grown on either complete (YE) medium or synthetic minimal (EMM2) medium supplemented as required (44). A diploid strain heterozygous for the null allele of Sptbf1 (h+/h+; SPBC19G7.13/SPBC19G7.13::KanMX4; ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32) was purchased from the Bioneer Corporation, Korea. The strain was transformed with the plasmid pON177 (59) to induce sporulation. Colonies that formed on EMM2 medium supplemented with leucine at 29°C were examined for the presence of azygotic asci. Tetrads were dissected using a Singer MSM micromanipulator, and spores were germinated on complete (YE) medium. The genotypes of colonies were assessed by replica plating. To prepare spores for germination in liquid culture, approximately 10 large (>2-mm diameter) sporulating colonies were resuspended in sterile water containing 1 μg/ml lysing enzymes (Sigma; L-1412) and incubated overnight at 29°C. The remaining vegetative cells were killed by incubation in 25% (vol/vol) ethanol for 30 min. The spores were washed twice in sterile water. To analyze the phenotype of the tbf1 null cells, spores were inoculated at 3 × 106 ml−1 in YE medium and incubated at 19°C for 18 h. At this time, when the majority of spores had swollen, indicating the resumption of vegetative growth, but had not yet divided, G418 was added to 100 μg/ml to stop further growth of the wild-type cells. In some cases the culture was incubated for 8 to 18 h at 29°C and cells were fixed and processed as described below. The 18-h timing represents four to five generation equivalents after spore germination, when the first differences between wild-type and null cells are observed. Genomic DNA was also prepared from cultures incubated for 24, 48, or 72 h, and telomere length was examined by Southern blotting.
To test complementation, the h+/h+ diploid heterozygous for the tbf1 null allele was crossed to a wild-type h−/h− ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 diploid. A tbf1::KMX4/+ h−/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 diploid was identified by replica plating of tetrads. This diploid was used for complementation, as it does not require the pON177 plasmid to induce meiosis upon starvation.
The diploid S. cerevisiae strain Y22124, heterozygous for the ScTBF1 null allele (Mata/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YPL128c::KanMX4/YPL128c), was supplied by Euroscarf (Institute of Microbiology, Frankfurt, Germany) and used for complementation assays. Transformations and complementation assays in fission and budding yeasts were performed as described previously (36).
GFP tagging of Tbf1p.
The Sptbf1, Pctbf1, and ScTBF1 ORFs were also subcloned in frame with green fluorescent protein (GFP) in the vector pINT41-GFP, which is based on pINT5 (17). In this vector, the full-strength nmt1 promoter has been replaced by the medium-strength nmt1-41 promoter (5). Inserts are expressed as N-terminal fusions with GFP. The leu1-nmt1-41 promoter-GFP-tbf1-(ura4+)-leu1 cassettes (S. pombe, S. cerevisiae, and P. carinii) were excised from the vector by digestion with NotI and gel purified. S. pombe ura4-D18 tbf1 wild-type cells were separately transformed with each of the fragments and cells were plated on EMM2-leucine plates. Transformants in which a tbf1 orthologue had replaced the leu1 gene were identified by replica plating. The tagged tbf1 alleles were introduced into the tbf1 null background by crossing the integrant strains with a haploid that carries the tbf1::KMX4 null allele and expresses the Sptbf1 or Pctbf1 gene from a plasmid that contains a LEU2 gene, which complements leu1-32. As the plasmid is not transmitted efficiently through meiosis (23), dissection of tetrads readily allows identification of haploids bearing the tbf1::KMX4 allele and the integrated expression cassette, which is tagged with the ura4+ gene. In tbf1 null strains rescued by either integrated GFP-SpTbf1 or GFP-PcTbf1, G418-resistant colonies grew after replica plating to plates containing leucine and thiamine or to complete YE medium, indicating that both the uninduced and induced levels of expression of either orthologue from the nmt1-41 promoter permit cell proliferation. Cells were photographed on a Zeiss Axioplan microscope, with a 100× numerical aperture 1.4 lens. Levels and contrast were adjusted in Adobe Photoshop and Image J.
DNA, tubulin, and actin localization in fixed cells.
Cells were fixed and processed for detection of tubulin (22) and F-actin (37) by immunofluorescence as described previously. Images were captured and processed as described previously (34). DNA was visualized by DAPI (4′,6′-diamidino-2-phenylindole) staining as described previously (45).
Southern blotting and telomere length comparisons.
Genomic DNAs were prepared according to standard protocols (45). Twenty micrograms of each DNA was digested overnight with ApaI. Telomere length was assessed by Southern blotting (12) using a synthetic telomere fragment (42) probe labeled by random priming.
Nucleotide sequence accession numbers.
The genomic and cDNA sequences for Pctbf1 have been deposited in GenBank of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) under accession numbers EU617328 and EU617329.
RESULTS
Identification of P. carinii tbf1.
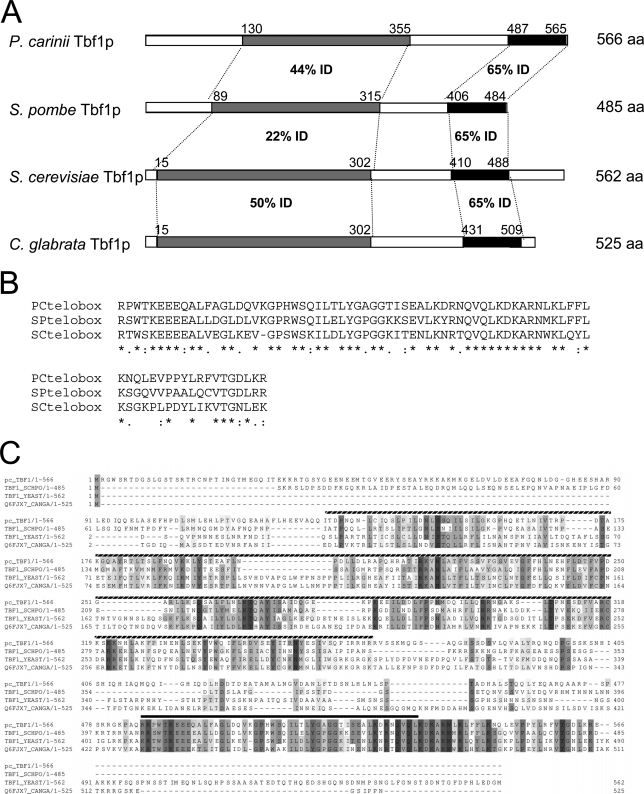
Searches of the S. cerevisiae, S. pombe, and vertebrate proteomes for homologues of the 1,042 unique P. carinii ESTs (http://pgp.cchmc.org) (14) were performed under low-stringency conditions (E value of 1E−3) using the local alignment tool BLASTx. Several of the ESTs had homology with the C-terminal regions of ScTBF1 (YPL128C) and Sptbf1 (SPBC19G7.13), with maximum homology values of 1E−21 and 8E−23, respectively. Contiguous alignment of these overlapping P. carinii ESTs aligned with the C-terminal portion (110 amino acids) of a putative ORF within the genomic sequence of P. carinii. PCR amplification was used to clone full-length genomic and cDNA clones of Pctbf1 (see Materials and Methods) (Table 1). Automated sequence alignment of the Sptbf1 and ScTBF1 coding regions with putative homologues from several other fungal species indicated that in addition to containing a C-terminal Myb-like domain, all the fungal family members are characterized by the presence of a 230-amino-acid domain (Pfam-B PB010333, Pfam v23.0.) in the N-terminal half which shows weaker but significant evidence of conservation (18). Pairwise sequence comparisons of the 79-amino-acid domains that encompass the Myb-like domain or telobox in the ScTbf1p (562 amino acids), SpTbf1p (485 amino acids), and pcTbf1p (566 amino acids) each show comparable degrees of identity and similarity (Fig. 1). The regions of S. pombe and P. carinii Tbf1p which correspond to the Pfam-B PB010333 domain align to each other with sequence identity of 44%, while their best alignments to the Pfam-B PB010333 domains of S. cerevisiae and C. glabrata Tbf1p each give sequence identities of approximately 22% (Fig. 1A and C).
TABLE 1.
Oligonucleotide primers used for PCR amplifications and DNA sequencing analysis
| Primer | Nucleotide sequence (5′→3′)a | Description |
|---|---|---|
| MC01 | GCGCGCGCGTCGACAAAAATGAGAGGATGGAGTAGAAC | First 20 ntb of Pctbf1 with SalI linker |
| MC02 | CGCCCCGGGTTACTCGCGCTTTAAATCTC | Last 20 nt of Pctbf1 with SmaI linker |
| MC04 | GCGCGCGCGTCGACAAAAATGTCAAAACGATCCTTAGACCCTA | First 25 nt of Sptbf1 with SalI linker |
| MC05 | CGCGGATCCTTAATCTCGCCGAAGATCGC | Last 20 nt of Sptbf1 with BamHI linker |
| MC17 | GCGCGCGCGCTAGCAAAATGTCAAAACGATCCTTAGACCCTA | First 25 nt of Sptbf1 with NheI linker |
| MC18 | CGCGGAGCTCTTAATCTCGCCGAAGATCGC | Last 20 nt of Sptbf1 with SacI linker |
| MC42 | GCGCGCGCGCTAGCAAAATGAGAGGATGGAGTAGAAC | First 20 nt of Pctbf1 with NheI linker |
| MC43 | CGCGGAGCTCTTACTCGCGCTTTAAATCTC | Last 20 nt of Pctbf1 with SacI linker |
| MC44 | GCGCGCGCGCTAGCAAAATGGATTCGCAAGTGCCCAAT | First 21 nt of ScTBF1 with NheI linker |
| MC45 | CGCGGAGCTCTTACTACATCCCATCTTCTAAATG | Last 24 nt of ScTBF1 with SacI linker |
| MC20 | ACCTGAAAACTCGCATTTGG | nt 225-205 upstream of the Sptbf1 ORF (SPBC19G7.13) |
| MC21 | GCCTCCGAGTATTAGCACCA | nt 121-142 downstream of the Sptbf1 ORF (SPBC19G7.13) |
| MC40 | GCGCGCGCGTCGACAAAAATGGATTCGCAAGTGCCCAAT | First 21 nt of ScTBF1 SalI linker |
| MC41 | CGCGGATCCCTACATCCCATCTTCTAAATG | Last 21 nt of ScTBF1with BamHI linker |
| pQ02457StartBamHI | GCGCGGGATCCATGGATTCGCAAGTGCCC | First 18 nt of ScTBF1with BamHI linker |
| pQ02457EndSalI | CCCCCCCCGTCGACCTACATCCCATCTTCTAAATG | Last 21 nt of ScTBF1with SalI linker |
| ScTBF1promStartSacI | CGACCGAGCTCTGATAATGCTTGGCAGCG | nt 1432-1414 upstream of the ScTBF1 ORF |
| ScTBF1promEndSpeI | AAGAACACTAGTGACAAATGGGGAAAGAAGTG | nt 20-1 upstream of the ScTBF1 ORF (1432) |
| CgTBF1StartBamHI | GCGCGGGATCCATGACAGGTGATATTCAGATGG | First 22 nt of CgTBF1 with BamHI linker |
| CgTBF1EndSalI | CCCCCCCCGTCGACTCAATTTGGGGGAATACTCC | Last 20 nt of CgTBF1 (1,578 bp) with SalI linker |
| PcCont609BegSeq | GGCATCGACGCCATTAAG | To extend EST 609 (Pctbf1) |
| PcCONTIG609_5ter | CGATGTTGTGGCGCGCGTAA | to extend EST 609 (Pctbf1) |
| p609StartEcoRI | CCTCGAATTCATGAGAGGATGGAGTAGAAC | First 20 nt of Pctbf1 with EcoRI linker |
| p609EndSalI | CCCCCCCCGTCGACTTACTCGCGCTTTAAATCT | Last 19 nt of Pctbf1 (1,701 bp) with SalI linker |
| T7 | ATTACGACTCACTATAGGG | In p416GPD, pRS405, pGEM-T Easy vector |
| T3 | ATTAACCCTCACTAAAGGG | Upstream of multicloning site in UniZAP XR vector, pRS405, p416GPD |
Restriction enzyme sites are underlined and flanked on the 5′ side by 5 to 8 random nucleotides to allow effective digestion. The fragments of each oligonucleotide that will anneal to the relevant genomic template and act as PCR amplification primers are in bold.
nt, nucleotides.
FIG. 1.
Multiple alignment of P. carinii Tbf1p with other fungal homologues indicates a higher degree of similarity to S. pombe Tbf1p than to S. cerevisiae Tbf1p in the N-terminal domain. (A) A stick diagram illustrating the principal domains of conservation between P. carinii, S. pombe, S. cerevisiae, and C. glabrata Tbf1p. Black boxes indicate the telobox domains: Gray boxes indicate regions corresponding to the less strongly conserved Pfam-B PB010333 domain. (B) Amino acid alignment showing degree of conservation between the telobox domains of P. carinii, S. pombe, and S. cerevisiae Tbf1 proteins. Asterisks indicate positions (65% overall) conserved between all three species. Double dots represent positions containing similar residues. Single dots represent positions containing similar residues in at least two of the sequences. (C) Amino acid alignment showing the conserved domains of Pctbf1, and the S. pombe, S. cerevisiae, and C. glabrata Tbf1 proteins from the Uniprot database. The hatched box indicates the Pfam-B PB010333 domain. The filled box represents the Pfam-A minimal Myb DNA-binding domain. A multiple alignment of Tbf1p sequences from 11 fungal species is shown in Fig. S1 in the supplemental material.
The use of multiple alignment comparison algorithms to analyze Tbf1-like proteins from P. carinii and a wide variety of other fungal species has allowed us to reevaluate a recent prediction of structural and functional conservation between the N termini of budding and fission yeast Tbf1 proteins and those of human telomere binding proteins TRF1 and TRF2. The predicted alignment of ScTbf1p and SpTbf1p based only on comparison with the human TRF proteins (51) is clearly different from and less accurate than the one arising from our multiple alignment of the Tbf1 proteins from many fungal species (Fig. 1C; see Fig. S1 in the supplemental material). We have found that pairwise profile alignments (HHalign) (7) between multiple alignments of fungal Tbf1 protein sequences and the full alignment of the human TRF domain (extracted from Pfam v22.0) produce significant scores only in the C-terminal Myb domain, while the N-terminal region produces an alignment with a very low significance. Pairwise profile alignments are a very powerful tool for detecting distant homologies. The lack of a clearly significant score indicates that there has been little evolutionary constraint between the N-terminal regions of fungal Tbf1 and the human TRF sequences, despite the presence of regions of low similarity at the secondary structure level.
Expression of P. carinii tbf1 rescues the S. pombe tbf1 null mutant but the S. cerevisiae tbf1 null mutant does not.
To determine whether the Pctbf1 gene could rescue the tbf1 deletion mutant, the heterozygous tbf1/tbf1::KanMX4 diploid was transformed with the empty vector or a plasmid expressing Sptbf1, Pctbf1, or ScTBF1 under the control of the thiamine-regulated nmt1-41 promoter and then starved to induce meiosis. Spores prepared from cells transformed with each of the plasmids were plated on media selective for the plasmid. Replica plating established that approximately 50% of the viable colonies that were recovered after transformation with the plasmids expressing either Sptbf1 or Pctbf1 were haploids that carried the tbf1::KanMX4 allele. As expected, the other half of the colonies carried the G418-sensitive, genomic copy of the wild-type allele. In contrast, all the colonies that arose after transformation with either empty vector or ScTBF1 carried the G418-sensitive, wild-type tbf1 allele (Table 2). We conclude that plasmid-borne copies of either Sptbf1 or Pctbf1 can rescue the fission yeast genomic tbf1 null allele.
TABLE 2.
The essential function of S. pombe tbf1 can be provided by expression of P. carinii tbf1 but not S. cerevisiae TBF1a
| Plasmid transformed | No. of viable colonies plated on minimal medium + Leu | No. of G418-resistant haploid colonies on replica plating | % of viable colonies that carry tbf1::Kanr |
|---|---|---|---|
| Vector | 580 | 0 | 0 |
| Sptbf1 | 545 | 211 | 39 |
| Pctbf1 | 129 | 64 | 50 |
| ScTBF1 | 196 | 0 | 0 |
Spores from the tbf1/tbf1::KanMX4 diploid transformed with the indicated plasmids were plated at approximately equivalent densities and allowed to germinate on medium selective for the presence of plasmid. The number of haploid colonies (adenine prototrophs), able to grow on medium supplemented with G418, was established by replica plating and calculated as a percentage of the total number of viable colonies.
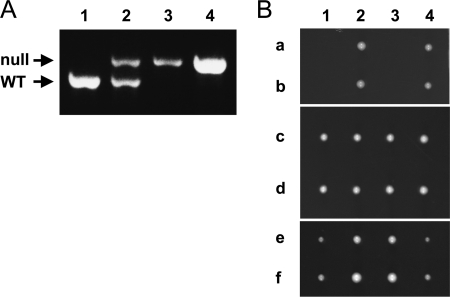
To confirm the absence of the wild-type tbf1 allele, genomic DNA was prepared from the wild-type and rescued haploid strains, as well as from the tbf1/tbf1::KanMX4 diploid. PCR using primers flanking the tbf1::KanMX4 deletion amplified a single fragment corresponding to the expected size of the KanMX4 substitution in the G418-resistant, Leu+ colonies that grow after transformation with Pctbf1 or Sptbf1. The same primers amplified fragments containing both wild-type and null alleles in the heterozygous diploid (Fig. 2A). Similar numbers of colonies were obtained whether the nmt1-41 promoter was induced or not. Furthermore, replica plating to conditions that altered expression of the nmt1-41 promoter after the initial selection for viable colonies did not affect viability or growth rate (data not shown). We conclude that even the repressed levels of expression of Sptbf1 and Pctbf1 are sufficient to rescue viability of the Sptbf1 null strain, while higher levels of their expression do not significantly impair the growth of the cells.
FIG. 2.
The essential function of S. pombe tbf1 can be provided by expression of P. carinii tbf1 but not S. cerevisiae TBF1. (A) The presence of the tbf1::KanMX4 allele and the absence of the wild-type (WT) Sptbf1 allele in the haploid colonies expressing ectopic Sptbf1 and Pctbf1 was confirmed by PCR using locus-specific primers on genomic DNA from the following strains: 1, wild-type 972 h− haploid strain; 2, heterozygous tbf1/tbf1::KanMX4 diploid; 3, tbf1::KanMX4 haploid expressing pREP41-Sptbf1; 4, tbf1::KanMX4 haploid expressing pREP41-Pctbf1. (B) The lethal phenotype of the S. cerevisiae TBF1::KanMX4 strain is rescued by heterologous expression of S. cerevisiae TBF1 or C. glabrata TBF1 but not by that of P. carinii tbf1. S. cerevisiae diploid cells heterozygous for the TBF1::KanMX4 null allele were transformed with plasmids p416ScTBF1pro-PcTBF1 (rows a and b), p416ScTBF1pro-ScTBF1 (rows c and d), or p416ScTBF1pro-CgTBF1 (rows e and f), and allowed to sporulate. Tetrads from each transformant were separated (columns 1 to 4), grown on rich medium, and then replica plated to verify that viable colonies were dependent on the plasmid.
We also examined whether Pctbf1 could rescue the lethality of the TBF1 null mutant in S. cerevisiae. Constructs that carry either ScTBF1, the homologous ORF from C. glabrata (CAGL0M02761g; CgTBF1), or Pctbf1 were cloned into S. cerevisiae expression vectors and expressed in the heterozygous TBF1/tbf1::KanMX4 S. cerevisiae diploid strain. Upon sporulation and germination, heterologous expression of either ScTBF1 or CgTBF1 under the control of the S. cerevisiae TBF1 promoter, rescued viability of the S. cerevisiae TBF1::KanMX4 haploids; however, expression of Pctbf1 in the same vector did not (Fig. 2B). Heterologous expression of Pctbf1 under the control of the strong constitutive S. cerevisiae GPD promoter was also unable to rescue viability (data not shown). Together, the results of transspecies complementation analyses in S. pombe and in S. cerevisiae demonstrate that some essential function of this family of related proteins has diverged over the course of evolutionary differentiation between the Taphrinamycotina and Saccharomycotina branches of the fungal kingdom.
Ectopically expressed GFP-SpTbf1p or GFP-PcTbf1p rescues the tbf1 null phenotype and shows nuclear localization patterns similar to that of GFP-ScTbf1p expressed in wild-type S. pombe.
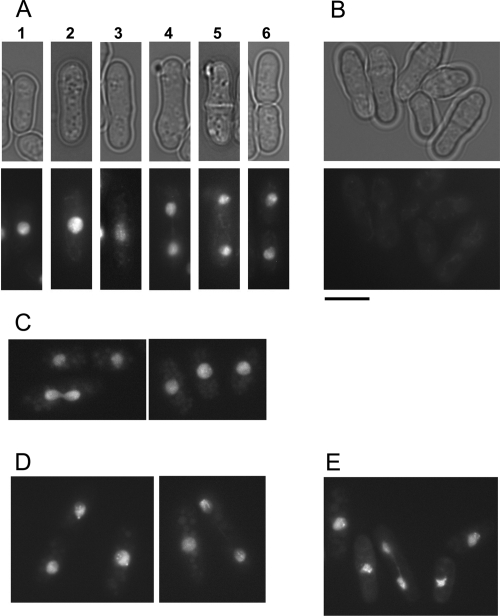
To localize the different orthologues of Tbf1p in S. pombe cells, we fused GFP tags to the N termini of the Sptbf1, Pctbf1, and ScTBF1 ORFs, which were then integrated into the leu1 gene. When expression was induced from the medium-strength nmt1-41 promoter (see Materials and Methods) in cells carrying the endogenous copy of tbf1, GFP-SpTbf1p, GFP-PcTbf1p, and GFP-ScTbf1p were each located in the nucleus, with enrichment throughout the euchromatic hemisphere, at all stages of the cell cycle (Fig. 3A and data not shown). In some cells (both mitotic and interphase), a few intense foci were also observed (Fig. 3A, panels 5 and 6, and E). When the GFP-SpTbf1p and GFP-PcTbf1p fusions were introduced into the tbf1::KMX4 null strain (see Materials and Methods), a similar pattern was observed (Fig. 3C and D), indicating that the GFP-tagged protein does not simply enter the nucleus by dimerizing with untagged Tbf1p. As observed with the native versions of SpTbf1 and PcTbf1, the GFP fusions of both proteins were able to rescue the tbf1::KMX4 null strain whether the nmt1-41 promoter was repressed or induced, while the GFP fusion of ScTbf1 did not rescue even when strongly expressed. The GFP-ScTbf1 fusion could be expressed in wild-type cells for >100 generations without noticeable effects on viability, indicating that it does not act as a dominant negative allele. Taken together, our observations suggest that lack of complementation by the S. cerevisiae homologue is unlikely to be due to either instability, mislocalization, or lack of DNA binding of the GFP-ScTbf1 fusion protein.
FIG. 3.
GFP-SpTbf1 GFP-PcTbf1, and GFP-ScTbf1 have similar localization patterns. (A and B) S. pombe leu1::nmt1-41-GFP-Sptbf1(ura4+) cells were grown in the presence (B) or absence (A) of thiamine. At early times after induction (18 h; four generations) GFP-SpTbf1p is observed in the nucleus at all stages of the cell cycle, including early interphase (cell 1), late interphase (cell 2), early mitosis (cell 3), anaphase (cells 4 and 5), and at cytokinesis/G1 (cell 6). Note that in the absence of induction, though the amount of protein produced is sufficient to rescue the tbf1 null allele, the fluorescence of the GFP-tagged protein falls below the threshold of detection. (C) tbf1::KanMX4 leu1::nmt1-41-GFP-Sptbf1(ura4+) cells were grown for 18 h at 25°C in the absence of thiamine. (D) S. pombe tbf1::KanMX4 leu1::nmt1-41-GFP-Pctbf1(ura4+) cells were grown in the absence of thiamine for 18 h at 25°C. (E) leu1::nmt1-41-GFP-Sctbf1(ura4+) cells were grown in the absence of thiamine for 18 h at 25°C. The scale bar represents 10 μm.
When the cultures are grown in thiamine, the GFP signals from all three fusion proteins are undetectable by fluorescence microscopy (Fig. 3B and data not shown). However crude estimates from previous work suggest that endogenous SpTbf1p is a relatively abundant protein (62) and is likely to be more closely approximated by the induced levels of the GFP fusions than by repressed levels.
Immunolocalization studies with S. cerevisiae have shown that ScTbf1p localizes to foci distributed throughout the nonnucleolar chromatin of interphase nuclei (33). Thus, although the two proteins are unable to functionally complement, both SpTbf1p and ScTbf1p appear to have broad distributions throughout chromatin.
Analysis of the phenotype of S. pombe cells lacking Tbf1p.
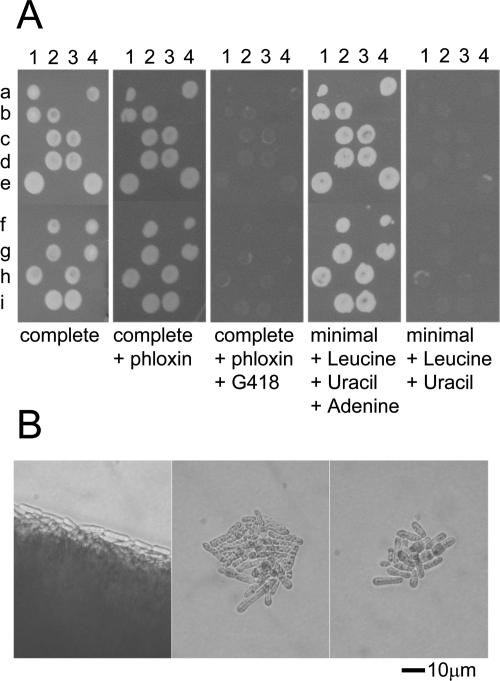
In the course of the complementation experiments described above, we examined the phenotype of the null mutant of Sptbf1 in detail. In agreement with another study (51), we found that the gene is essential in S. pombe (Fig. 4A); however, in contrast to that report, we observed that the two spores carrying the tbf1::KMX4 allele routinely gave rise to microcolonies of up to 50 cells, which were elongated compared to wild-type cells. (Fig. 4B). This was not influenced by temperature, in the range 19°C to 32°C. Examination at intervals of germinating spores on plates at 25°C revealed that the null and wild-type colonies divided at similar rates for the first three divisions, with the null mutant undergoing several additional, significantly retarded divisions thereafter (not shown). These data are consistent with the hypothesis that the SpTbf1p inherited from the spore is diluted among the progeny in the course of the early division cycles and that the phenotype does not become apparent until its level decreases below a critical threshold.
FIG. 4.
Characterization of the phenotype of Sptbf1 null haploids. An S. pombe diploid heterozygous for the tbf1::KanMX4 null allele was allowed to undergo meiosis, and spores were dissected onto YE medium at 29°C. Colonies were allowed to form and were then replicated to the indicated media. (A) nine tetrads (a to i) are shown; the four spores are at positions 1 to 4. Note that all the surviving colonies are sensitive to G418 and are adenine auxotrophs, indicating that they are haploid, and that cells carrying the tbf1::KanMX4 allele cannot form a visible colony (B). The center and right panels show microcolonies derived from the germinating of tbf1::KanMX4 spores. The null mutant stopped dividing after forming a microcolony of 50 cells or fewer. The cells become elongated compared with cells in a tbf1+ colony, indicative of a cell cycle arrest or delay. For comparison, the left panel shows the edge of a colony of G418-sensitive cells from the same tetrad.
S. pombe tbf1 null haploids arrest in interphase, prior to NETO (new-end takeoff), and have an aberrant chromatin structure.
To examine the phenotype of the tbf1 null mutant cells in more detail, spores were first allowed to germinate and then grown for the equivalent of five generation times in medium containing G418 to prevent propagation of all germinated spores that carry the wild-type Sptbf1 allele (see Materials and Methods). The liquid cultures were then fixed and stained to analyze the state of the DNA, microtubules, and F-actin in the tbf1 null mutant cells.
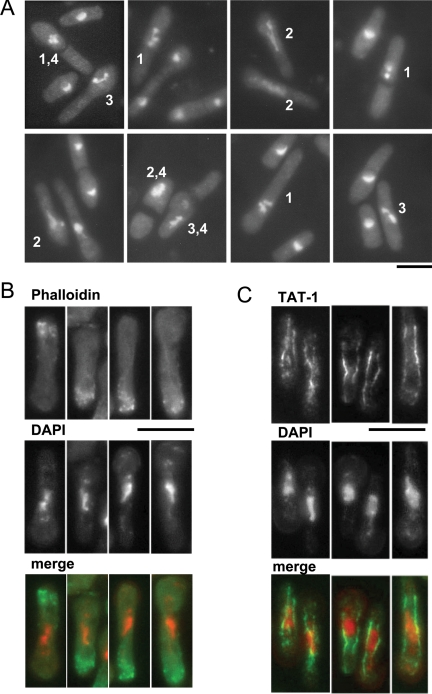
The null mutant cells were swollen and elongated, consistent with a delay in cell cycle progression. DAPI staining showed that most contained a single irregular mass of chromatin, very different in appearance from the chromatin in wild-type cells at any stage of the cell cycle (Fig. 5A).
FIG. 5.
The Sptbf1 null haploids arrest in interphase. Spores prepared from a diploid heterozygous for the tbf1::KanMX4 null allele were grown in complete medium overnight at 19°C, when G418 was added to block further growth and division of tbf1+ cells. At a time corresponding to four generations (see text for details), cells were fixed and the cytoskeleton was analyzed by indirect immunofluorescence. (A) The fixed cells were stained with DAPI. Note the presence of condensed chromosomes (cells marked 1), aberrant nuclear structure (cells marked 2), and problems in chromosome segregation (cells marked 3). Note also the presence of division septa that bisect the cell to generate an anucleate compartment (cells marked 4), indicating that cytokinesis has occurred without nuclear division. (B). The fixed cells were permeabilized and stained with DAPI and rhodamine-conjugated phalloidin. The right panels show tbf1 null cells. Note that F-actin patches are located only at one end of the cell, consistent with a delay early in the cell cycle. (C) The fixed cells were digested, and the TAT-1 antibody was used to analyze the microtubules, as described in Materials and Methods. DAPI is shown in red and microtubules in green. The right panels show the phenotype of elongated tbf1 null cells. Note that all of them have an interphase microtubule array. The scale bars for each panel represent 10 μm.
To determine whether or not the irregularly shaped DAPI-stained structures might reflect a problem of chromosome segregation or a delay in mitosis, we examined the distribution of tubulin and F-actin. We observed that the tbf1 null cells with aberrant DNA had microtubule structures characteristic of interphase, with long microtubules running the length of the cell; no mitotic spindles were observed (Fig. 5B; see Fig. S2a in the supplemental material) (21, 22).
Staining of cells with rhodamine-conjugated phalloidin, DAPI, and Calcofluor revealed that the tbf1 null cells all had F-actin patches at one end (Fig. 5C). In wild-type cells, F-actin redistribution from the “old” (preexisting) end of a cell to both ends (NETO) occurs when cells have reached a characteristic size and have completed S phase (43) (for wild-type images, see Fig. S2b in the supplemental material). The absence of any medially placed contractile F-actin rings in the elongated cells and the unipolar F-actin distribution are both consistent with an interphase arrest or delay early in the cell cycle.
Since the tbf1 null mutant cell cycle is delayed before NETO, we wondered if a DNA damage checkpoint was essential for the delay. To test this, we deleted the ATR-related Rad3p, which is required for the DNA structure checkpoint signaling engaged by abnormal telomeres (40). We crossed a tbf1::KMX4 haploid, carrying a plasmid expressing the Pctbf1 gene, with a rad3::ura4+ mutant. In the vast majority of cells the rescuing plasmid is lost in meiosis, as it does not contain a functional centromere. The genotypes of viable colonies were determined, and tetrads in which both viable colonies were ura− (that is, the Sptbf1 null must be rad3−) or ura+ (where both the Sptbf1 null mutants must be rad3+) were examined. If the later, retarded divisions of the tbf1 null mutant can occur because activation of the DNA structure checkpoint permits time to repair defects in chromosome structure, then in the rad3 tbf1 double mutant one would expect to see fewer cells in the microcolony, since a division delay could no longer be imposed. However, the microcolonies with rad3+ or rad3Δ backgrounds were indistinguishable by either cell number or phenotype. These data suggest that the cell cycle delay of tbf1 null cells is not imposed by a rad3-dependent signaling pathway.
S. pombe telomeres are mildly elongated both in the tbf1 null and the rescued strains.
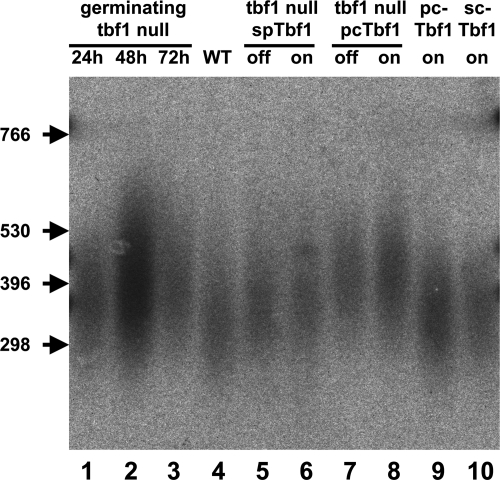
S. pombe Tbf1p overexpression is reported to slightly increase mean telomere length, and it has been speculated that it could have an essential role at telomeres (51). Therefore, we examined whether telomeres had undergone any changes in tbf1 null cells that had ceased to divide. We found that the mean telomere repeat length was some 50 to 80 bp greater in tbf1 null cells than in the isogenic haploid wild-type strain (Fig. 6, lanes 1 to 3 compared with lane 4). There was no sign of end-to-end fusion events having occurred, and telomeres appeared similar in cultures maintained for up to 72 h under G418 selection. Therefore, we conclude that tbf1 null cells do not stop dividing because of rapid telomere shortening or chromosome end fusion.
FIG. 6.
Telomere length in both tbf1 null cells and cells rescued by GFP-SpTbf1 or GFP-PcTbf1 is slightly longer than wild type. Genomic DNA was extracted from the indicated strains and digested overnight with ApaI. The DNA fragments were separated by electrophoresis overnight at 20 V (1 V/cm) through a 1.5% (wt/vol) agarose gel. The filter was probed with a synthetic telomere fragment as described in Materials and Methods. A portion of the filter is shown. The arrowheads indicate the positions of DNA markers visualized by ethidium bromide staining prior to transfer. The lane numbers are referred to in the text. Lanes 1 to 3, DNA from germinating tbf1 null spores at 24, 48, and 72 h after germination. Note that wild-type (WT) cells do not contribute significantly to the culture, as they are unable to divide in the presence of G418, while null spores undergo multiple divisions. Lane 4, wild-type cells. Lanes 5 and 6, DNA from tbf1::KanMX4 leu1::nmt1-41-GFP-Sptbf1(ura4+) cells grown in the absence (on) or presence (off) of thiamine. Lanes 7 and 8, DNA from tbf1::KanMX4 leu1::nmt1-41-GFP-Pctbf1(ura4+) cells grown in the absence (on) or presence (off) of thiamine. Lane 9, DNA from leu1::nmt1-41-GFP-Pctbf1(ura4+) cells grown in the absence (on) of thiamine. Lane 10, DNA from leu1::nmt1-41-GFP-Sctbf1(ura4+) cells grown in the absence (on) of thiamine. Note that lane 2 contains approximately three times as much DNA as the others.
We also compared the lengths of telomeres in cells rescued by the Pneumocystis and S. pombe GFP-Tbf1 fusions (Fig. 6, lanes 5 to 8). Telomere length in the tbf1 null cells rescued by GFP-SpTbf1 was also found to be slightly greater than wild type (approximately 50 to 80 bp) whether the nmt1-41 promoter was induced or not (Fig. 6, compare lanes 4, 5, and 6). This is consistent with the previous studies by Pitt et al., who found that prolonged expression of Tbf1 from the highly attenuated nmt81 promoter caused a discrete increase in telomere length (51). Interestingly, the increase in telomere length in the tbf1 null cells rescued by GFP-PcTbf1 (approximately 100 to 150 bp) was somewhat larger than that in tbf1 null cells rescued by GFP-SpTbf1 but also did not change upon induction of the promoter (Fig. 6, compare lanes 4, 7, and 8). Prolonged overexpression of either GFP-PcTbf1 or noncomplementing GFP-ScTbf1 in the wild-type strain had no dominant effects on telomere length (Fig. 6, lanes 9 and 10).
The telobox motif is also found in the S. pombe telomere binding protein Taz1p (13). Loss of Taz1p function leads to a chromosome segregation defect at low temperatures, which can be rescued by the top2-191 mutant. To test whether the nuclear structure defect of the tbf1 null mutant might reflect chromosome entanglements that could be rescued by top2-191, we crossed top2-191 ura4-D18 with tbf1::KanMx4 leu1::nmt41-GFP-Tbf1 (ura4+) ura4-D18 (see above). Since top2 is very closely linked (<1 cM) to leu1, almost 50% of the cells that inherit the tbf1::KanMX4 allele also inherit top2-191. Dissection of tetrads at either 19°C or 25°C, followed by replica plating, demonstrated that the top2-191 mutant was unable to rescue the essential function of Tbf1p. Taken together, our observations suggest that tbf1 null cells are unlikely to die because of a catastrophic loss of telomere capping function and exhibit no symptoms of either telomere erosion or entanglement.
DISCUSSION
Sequence divergence at interfaces mediating essential functions of Tbf1p in S. cerevisiae and S. pombe/P. carinii.
Our transspecies complementation assays indicate that the fission and budding yeast Tbf1 proteins have diverged significantly. We propose that this functional divergence is most likely to be correlated with the divergence in sequence and secondary structure observed at their N termini. Though not required for sequence-specific DNA binding, part of the equivalent region of ScTbf1p still confers telomere length regulation properties in S. cerevisiae when tethered there by means of a heterologous DNA binding motif (6). Thus, it could be important to gain a better understanding of the interactions mediated by this domain in the S. pombe and P. carinii orthologues. Proteins that participate in the essential function of Tbf1p via interactions with the N-terminal domain might also be conserved between the saprobe S. pombe and the mammalian pathogens P. carinii and P. jirovecii. However, the absence of a system to cultivate Pneumocystis ex vivo precludes investigation of whether heterologous complementation is reciprocal. The S. pombe in vivo complementation assay for viability would permit screening for Sptbf1 and Pctbf1 point mutants with deficient interactions. Proteins from both S. pombe and P. carinii which specifically interact with functionally important regions of their Tbf1 orthologues could then be identified through two-hybrid approaches.
The Tbf1p family as global coordinators of gene expression.
It has recently been proposed that Tbf1p was an important contributor to the coordinated regulation of ribosomal protein genes in ancestral eukaryotes. This regulatory function was apparently taken over by Rap1p in the S. cerevisiae lineage but maintained in other Hemiascomycetes, including S. pombe (27). The essential role of ScTbf1p remains enigmatic. However, comparative genomics analysis of Tbf1p consensus binding sites in several closely related budding yeast species has revealed that they are highly conserved within many different regulatory regions (32). Tbf1p binding sites in these budding yeasts are often found adjacent to one of several stress response motifs (32), suggesting that many genes are regulated by combinatorial interactions between Tbf1p and the proteins that bind these stress response elements. In this context it is also noteworthy that ScTBF1 is one of only 17 genes in the S. cerevisiae genome with a profile of strong upregulation (>5-fold) in response to starvation for either phosphate, glucose, ammonium, or ethanol (64). This implies the existence of feedback loops linking ScTBF1 transcription to specific types of stress. It has been speculated that ScTbf1p and ScRap1p might, respectively, coordinate stress responses and global changes in protein synthesis levels with genome-wide epigenetic changes (32, 46).
The terminal phenotype observed in the Sptbf1 null mutant is consistent with the view that the S. pombe protein is also likely to be a global regulator of transcription, i.e., more similar in function to the pleiotropic Tbf1 and Rap1 proteins of S. cerevisiae (10, 33, 35, 46, 54) than to the telobox protein Taz1p of S. pombe (13, 40, 41). In their respective species, SpTbf1p and ScTbf1p localize widely throughout the euchromatic nuclear hemisphere, supporting the assumption that both do more than regulate telomeres. Consistent with this, we have been unable to obtain any survivors with circular chromosomes from plating out large numbers of tbf1 null haploids (data not shown), whereas this phenotype is characteristic of S. pombe mutants in which cell inviability is due principally to telomere erosion (48, 49). The null phenotype in S. pombe is also quite different from the phenotype produced by inhibiting protein synthesis with cycloheximide (52), suggesting that Sptbf1 probably controls other regulons in addition to those of the ribosome and the subtelomeres. The reason for the cell cycle-specific arrest of Sptbf1 null cells prior to NETO is still unclear. Perhaps genes required for this transition are among its regulatory targets.
A conserved role for Tbf1p in the regulation of gene expression at telomere-proximal domains in fungi?
Tbf1 proteins from both the Taphrinal and the Saccharomycetal fungal orders appear to mildly influence telomere length homeostasis (references 2 and 51 and this study). However our data clearly establish that the primary cause of death in the Sptbf1 null mutants is not rapid telomere shortening or chromosomal end fusions. In addition, entanglement at chromosome ends, such as has been noted for taz1 null mutants (40), does not seem to cause the tbf1 null phenotype. Taken together, our data provide strong evidence that tbf1 has an essential function in S. pombe, unrelated to telomere maintenance. If the role of tbf1 at S. pombe telomeres is also essential but is masked by the plieotropic effects of the null mutant, then the Pneumocystis carinii orthologue must be able to perform it too. If the role at S. pombe telomeres is nonessential, we cannot assume that it is carried out by Pctbf1, since S. pombe telomeres are slightly elongated both in tbf1 null cells and in the rescued strains.
Even if its telomeric role is not vital in S. pombe, a similar role for the Pctbf1 orthologue at Pneumocystis telomeres might have more critical consequences. In contrast to the degeneracy of telomeric repeats found in S. pombe (26), P. carinii telomeres consist of several kilobases of identical repeats of TTAGGG. The subtelomeric regions harbor tandemly repeated copies of several gene families (1, 30, 31, 53), including the abundantly expressed major surface glycoproteins (MSGs), which play an important role in the host-parasite interaction and the escape from host responses during lung infection (20, 58). Only a single copy of approximately 100 or so MSG genes is expressed at any one time; telomeric recombination appears to mediate the switching of different antigenic variants into the unique subtelomeric locus from which expression takes place (29, 56, 57, 60). Whether the integrity of P. carinii telomere ends is maintained through telomerase-based addition of repeats or entirely by recombination is still unclear. There are no obvious homologues to the catalytic or RNA subunits of S. pombe telomerase in the almost completed draft genomic sequence of P. carinii.
The global organization of P. carinii gene families such as MSG, and the related MSR and PRT1 families (30), into special chromatin domains is reminiscent of a feature that is common to many distantly related species. Virulence-related surface glycoproteins in other pathogenic fungi are frequently found encoded in subtelomeric clusters that are subject to control by regulators of chromatin structure (4, 15, 16). Even in the free-living S. cerevisiae, many variantly expressed cell surface adhesin genes and other genes regulating facultative use of different carbon sources are found in the subtelomeric regions (11, 24, 39, 61). This strategy may afford great flexibility for coordinating the regulated expression of such loci by modifications of their chromatin structure (38).
Unsurprisingly, the genes that lie in the subtelomeric regions of S. pombe bear no resemblance to the virulence factors and cell surface adhesins of its commensal and opportunistically pathogenic cousins. They nevertheless share two noteworthy general features: many are organized in multicopy families, and many encode proteins likely to be expressed at the cell surface (3, 63). Perhaps all Tbf1p family members have a general role at telomere-proximal regions to coordinate the facultative expression of such genes in response to different growth conditions.
Supplementary Material
Acknowledgments
We are indebted to Melanie Cushion for providing helpful information on the Pneumocystis genomic databases and to George Smulian for the gift of the P. carinii cDNA library. We are also grateful to Patrick Reichenbach (Lingner lab, ISREC-EPFL) for assistance with hybridizations and to Sandra Dischinger and the EPFL BioImaging platform for assistance with imaging. We are grateful to Julie Cooper (London, United Kingdom) and Paul Russell (San Diego, CA) for reagents to detect fission yeast telomeres.
L.C. was funded by Swiss National Science Foundation grant 315200-116864. This study was supported by Swiss National Science Foundation grant 310000-112360 to P.M.H. and V.S.
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ambrose, H. E., S. P. Keely, E. M. Aliouat, E. Dei-Cas, A. E. Wakefield, R. F. Miller, and J. R. Stringer. 2004. Expression and complexity of the PRT1 multigene family of Pneumocystis carinii. Microbiology 150293-300. [DOI] [PubMed] [Google Scholar]
- 2.Arneric, M., and J. Lingner. 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 81080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslett, M., and V. Wood. 2006. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100%. Yeast 23913-919. [DOI] [PubMed] [Google Scholar]
- 4.Barry, J. D., M. L. Ginger, P. Burton, and R. McCulloch. 2003. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 3329-45. [DOI] [PubMed] [Google Scholar]
- 5.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123131-136. [DOI] [PubMed] [Google Scholar]
- 6.Berthiau, A. S., K. Yankulov, A. Bah, E. Revardel, P. Luciano, R. J. Wellinger, V. Geli, and E. Gilson. 2006. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J. 25846-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biegert, A., C. Mayer, M. Remmert, J. Soding, and A. N. Lupas. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34W335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilaud, T., C. Brun, K. Ancelin, C. E. Koering, T. Laroche, and E. Gilson. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17236-239. [DOI] [PubMed] [Google Scholar]
- 9.Bilaud, T., C. E. Koering, E. Binet-Brasselet, K. Ancelin, A. Pollice, S. M. Gasser, and E. Gilson. 1996. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 241294-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brigati, C., S. Kurtz, D. Balderes, G. Vidali, and D. Shore. 1993. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol. Cell. Biol. 131306-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charron, M. J., E. Read, S. R. Haut, and C. A. Michels. 1989. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics 122307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385744-747. [DOI] [PubMed] [Google Scholar]
- 13.Cooper, J. P., Y. Watanabe, and P. Nurse. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392828-831. [DOI] [PubMed] [Google Scholar]
- 14.Cushion, M. T., A. G. Smulian, B. E. Slaven, T. Sesterhenn, J. Arnold, C. Staben, A. Porollo, R. Adamczak, and J. Meller. 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS One 2e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Las Penas, A., S. J. Pan, I. Castano, J. Alder, R. Cregg, and B. P. Cormack. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 172245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas, Jr., A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 12113-24. [DOI] [PubMed] [Google Scholar]
- 17.Fankhauser, C., and V. Simanis. 1994. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J. 133011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34D247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fourel, G., E. Revardel, C. E. Koering, and E. Gilson. 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 182522-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbe, T. R., and J. R. Stringer. 1994. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect. Immun. 623092-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagan, I. M. 1998. The fission yeast microtubule cytoskeleton. J. Cell Sci. 1111603-1612. [DOI] [PubMed] [Google Scholar]
- 22.Hagan, I. M., and J. S. Hyams. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89343-357. [DOI] [PubMed] [Google Scholar]
- 23.Hahnenberger, K. M., M. P. Baum, C. M. Polizzi, J. Carbon, and L. Clarke. 1989. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 86577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halme, A., S. Bumgarner, C. Styles, and G. R. Fink. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116405-415. [DOI] [PubMed] [Google Scholar]
- 25.Hauser, P. M., L. Lo Presti, M. Cockell, L. Cerutti, and V. Simanis. 2006. Analysis of Pneumocystis carinii gene function by complementation in yeast mutants. J. Eukaryot. Microbiol. 53(Suppl. 1)S149-S150. [DOI] [PubMed] [Google Scholar]
- 26.Hiraoka, Y., E. Henderson, and E. H. Blackburn. 1998. Not so peculiar: fission yeast telomere repeats. Trends Biochem. Sci. 23126. [DOI] [PubMed] [Google Scholar]
- 27.Hogues, H., H. Lavoie, A. Sellam, M. Mangos, T. Roemer, E. Purisima, A. Nantel, and M. Whiteway. 2008. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell 29552-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 303059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keely, S. P., M. J. Linke, M. T. Cushion, and J. R. Stringer. 2007. Pneumocystis murina MSG gene family and the structure of the locus associated with its transcription. Fungal Genet. Biol. 44905-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keely, S. P., H. Renauld, A. E. Wakefield, M. T. Cushion, A. G. Smulian, N. Fosker, A. Fraser, D. Harris, L. Murphy, C. Price, M. A. Quail, K. Seeger, S. Sharp, C. J. Tindal, T. Warren, E. Zuiderwijk, B. G. Barrell, J. R. Stringer, and N. Hall. 2005. Gene arrays at Pneumocystis carinii telomeres. Genetics 1701589-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keely, S. P., A. E. Wakefield, M. T. Cushion, A. G. Smulian, N. Hall, B. G. Barrell, and J. R. Stringer. 2001. Detailed structure of Pneumocystis carinii chromosome ends. J. Eukaryot Microbiol. Suppl. 118S-120S. [DOI] [PubMed]
- 32.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423241-254. [DOI] [PubMed] [Google Scholar]
- 33.Koering, C. E., G. Fourel, E. Binet-Brasselet, T. Laroche, F. Klein, and E. Gilson. 2000. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 282519-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krapp, A., S. Schmidt, E. Cano, and V. Simanis. 2001. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 111559-1568. [DOI] [PubMed] [Google Scholar]
- 35.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28327-334. [DOI] [PubMed] [Google Scholar]
- 36.Lo Presti, L., M. Cockell, L. Cerutti, V. Simanis, and P. M. Hauser. 2007. Functional characterization of Pneumocystis carinii brl1 by transspecies complementation analysis. Eukaryot. Cell 62448-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marks, J., I. M. Hagan, and J. S. Hyams. 1986. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. Suppl. 5229-241. [DOI] [PubMed] [Google Scholar]
- 38.Merrick, C. J., and M. T. Duraisingh. 2006. Heterochromatin-mediated control of virulence gene expression. Mol. Microbiol. 62612-620. [DOI] [PubMed] [Google Scholar]
- 39.Michels, C. A., E. Read, K. Nat, and M. J. Charron. 1992. The telomere-associated MAL3 locus of Saccharomyces is a tandem array of repeated genes. Yeast 8655-665. [DOI] [PubMed] [Google Scholar]
- 40.Miller, K. M., and J. P. Cooper. 2003. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell 11303-313. [DOI] [PubMed] [Google Scholar]
- 41.Miller, K. M., M. G. Ferreira, and J. P. Cooper. 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 243128-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, K. M., O. Rog, and J. P. Cooper. 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440824-828. [DOI] [PubMed] [Google Scholar]
- 43.Mitchison, J. M., and P. Nurse. 1985. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75357-376. [DOI] [PubMed] [Google Scholar]
- 44.Moreno, S., J. Hayles, and P. Nurse. 1989. Regulation of p34cdc2 protein kinase during mitosis. Cell 58361-372. [DOI] [PubMed] [Google Scholar]
- 45.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 46.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 1651-53. [DOI] [PubMed] [Google Scholar]
- 47.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156119-122. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282493-496. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 1611437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penkett, C. J., J. A. Morris, V. Wood, and J. Bahler. 2006. YOGY: a web-based, integrated database to retrieve protein orthologs and associated Gene Ontology terms. Nucleic Acids Res. 34W330-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pitt, C. W., L. P. Valente, D. Rhodes, and T. Simonsson. 2008. Identification and characterization of an essential telomeric repeat binding factor in fission yeast. J. Biol. Chem. 2832693-2701. [DOI] [PubMed] [Google Scholar]
- 52.Polanshek, M. M. 1977. Effects of heat shock and cycloheximide on growth and division of the fission yeast, Schizosaccharomyces pombe. With an Appendix. Estimation of division delay for S. pombe from cell plate index curves. J. Cell Sci. 231-23. [DOI] [PubMed] [Google Scholar]
- 53.Schaffzin, J. K., T. R. Garbe, and J. R. Stringer. 1999. Major surface glycoprotein genes from Pneumocystis carinii f. sp. ratti. Fungal Genet. Biol. 28214-226. [DOI] [PubMed] [Google Scholar]
- 54.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10408-412. [DOI] [PubMed] [Google Scholar]
- 55.Spink, K. G., R. J. Evans, and A. Chambers. 2000. Sequence-specific binding of Taz1p dimers to fission yeast telomeric DNA. Nucleic Acids Res. 28527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stringer, J. R. 2007. Antigenic variation in pneumocystis. J. Eukaryot Microbiol. 548-13. [DOI] [PubMed] [Google Scholar]
- 57.Stringer, J. R., and S. P. Keely. 2001. Genetics of surface antigen expression in Pneumocystis carinii. Infect. Immun. 69627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stringer, S. L., T. Garbe, S. M. Sunkin, and J. R. Stringer. 1993. Genes encoding antigenic surface glycoproteins in Pneumocystis from humans. J. Eukaryot Microbiol. 40821-826. [DOI] [PubMed] [Google Scholar]
- 59.Styrkarsdottir, U., R. Egel, and O. Nielsen. 1993. The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr. Genet. 23184-186. [DOI] [PubMed] [Google Scholar]
- 60.Sunkin, S. M., and J. R. Stringer. 1996. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol. Microbiol. 19283-295. [DOI] [PubMed] [Google Scholar]
- 61.Teunissen, A. W., and H. Y. Steensma. 1995. The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 111001-1013. [DOI] [PubMed] [Google Scholar]
- 62.Vassetzky, N. S., F. Gaden, C. Brun, S. M. Gasser, and E. Gilson. 1999. Taz1p and Teb1p, two telobox proteins in Schizosaccharomyces pombe, recognize different telomere-related DNA sequences. Nucleic Acids Res. 274687-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, A. Dusterhoft, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, V. Lelaure, S. Mottier, F. Galibert, S. J. Aves, Z. Xiang, C. Hunt, K. Moore, S. M. Hurst, M. Lucas, M. Rochet, C. Gaillardin, V. A. Tallada, A. Garzon, G. Thode, R. R. Daga, L. Cruzado, J. Jimenez, M. Sanchez, F. del Rey, J. Benito, A. Dominguez, J. L. Revuelta, S. Moreno, J. Armstrong, S. L. Forsburg, L. Cerutti, T. Lowe, W. R. McCombie, I. Paulsen, J. Potashkin, G. V. Shpakovski, D. Ussery, B. G. Barrell, and P. Nurse. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415871-880. [DOI] [PubMed] [Google Scholar]
- 64.Wu, J., N. Zhang, A. Hayes, K. Panoutsopoulou, and S. G. Oliver. 2004. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc. Natl. Acad. Sci. USA 1013148-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.