Abstract
For Candida albicans, evidence has suggested that the mating pheromones activate not only the mating response in mating-competent opaque cells but also a unique response in mating-incompetent white cells that includes increased cohesion and adhesion, enhanced biofilm formation, and expression of select mating-related and white cell-specific genes. On the basis of a recent microarray analysis comparing changes in the global expression patterns of white cells in two strains in response to α-pheromone, however, skepticism concerning the validity and generality of the white cell response has been voiced. Here, we present evidence that the response occurs in all tested media (Lee's, RPMI, SpiderM, yeast extract-peptone-dextrose, and a synthetic medium) and in all of the 27 tested strains, including a/a and α/α strains, derivatives of the common laboratory strain SC5314, and representatives from all of the five major clades. The white cell response to pheromone is therefore a general characteristic of MTL-homozygous strains of C. albicans.
For a/a and α/α strains of Candida albicans to mate, they must first switch from the mating-incompetent white phenotype to the mating-competent opaque phenotype (7, 9). Opaque cells of opposite mating types then signal each other through the release of pheromones to form conjugation projections and fuse in the mating process, which is similar to that of Saccharomyces cerevisiae (2, 8). Haploid cells of S. cerevisiae, however, do not have to undergo the highly complex white-opaque transition to mate. This species difference raises two questions. First, why must C. albicans switch to mate, when S. cerevisiae has no such requirement? And second, what is the role of the white phenotype? Why are C. albicans cells not mating competent immediately after they undergo MTL homozygosis? In studying the role of switching in the mating process, we made an observation which shed light on these questions (5). We found that the same pheromones that signaled opaque cells in the mating process also signaled white cells to become adhesive and cohesive and to form thicker biofilms, and the same pheromones upregulated both select mating-related and white cell-specific genes (5, 8, 16). We hypothesized that by forming a protective biofilm, this unique white cell response facilitated mating between minority opaque cells that appeared spontaneously (5).
Although we demonstrated that the white cell response to α-pheromone occurred in two natural a/a strains, P37005 and L26, and to a-pheromone in one natural α/α strain, P57072 (5, 16), skepticism concerning the generality of the white cell response among all C. albicans strains has been voiced formally and informally. This has arisen in part from observations reported by Bennett and Johnson (3) in a microarray comparison of α-pheromone-induced gene expressions in white cells of two strains, RBY717, an a/a derivative of the laboratory strain SC5314 (2), and P37005, a natural a/a strain (7). Although the focus of their study was on the effects of pheromone on global expression patterns of opaque cells in the mating process, they found by microarray analysis that the global response of gene expression to α-pheromone by white cells of strain RBY717 was weaker than that of strain P37005 (3). They also found that the effect of α-pheromone on the global expression patterns of white cells was affected by the composition of the supporting medium (3). These results suggested that strain backgrounds and the supporting nutrient media impacted the intensity of the white cell response and led to the notion that the white cell response to pheromone did not occur in all strains or in all media. Since we believe that the white cell response to pheromone may provide a key to understanding the essential role that white-opaque switching plays in C. albicans mating (5, 13), we found it imperative to test the generality of the white cell response in a wide variety of strains representing all of the major clades of C. albicans (11) and in a variety of common media. We assayed two characteristics of the white cell response that were previously shown to be clear indicators of the white cell response (5, 16), an increase of more than 100-fold in cell adhesion to a plastic surface and upregulation of the pheromone receptor genes and several white cell-specific genes.
α-Pheromone induction of adhesion.
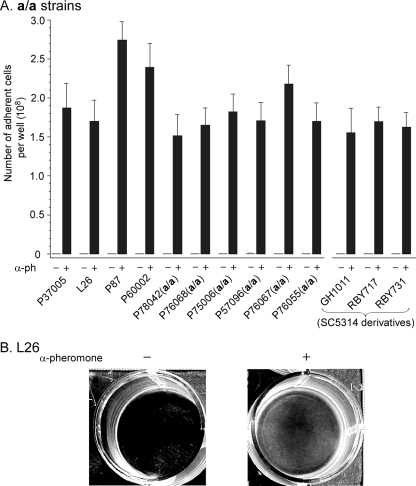
α-Pheromone induces adhesion and cohesion in white, but not opaque, a/a cells (5). White cells of 13 a/a strains (see Table S1 in the supplemental material) were tested for α-pheromone-induced adhesion to the plastic bottoms of wells in a cluster well plate according to the methods of Daniels et al. (5) (see the supplemental material). Adhesion to well bottoms was negligible in the absence of α-pheromone (<106 cells per well bottom) but high in its presence (1.5 × 108 to 2.7 × 108 per well bottom) for the 13 tested strains (Fig. 1A). The increase was well over 100-fold for every tested strain (Fig. 1A). The well bottoms for white cells of the natural a/a strain L26 (7) in the absence and presence of α-pheromone were representative of all tested a/a strains (Fig. 1B). In control experiments, we found that α-pheromone did not stimulate adhesion in white cells of five tested α/α strains or in cells of five tested a/α strains; in all of these controls, levels of adhesion to the well bottom were comparable in the absence and presence of α-pheromone (data not shown).
FIG. 1.
α-Pheromone induces a dramatic increase (>100-fold) in adhesion in white cells of all tested a/a strains of C. albicans. The methods of Daniels et al. (5) were employed (see the supplemental material). In brief, white cells were incubated for 16 h at 25°C in supplemented Lee's medium (1) in the wells of cluster well plates in the absence (−) or presence (+) of 10−6 M α-pheromone (13-mer). Well bottoms were then gently rinsed, photographed, and then scraped and the suspended cells counted. (A) Histogram of the average number of cells adhering to the well bottom for each of 10 a/a strains. Those strains with a/a in parenthesis were obtained by treating the noted wild-type a/α strain with sorbose (6), screening for opaque sectors (MTL-homozygous offspring), and genotyping for a/a strains by PCR (9, 15). Three a/a derivatives of the laboratory strain SC5314 were also tested. The origins and genotypes of the tested a/a strains are provided in Table S1 in the supplemental material. The mean of results from three well bottoms plus the standard deviation (error bar) are presented for each strain. α-ph, α-pheromone. (B) The well bottoms for strain L26 in the absence (−) or presence (+) of α-pheromone were representative of all a/a strains tested.
a-Pheromone induction of adhesion.
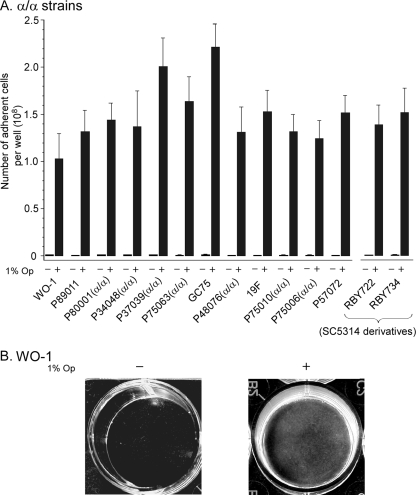
a-Pheromone induces adhesion and cohesion in white, but not opaque, α/α cells (5). White cells of 14 α/α strains were tested for a-pheromone-induced adhesion to a plastic surface by a modification of the procedure of Daniels et al. (5) (see the supplemental material). a-Pheromone was generated by adding 1% opaque cells, consisting of a 50:50 mixture of opaque α/α cells (strain WO-1) and opaque a/a cells (strain P37005), to 99% white test cells. Presumably, the release of α-pheromone by minority opaque α/α cells upregulated a-pheromone production by minority opaque a/a cells, which in turn signaled majority white α/α cells. Adhesion of white cells to the substrate was negligible in the absence of the opaque cell mixture (<106 cells per well bottom) but high in its presence (1.0 × 108 to 2.2 × 108 per well bottom) for all 14 strains (Fig. 2A). The increase was well over 100-fold for every tested strain (Fig. 2A). The well bottoms for white cells of the natural α/α strain WO-1 (10) in the absence and presence of the minority opaque cell mixture were representative of all tested α/α strains (Fig. 2B). In control experiments, we found that the 1% mixture of opaque cells did not stimulate adhesion in five tested a/α strains; levels of adhesion to the dish bottom in the absence and presence of minority opaque cells were comparable. We could not test the effects of a-pheromone on white a/a cells, since the opaque cell mixture, which is the source of a-pheromone, also produces α-pheromone.
FIG. 2.
a-Pheromone induces a dramatic increase (>100-fold) in adhesion in white cells of all tested α/α strains of C. albicans. A modified version of the methods of Daniels et al. (5) was employed. In brief, majority white cells (99%) of each strain were mixed with minority opaque cells (1%), the latter composed of a 50:50 mixture of opaque a/a (P37005) and opaque α/α (WO-1) cells. Presumably, the α/α opaque cells in the mixture produced α-pheromone, which upregulated a-pheromone production in the opaque a/a cells (5). White cells of each test strain, alone or mixed with the minority opaque cell mixture, were then assayed for adhesion as described in the legend to Fig. 1 for the α-pheromone response of a/a cells. (A) Histogram of the average number of cells adhering to the well bottom for each of 14 α/α strains. The three α/α strains P48076, P75010, and P75006 were obtained by growing the natural a/α strains with sorbose (6), screening for opaque sectors (MTL-homozygous offspring), and genotyping for α/α strains by PCR (9, 15). The four α/α strains P80001, P34048, P37039, and P75063 were spontaneous α/α derivatives of natural a/α strains. The mean of results from three well bottoms plus the standard deviation are presented for each strain. 1% Op, 1% opaque cell mixture. (B) The well bottoms for strain WO-1 in the absence (−) or presence (+) of the mixture of minority opaque cells were representative of all α/α strains tested.
Pheromone induction of gene expression.
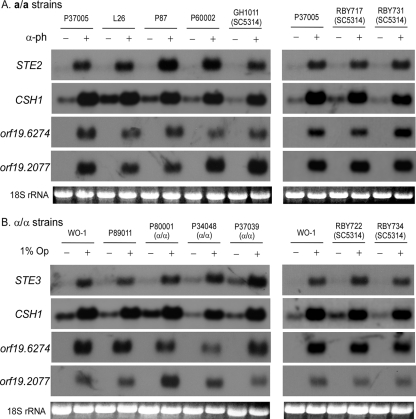
We next tested whether α-pheromone upregulated STE2, the α-pheromone receptor gene, and three white cell-specific genes, CSH1 (16), orf19.2077 (N. Sahni, S. Yi, K. J. Daniels, T. Srikantha, and D. K. Soll, submitted for publication), and orf19.6274 (Sahni et al., submitted), in white cells of seven a/a strains and whether a-pheromone upregulated STE3, the a-pheromone receptor gene, and the same three white cell-specific genes in white cells of seven α/α strains. α-Pheromone upregulated STE2 and the three white cell-specific genes in all tested a/a strains (Fig. 3A), and a-pheromone upregulated STE3 and the three white cell-specific genes in all α/α strains (Fig. 3B). α-Pheromone upregulated STE2 but not the three white cell-specific genes in opaque cells of the tested a/a strains (data not shown), and a-pheromone upregulated STE3, but not the three white cell-specific genes, in opaque cells of the seven α/α test strains (data not shown). Strain variation was observed in the expression levels of the assayed genes upon pheromone induction, but most importantly, pheromone upregulated every tested gene in every tested strain.
FIG. 3.
Pheromone induces the expression of the pheromone receptor genes and white cell-specific genes in white cells of all tested strains. Saturation phase white cells of a/a strains were released into supplemented Lee's medium in the absence or presence of 10−6 M α-pheromone (13-mer) and incubated for 4 h. Saturation phase white cells of α/α strains were mixed 99:1 with a 50:50 mixture of opaque a/a P37005 and opaque α/α WO-1 cells and incubated for 4 h. Northern analyses were performed as previously described (14, 16). (A) Northern analysis of the expressions of STE2 and the white cell-specific, pheromone-induced genes CSH1, orf19.6274, and orf19.2077 in white cells of seven a/a test strains in the absence (−) or presence (+) of α-pheromone (α-ph). Three a/a derivatives of the laboratory strain SC5314 were tested. (B) Northern analysis of the expressions of STE3 and the white cell-specific genes CSH1, orf19.6274, and orf19.2077 in white cells of seven α/α strains in the absence (−) or presence (+) of 1% opaque cell mixture (1% Op). The patterns to the left and right of each panel represent independent experiments, and therefore, each has one common strain (P37005 for a/a strains and WO-1 for α/α strains). The ethidium bromide-stained 18S rRNA patterns are provided to demonstrate uniform loading.
White cell response of SC5314 derivatives.
In the Bennett and Johnson (3) study, the α-pheromone-induced pattern of gene expression was stronger in white cells of the natural a/a strain P37005 than in the SC5314 a/a derivative RBY717. We therefore compared the white cell responses to α-pheromone in the RBY717 and natural a/a strains. We also compared a second SC5314 a/a derivative generated by Bennett and Johnson (3), RBY731. Similarly, we compared the white cell responses to a-pheromone in two SC5314 α/α derivatives also generated by Bennett and Johnson (3), RBY722 and RBY734. All of these strains were generous gifts from Richard Bennett of Brown University. Finally, we tested the white cell response to α-pheromone of the SC5314 a/a derivative GH1011, which we independently generated (G. Huang, T. Srikantha, and D. R. Soll, submitted for publication). α-Pheromone induced adhesion (Fig. 1A) and upregulated gene expression (Fig. 3A) in white cells of the three SC5314 a/a derivatives, and a-pheromone induced adhesion (Fig. 2A) and upregulated gene expression (Fig. 3B) in white cells of the two SC5314 α/α derivatives. All of the SC5314 derivatives responded to pheromone as robustly, on average, as the other MTL-homozygous strains (Fig. 1, 2, and 3). No significant difference was observed between P37005 and the SC5314 a/a derivatives, including strain RBY717, in level of adhesion (P > 0.05) or levels of gene expression induced by pheromone.
White cell responses in different media.
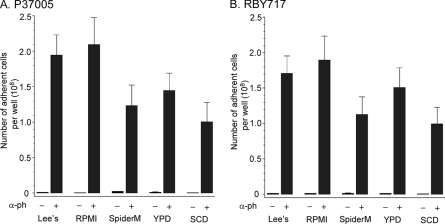
Bennett and Johnson (3) observed marked differences in the effects of α-pheromone on the global expression patterns of white cells of strain P37005 and RBY717 in different nutrient media and noted that the effect of medium composition was more pronounced for strain RBY717 than for strain P37005. They found that Lee's medium was better than SpiderM medium for the white cell response (3). We therefore tested whether growth medium influenced the white cell response to pheromone in the natural a/a strain P37005 and SC5314 a/a derivative RBY717. In five test media (Lee's, RPMI, SpiderM, yeast extract-peptone-dextrose [YPD], and synthetic complete medium with 2% dextrose [SCD]), α-pheromone induced adhesion over 100-fold for both P37005 (Fig. 4A) and RBY717 (Fig. 4B). Differences in the increases induced by α-pheromone in the different media ranged from 1.0 × 108 to 2.1 × 108 cells per well bottom (Fig. 4A and B). Pheromone induction was higher in Lee's and RPMI media than in SpiderM, YPD, and SCD media for both P37005 and RBY717 white cells (Fig. 4A and B, respectively). We found, as did Bennett and Johnson (3), that Lee's medium was better in supporting the pheromone response than SpiderM medium. More importantly, a robust white cell response to α-pheromone occurred in both strains in all tested media.
FIG. 4.
The increase in white cell adhesion induced by pheromone occurs in five common media used in C. albicans research. White a/a cells of natural strain P37005 (A) and strain RBY717 (B), an a/a derivative of laboratory strain SC5314, were grown and tested with α-pheromone (α-ph) as described in the legend to Fig. 1, but the media in which they were tested included one of the following: supplemented Lee's (1), RPMI (5), SpiderM (3), YPD (3, 14), or SCD (3). The increases in adhesion induced by pheromone varied between >100-fold (in SCD) and >200-fold (in RPMI).
Distribution between clades.
DNA fingerprinting studies with the complex probe Ca3 have separated the majority of C. albicans isolates into five major clades, I, II, III, SA, and E (11). To assess the generality of the white cell response, we selected a minimum of two a/a and two α/α test strains from each of the major clades in testing for the response. Four of the a/a strains (P37005, L26, P87, and P60002) possessed this genotype at the time of collection, six (P78042, P76068, P75006, P57096, P76067, and P76055) were natural a/α strains that were induced by sorbose treatment (2, 6) to undergo MTL homozygosis, and three (GH1011, RBY717, and RBY731) were derived from the laboratory strain SC5314 by sorbose treatment (3). Five of the α/α strains (WO-1, P89011, P57072, GC75, and 19F) possessed this genotype at the time of collection, four (P80001, P34048, P37039, and P75063) were natural a/α strains that underwent spontaneous MTL homozygosis, three (P48076, P75010, and P75006) were natural a/α strains induced to undergo MTL homozygosis by sorbose treatment, and two (RBY722 and RBY734) were derived from laboratory strain SC5314 by sorbose treatment (3). As noted, white cells of every a/a and α/α test strain, representing all of the five major clades, responded to pheromone with an increase in adhesion of greater than 100-fold (Fig. 1 and 2). A dendrogram was generated on the basis of the similarity coefficients (4, 11, 12) computed among 25 DNA fingerprinted test strains and 50 other previously DNA fingerprinted strains that were distributed among the five major clades in order to emphasize the generality of the white cell response (Fig. 5).
FIG. 5.
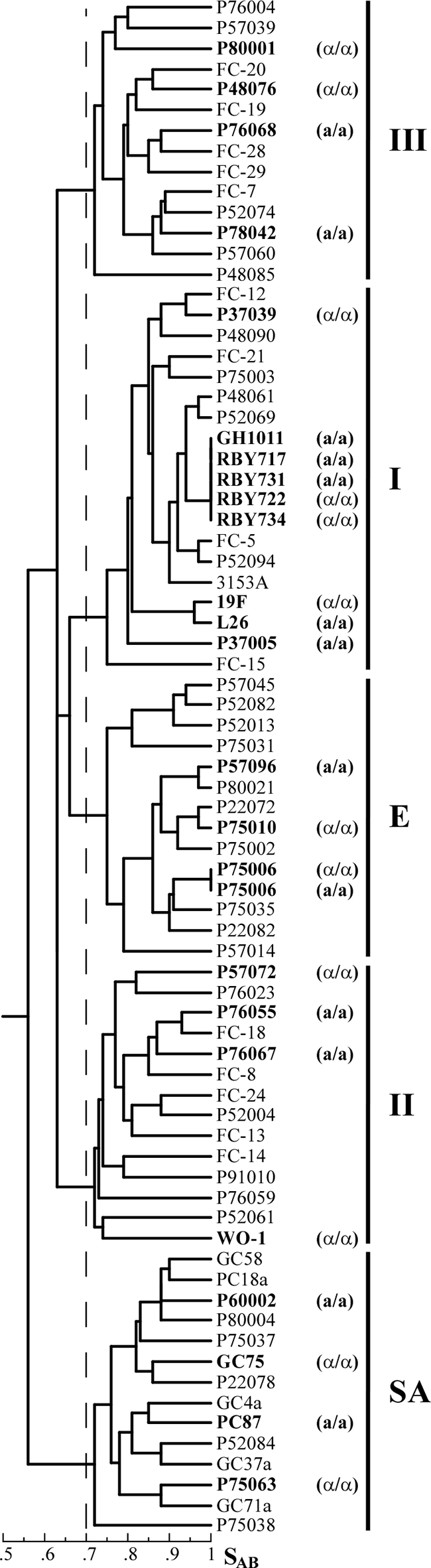
Strains exhibiting the white cell response to α-pheromone or a-pheromone are distributed throughout the major clades of C. albicans. Two or more strains from each of the five major clades of C. albicans, I, II, III, E, and SA (11), were tested for and found to exhibit the white cell response to pheromone. In the dendrogram generated, the Ca3 hybridization patterns of 25 of the test strains or substrains that exhibited the white cell response and had been genetically fingerprinted with the DNA fingerprinting probe Ca3 (12) were compared to the patterns of 50 strains representing the five major clades by com- puting similarity coefficients (SAB), using the DENDRON software program (12). The MTL-homozygous strains that were tested for the white cell response are presented in bold print. The five major clades are labeled to the right of the dendrogram. The dashed line represents the threshold for clades (11).
Concluding remarks.
We have, therefore, found that a robust white cell response to pheromone occurred in all of the 27 MTL-homozygous strains tested, both in a/a and in α/α representatives of the five major clades of C. albicans. We have found that white a/a cells responded to α-pheromone similarly to white α/α cells responding to a-pheromone and that the basic white cell response occurred in a variety of nutrient media. Moreover, we found that all of the tested a/a and α/α derivatives of the common laboratory strain SC5314 underwent the white cell response to their respective pheromones and did so with a robustness similar to that of white cells of strain P37005 and the other strains tested. Variation was observed in strength of response among strains and media, but the level of induction by pheromone was still robust for each tested strain and in every medium. The changes in the global expression pattern induced by α-pheromone in white cells of laboratory strain RBY717 were demonstrated by Bennett and Johnson (3), using microarray technology, to be weaker than the changes induced in the global expression pattern of the natural strain P37005 in each of the two media. The expression patterns of RBY717 were also demonstrated to be sensitive to the composition of the supporting medium. The combined results, however, suggest that the increase in adhesion and upregulation of receptor genes and white cell-specific genes provide more-specific indicators of the white cell response to pheromone than changes in global expression patterns assessed by microarrays. More importantly, our results demonstrate that the white cell response to pheromone is a general characteristic of C. albicans, as is the opaque cell response to pheromone (2, 8).
Supplementary Material
Acknowledgments
We are indebted to Richard Bennett for generously supplying us with strains.
This research was funded by NIH grant AI2393 and the Developmental Studies Hybridoma Bank-Microbe, a National Resource under the auspices of NIH.
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bedell, G. W., and D. R. Soll. 1979. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect. Immun. 26348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. J., M. A. Uhl, M. G. Miller, and A. D. Johnson. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 238189-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, R. J., and A. D. Johnson. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 62100-119. [DOI] [PubMed] [Google Scholar]
- 4.Blignaut, E., C. Pujol, S. Lockhart, S. Joly, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa. J. Clin. Microbiol. 40826-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniels, K. J., T. Srikantha, S. R. Lockhart, C. Pujol, and D. R. Soll. 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 252240-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 955150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lockhart, S. R., C. Pujol, K. J. Daniels, M. G. Miller, A. D. Johnson, M. A. Pfaller, and D. R. Soll. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockhart, S. R., K. J. Daniels, R. Zhao, D. Wessels, and D. R. Soll. 2003. Cell biology of mating in Candida albicans. Eukaryot. Cell 249-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110293-302. [DOI] [PubMed] [Google Scholar]
- 10.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soll, D. R., and C. Pujol. 2003. Candida albicans clades. FEMS Immunol. Med. Microbiol. 391-7. [DOI] [PubMed] [Google Scholar]
- 12.Soll, D. R., C. Pujol, and S. R. Lockhart. 2007. Laboratory procedures for the epidemiological analysis of microorganisms, p. 129-151. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 13.Soll, D. R. 2008. Candida biofilms: is adhesion sexy? Curr. Biol. 18R717-R720. [DOI] [PubMed] [Google Scholar]
- 14.Srikantha, T., A. R. Borneman, K. J. Daniels, C. Pujol, W. Wu, M. R. Seringhaus, M. Gerstein, S. Yi, M. Snyder, and D. R. Soll. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 51674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, W., C. Pujol, S. R. Lockhart, and D. R. Soll. 2005. Chromosome loss followed by duplication is the major mechanism of spontaneous mating-type locus homozygosis in Candida albicans. Genetics 1691311-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi, S., N. Sahni, K. J. Daniels, C. Pujol, T. Srikantha, and D. R. Soll. 2008. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol. Biol. Cell 19957-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.