Abstract
Leptospira interrogans is responsible for leptospirosis, a zoonosis of worldwide distribution. LipL32 is the major outer membrane protein of pathogenic leptospires, accounting for up to 75% of total outer membrane protein. In recent times LipL32 has become the focus of intense study because of its surface location, dominance in the host immune response, and conservation among pathogenic species. In this study, an lipL32 mutant was constructed in L. interrogans using transposon mutagenesis. The lipL32 mutant had normal morphology and growth rate compared to the wild type and was equally adherent to extracellular matrix. Protein composition of the cell membranes was found to be largely unaffected by the loss of LipL32, with no obvious compensatory increase in other proteins. Microarray studies found no obvious stress response or upregulation of genes that may compensate for the loss of LipL32 but did suggest an association between LipL32 and the synthesis of heme and vitamin B12. When hamsters were inoculated by systemic and mucosal routes, the mutant caused acute severe disease manifestations that were indistinguishable from wild-type L. interrogans infection. In the rat model of chronic infection, the LipL32 mutant colonized the renal tubules as efficiently as the wild-type strain. In conclusion, this study showed that LipL32 does not play a role in either the acute or chronic models of infection. Considering the abundance and conservation of LipL32 among all pathogenic Leptospira spp. and its absence in saprophytic Leptospira, this finding is remarkable. The role of this protein in leptospiral biology and pathogenesis thus remains elusive.
Leptospira interrogans is a zoonotic spirochete with a worldwide distribution. In the chronic carrier state, host animals such as rats do not exhibit overt disease but are colonized by Leptospira in their renal tubules and shed bacteria in their urine. Humans are incidental hosts that become infected through exposure to contaminated water, soil, or urine. In the acute form of leptospirosis, disease severity ranges from asymptomatic infection to multiple organ failure, pulmonary hemorrhage, and death (13).
The cellular and molecular mechanisms of leptospiral pathogenesis remain unclear. One of the major sites of interaction with the host is the bacterial outer membrane (OM). Analysis of the L. interrogans OM has identified a number of proteins (11, 12), the most abundant of which is LipL32, a 32-kDa lipoprotein estimated to account for a remarkable 75% of the OM proteome (11). LipL32 is also the most abundant surface-exposed protein (12).
LipL32 is found in all pathogenic species tested to date and is highly conserved, with average amino acid identity over 98%, but it is not found in saprophytic species (16, 17). LipL32 is expressed during both chronic and acute infection and is highly immunogenic (15, 16, 27). These features have generated interest in LipL32 as a potential diagnostic reagent in both PCRs (21) and enzyme-linked immunosorbent assays (14). There has also been much interest in the potential of LipL32 to generate heterologous immunity, overcoming the limitations of serovar-specific immunity. However, to date LipL32-based vaccines have met with limited success (5, 6).
The abundance, conservation, unique presence in pathogenic species, and immunogenicity of LipL32 are consistent with an important role in pathogenesis. Available microarray data provide little insight into the role of LipL32 since gene expression is unchanged under conditions of different temperatures (22). Recent studies have shown that LipL32 may act as an adhesin binding to collagen, laminin, and fibronectin (18, 19) while LipL32 has also been associated with hemolysis (20). However, the precise role of LipL32 in pathogenesis remains unknown.
In this study an lipL32 mutant was constructed by transposon mutagenesis. To our surprise, analysis of this mutant in the hamster model of acute infection and rat model of chronic infection showed that LipL32 is not required for causing either acute leptospirosis or renal colonization.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. interrogans serovar Manilae strain L495 was obtained from N. Koizumi, National Institute of Infectious Diseases, Tokyo, Japan. Mutant M777 is a TnSC189 mutant constructed from strain L495 with an insertion after base 3979065 (L. interrogans serovar Lai genome) and retains virulence (25). Bacteria were grown in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (Becton Dickinson) at 30°C without aeration. Plates were made by solidification of medium with 1.5% agar. Kanamycin (25 μg/ml) was added where appropriate. To examine survival in water, mid-log-phase leptospires were centrifuged to remove culture medium, resuspended in sterile deionized water, and then inoculated into 5 ml of sterile deionized water in triplicate at a concentration of 108 leptospires/ml. Samples were removed at regular intervals for enumeration using a Petroff-Hausser chamber.
Transposon mutagenesis.
Transposon mutagenesis was conducted as described previously (4). In brief, leptospires were grown to late log phase and made electrocompetent by repeated washing in ultrapure water. Plasmid pSC189 (8) containing transposon TnSC189 was introduced into cells by electroporation, and transformants were selected on EMJH plates containing kanamycin. The location of transposon insertion was determined by sequencing directly from the chromosome (24). PCR primers used to confirm the mutant genotype were 3453 (5′-CGTCATGGTCTTTGTAGTCTATGG-3′), annealing to the 5′ end of the transposon; 4777 (5′-AAAAGATCTTTAACCAACAGATGCAACGAAAGA-3′), annealing to the 3′ end of lipL32; and 4956 (5′-TTTTGATATCTAAGAATTTCTGATGTTTCAATCGT-3′), annealing to the intergenic region proximal to lipL32.
Protein analysis techniques and leptospiral adhesion assays.
Preparation of total membranes was carried out as described previously (19). Western blotting and one-dimensional and two-dimensional gel electrophoresis were carried out by standard methods (11). Spots from two-dimensional gels were excised and analyzed by mass spectrometry or identified through Western blotting with anti-LipL32 serum kindly provided by D. Haake (16). Adhesion of leptospires to Matrigel (Becton Dickinson), laminin, and collagen (Sigma) and the laminin binding blot were assayed as described previously (19).
Microarray studies.
Microarray analysis comparing M933 and intergenic mutant M777 was performed using three biological replicates, each with a dye swap, resulting in six arrays. RNA purification, probe preparation, hybridizations, and analysis were carried out as described previously (22).
Hamster infection model.
Golden hamsters were infected by one of two routes. For systemic infection, groups of eight hamsters were injected intraperitoneally with 103 leptospires in 100 μl of EMJH medium and monitored for 14 days. Tissue was fixed in 10% formalin for histopathology analysis. For the mucosal route of infection, groups of 10 hamsters were inoculated with 106 leptospires instilled onto the eye in a volume of 10 μl in EMJH medium. In accordance with animal ethics requirements, moribund animals were euthanized.
Rat infection model.
Groups of six-week-old Wistar rats (eight per group) were inoculated intraperitoneally with 108 leptospires in 1 ml of EMJH medium as described previously (1). Control animals were injected with 1 ml of sterile EMJH medium. We have shown previously that colonization is well established by 15 days and correlates with long-term carriage (1). After 15 days animals were euthanized. Kidney tissue was passed through a sterile syringe, homogenized in 5 ml of EMJH liquid medium, and reinoculated into 5 ml of liquid EMJH medium. Cultures were maintained at 30°C and examined weekly for growth by dark-field microscopy for up to 6 weeks postinoculation.
Touch preparations of sectioned kidney were used for the direct visualization of leptospires. Kidney fragments were spotted onto poly-l-lysine (Sigma-Aldrich)-coated glass slides, which were then dried and fixed in acetone for 3 min. Leptospires were visualized by immunofluorescence as described below. Additional kidney fragments were fixed in 4% formalin, embedded in paraffin, and cut into 4- to 5-μm sections for conventional histology and immunohistochemical studies. Immunofluorescence labeling was performed using a protocol modified from Cullen et al. (12). Slides were washed twice with phosphate-buffered saline (PBS) after fixation, blocked twice with 2% bovine serum albumin (Sigma) (PBS-BSA) for 20 min each at 30°C, and incubated for 1 h with rabbit antiserum to whole L. interrogans serovar Icterohaemorrhagiae (diluted 1:200 in PBS-BSA) or control normal rabbit serum at 30°C. Slides were washed with PBS-BSA and incubated with fluorescein-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories) for 1 h at 30°C. Slides were washed, mounted in antifading solution (Prolong-Molecular Probes), and visualized by fluorescence microscopy (Olympus BX51).
Kidney tissue sections were treated according to routine histological procedures, and sections were then stained with hematoxylin and eosin. For immunohistochemical analyses, paraffin was removed from tissue sections with xylene and ethanol. Slides were blocked with 10% powdered milk at room temperature for 20 min and then incubated with a 1:1,000 dilution of rabbit antiserum against LipL32 or whole leptospires at room temperature for 1 h. Slides were treated with 0.3% hydrogen peroxide for 15 min at room temperature and then incubated at room temperature for 30 min with peroxidase-conjugated anti-rabbit immunoglobulin (Histostain-Bulk Kit; Invitrogen). Enzyme reactions were developed using 3,3′-diaminobenzidine (Sigma). Slides were examined in a blinded manner to prevent bias in the interpretation of the results.
RESULTS
Construction of an L. interrogans lipL32 mutant.
Random transposon mutagenesis was performed on L. interrogans serovar Manilae as described previously (4), and the location of transposon insertion was determined by direct sequencing from the chromosome (24). In mutant M933 the transposon was determined to have inserted after base 2620245 of the Lai genome, 177 bp into the 816-bp gene encoding LipL32 (also known as LA2637 or LIC11352). An intergenic mutant (M777) constructed from the same parent strain with TnSC189 was also used as a control in some experiments (25). The growth rate of M933 in EMJH medium with kanamycin was slightly slower than that of wild-type L. interrogans or M777 (data not shown). By dark-field microscopy, M933 had a slightly less uniform cell size, but otherwise bacteria were motile and appeared normal (data not shown). The mutation was confirmed by PCR.
Characterization of the OM of the lipL32 mutant.
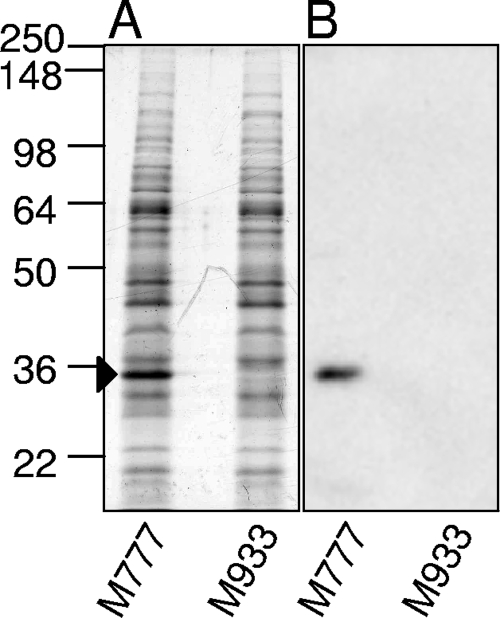
LipL32 is the major protein of the OM, accounting for an estimated 75% of OM protein (11). Immunoblotting with specific LipL32 antiserum showed that a band of approximately 32 kDa was absent in the mutant (Fig. 1). Two-dimensional gel electrophoresis analysis of membrane preparations showed no difference in protein profiles between L. interrogans serovar Manilae wild type and strain M933 except for the absence of LipL32 (data not shown).
FIG. 1.
Confirmation of the lipL32 mutant phenotype. Whole-cell lysates of M933 (lipL32 mutant) and M777 (control) were analyzed by SDS-PAGE with Coomassie blue staining (A) and by Western blotting with anti-LipL32 serum (B). The band corresponding to LipL32 is indicated by an arrow. Numbers on the left correspond to approximate molecular sizes (kDa).
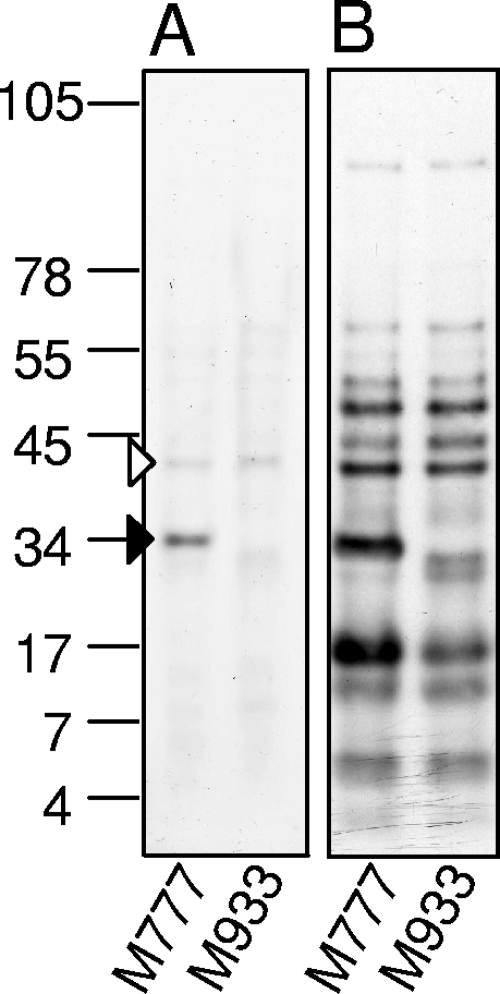
As LipL32 is the major OM protein, surface biotinylation was performed to determine the major surface-exposed proteins in the absence of LipL32. Biotinylated proteins were purified on a streptavidin column and examined on a large-format 7.5 to 15% gradient Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel with Coomassie blue staining or by streptavidin blotting. LipL41 was found to be the major OM protein in mutant M933 (Fig. 2).
FIG. 2.
Detection of surface-exposed proteins in the lipL32 mutant. M933 (lipL32 mutant) or M777 (control) cells were surface labeled with biotin, and then surface-exposed proteins were purified on a streptavidin column and separated on a gel. Detection was performed by Coomassie blue staining or streptavidin-horseradish peroxidase detection. The dominant bands correspond to LipL32 (solid arrow head) and LipL41 (open arrow head). Numbers on the left correspond to approximate molecular sizes (kDa).
Microarray analysis of the lipL32 mutant.
Loss of the major membrane protein may have potential consequences for the expression of genes encoding other membrane components, potentially providing an indication of function for LipL32. Microarray analysis was conducted to compare gene expression of M933 and the control strain M777. The same samples were used for both two-dimensional gel analysis and microarray analysis. A total of 34 genes (0.96% of total) were upregulated in M933 compared to M777, while 12 genes (0.34%) were downregulated (1.5-fold cut off; P < 0.01) (see Table S1 in the supplemental material).
Of the 34 upregulated genes, 18 (53%) had no known function. There was an overrepresentation of genes from cluster of orthologous groups functional category H (coenzyme transport and metabolism), accounting for 21% of upregulated genes, compared to a 3% total in the genome. These genes were from the heme and vitamin B12 (cobalamin) synthesis operons. The strongest upregulated protein was LA3881, a hypothetical lipoprotein with alpha integrin-like repeat domains (2.6-fold).
Of the 12 downregulated genes, 7 (58%) had no function attributed. Expression of lipL32 was very low and likely to be a background signal (0.035-fold). The most strongly downregulated gene was that encoding a putative TonB-dependent receptor, LA2641 (0.072-fold). LA2641 is located on the large chromosome and unlinked to the genes for B12 and heme synthesis that are found on the small chromosome.
Adherence to extracellular matrix.
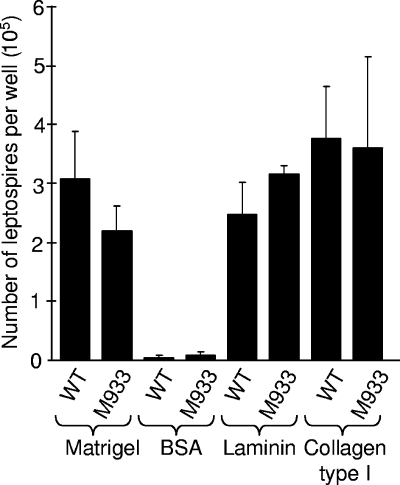
It has previously been shown that LipL32 is an adhesin that binds to extracellular matrix (18, 19). A leptospiral adhesion assay (19) was used to determine the adhesion of M933 to a commercially available extracellular matrix preparation, Matrigel. M933 and M777 bound equally to Matrigel-coated microtiter tray wells (Fig. 3). There was likewise no difference in binding to trays coated with collagen type I or laminin.
FIG. 3.
Leptospiral cell adhesion assay. Leptospires were incubated in microtiter tray wells coated with the indicated substrate for 1 h. After a washing step, adherent leptospires were released by trypsin for direct counting by microscopy. A subset of these data appeared in a previous study (19). Error bars correspond to 1 standard deviation. WT, wild type.
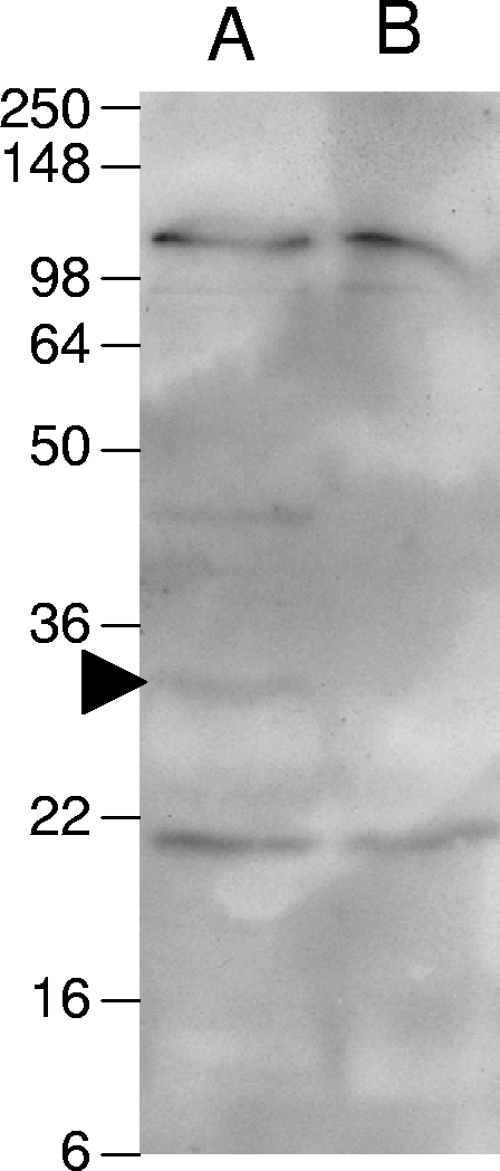
To investigate the presence in the mutant of other molecules that adhere to extracellular matrix, blots of total membrane were probed with biotinylated laminin. Several bands were found to bind to laminin in both the mutant and the wild type, except for the band at approximately 32 kDa corresponding to LipL32 (Fig. 4). This supports the above data indicating loss of LipL32 in mutant M933 and also demonstrates the presence of additional laminin-binding proteins in M933. An additional band between the 36- and 50-kDa markers was observed in the wild-type sample but not in the mutant. It is possible that this corresponds to a dimer of LipL32.
FIG. 4.
Laminin binding blot. Bacterial membrane preparations were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was probed with biotinylated laminin and then detected with streptavidin-horseradish peroxidase. The arrowhead indicates the band corresponding to LipL32. The positions of molecular mass markers (kDa) are indicated on the left.
LipL32 is not required for acute infection in hamsters.
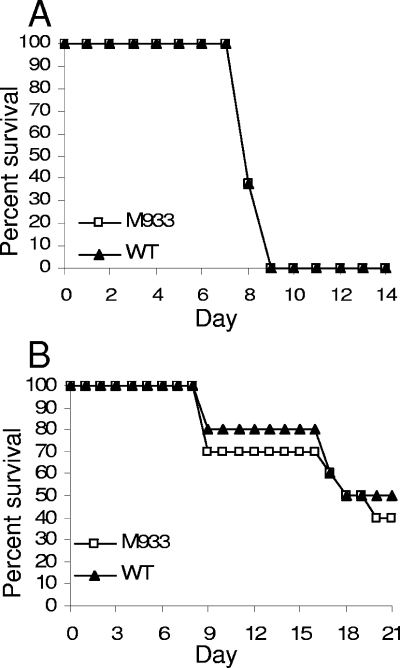
To analyze the role of LipL32 in the acute model of infection, hamsters were injected intraperitoneally with 103 leptospires. The courses of infection for wild-type serovar Manilae and M933 were indistinguishable, with all hamsters succumbing to infection between days 7 and 9 (Fig. 5 A). Infection with a lower dose of 102 leptospires also resulted in no survival for either the wild-type strain (n = 8 hamsters) or M933 (n = 4), indicating a 100% lethal dose of <102 leptospires for both strains and showing that there was no subtle attenuation of the lipL32 mutant.
FIG. 5.
Survival of hamsters inoculated with M933 (lipL32 mutant) or wild-type (WT) Manilae. (A) Groups of eight hamsters were inoculated with 103 leptospires intraperitoneally and monitored for 14 days. (B) Groups of 10 hamsters were inoculated with 106 leptospires via the ocular route and monitored for 21 days.
Histopathological examination of tissues found no difference between the mutant and the wild type. The lungs showed mild to moderate multifocal alveolar hemorrhage while the livers showed diffuse dissociation and single-cell necrosis of hepatocytes with abundant leptospires in the sinusoids. Kidneys from both wild-type- and M933-infected hamsters showed multifocal tubular necrosis, mild tubular proteinuria, and rare hemoglobin casts with numerous leptospires in affected areas. The genotype of M933 reisolated from hamster kidneys was confirmed by PCR using transposon- and gene-specific primers.
Although the loss of LipL32 did not affect the ability of leptospires to cause acute systemic disease after intraperitoneal inoculation, LipL32 could play a role early during natural infection when the pathogen penetrates host skin or mucosa prior to dissemination. To evaluate this possibility, hamsters were inoculated with 106 leptospires via the conjunctiva. The survival curve for M933 again closely resembled that for wild-type serovar Manilae, indicating no difference in virulence (Fig. 5B).
LipL32 is not required for chronic infection in the rat.
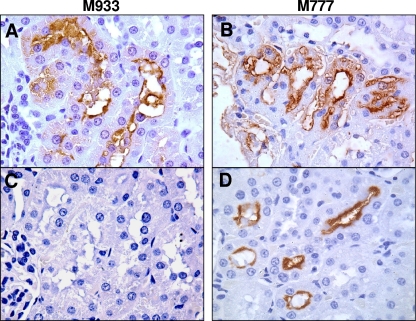
In carrier hosts such as rats, L. interrogans can establish chronic infections where bacteria colonize the proximal renal tubules and are subsequently shed in the urine. To examine the role of LipL32 in this process, M933, M777, and L. interrogans serovar Copenhageni strain Fiocruz L1-130 were injected into groups of eight rats at 108 leptospires per animal. After 14 days the kidneys were removed and examined for colonization by culture isolation and immunofluorescence detection of leptospires in kidney touch preparations. In two experiments all eight animals infected with M933 were colonized (Table 1). This was equal to or greater than the colonization rate observed in rats inoculated with control strains. Histopathological analysis was also conducted on rat kidneys to visualize colonization. Immunohistochemical staining using antiserum to whole bacteria detected leptospires in the lumen of tubules from rats infected with both M933 and M777 (Fig. 6, top row). When anti-LipL32 serum was used, leptospires were detected only in the kidneys of animals infected with the M777 control (bottom row). All reisolated bacteria were shown by PCR to retain the transposon in the original chromosomal location (data not shown).
TABLE 1.
Colonization of rat kidneys in the chronic infection model
| Strain | Colonization of rat kidneys (no. of positive animals/total no. of animals tested)
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| IFA (%)a | Culture (%)b | IFA (%)a | Culture (%)b | |
| M933 | 8/8 (100) | 8/8 (100) | 8/8 (100) | 8/8 (100) |
| M777 | 8/8 (100) | 5/6 (83) | 6/8 (75) | 6/7 (86) |
| Fiocruz L1-130 | 8/8 (100) | 8/8 (100) | ||
IFA, immunofluorescence assay on kidney touch preparations.
Not all kidney cultures could be interpreted due to contamination.
FIG. 6.
Rat colonization by M933 (lipL32 mutant) and M777 (control). Representative photomicrographs of immunohistochemically stained sectioned kidneys from rats infected with the indicated strain. Detection was performed with antiserum to whole Leptospira organisms (A and B) or to LipL32 (C and D). Photomicrographs were taken at a magnification of ×1,000.
LipL32 is not required for survival in water.
Since LipL32 did not appear to play a role in infection, it was hypothesized that the protein may be required for survival in the environment. To examine survival in water, the mutant M933, M777 (control), and Manilae wild-type strains were centrifuged to remove culture medium and inoculated into sterile deionized water. There was no difference in survival curves over 7 days of counting. After this time all strains lost structural integrity, making identification of live organisms for counting impossible. After 2 weeks of incubation, 100-μl samples were inoculated into 5 ml of EMJH medium; motile leptospires were observed in these cultures after 1 week of incubation, indicating that viability was retained after 2 weeks in water.
DISCUSSION
This study describes the construction and characterization of a lipL32 mutant of L. interrogans. A large body of evidence suggests that LipL32 is an important protein in Leptospira; LipL32 is by far the most abundant protein in the OM and on the cell surface of pathogenic leptospires, and LipL32 is expressed in vivo and in vitro and stimulates a strong immune response. LipL32 is also absent in saprophytic Leptospira spp. while being highly conserved among different pathogenic isolates. It was therefore surprising that the protein is not essential for the survival of L. interrogans: a LipL32 mutant exhibits only a slightly delayed growth rate and relatively normal morphology. The slower growth and change in uniformity of cell size might suggest a degree of stress in the mutant; however, this was not supported by the microarray data, which failed to show a general stress response. Taken together, these data suggest that the integrity of the OM was not compromised by the loss of LipL32.
Animal infection experiments showed unequivocally that LipL32 is not required for acute infection in hamsters; mortality in M933-infected hamsters and the pathology of lesions in target organs were found to be the same as those observed in wild-type-infected hamsters. When the mutant strain was administered through a more natural route of infection, the conjunctiva, it was still virulent, indicating that LipL32 is not required for infection by the mucosal route. L. interrogans can cause chronic infection in carrier hosts such as rats and dogs, where organisms colonize the renal tubules of the kidneys and are shed in the urine. Further animal studies found that LipL32 is also not required for chronic infection of rats since animal kidneys were colonized when infected with either M933 or the control mutant M777. It is possible that LipL32 is required for virulence in host species not tested in this study or in mechanisms of pathogenesis that were not stringently tested in our model systems. Alternatively, the role of LipL32 may be compensated for by other proteins in the OM although we observed no significant change in the profile of the OM subproteome. To date, only two potential L. interrogans virulence factors have been characterized by transposon mutagenesis, the OmpA-like protein Loa22 (29) and the heme oxygenase enzyme, HemO (26).
Recent studies have shown that LipL32 binds to extracellular matrix proteins (18, 19). In this study the lipL32 mutant showed the same level of adhesion to extracellular matrix as wild-type L. interrogans serovar Manilae. A number of other potential leptospiral adhesins were highlighted in an extracellular matrix binding blot, indicating that the adhesin function of LipL32 may be redundant. Additionally, the Lig and Len families of proteins and Lsa21 have all been assigned an adhesin role (2, 3, 9, 32). This study adds to a growing body of evidence that L. interrogans has a high degree of redundancy in virulence mechanisms. Recently, several other potential virulence factors with putative virulence roles (such as adhesion or factor H binding) were determined not to be essential for virulence including ligB (10) and ligC, lenB, and lenE (25). Redundancy is common in bacterial systems. For example, Haemophilus influenzae isolates have up to four hgp hemoglobin binding proteins, each of which is sufficient alone for normal hemoglobin-haptoglobin complex utilization (23).
It is a reasonable conclusion that the absence of LipL32 in the mutant would be compensated for by an increase in other OM proteins. Interestingly, the OM protein profile of the LipL32 mutant was largely the same as that for a control strain. LipL41 was found to be the most abundant surface protein in the absence of LipL32. In Borrelia burdorferi OspC is a major OM protein that is required for certain phases of virulence. Mutation of ospC can be largely compensated for by the overexpression of other lipoproteins (33). There was no evidence for compensation for the loss of LipL32 in L. interrogans. It is possible that existing proteins in the OM can assume the function of LipL32 without a noticeable increase in expression.
Although LipL32 is unlikely to have a transcriptional role in L. interrogans, microarray analysis was conducted to identify transcriptional changes induced by the loss of such an abundant lipoprotein. Results did not indicate a general stress response in the mutant. There were few upregulated lipoproteins that may compensate for the loss of LipL32. In general, results indicate a lack of obvious compensation for loss of LipL32. However, there was an upregulation of heme and vitamin B12 synthesis pathways, suggesting an association between LipL32 in the uptake or metabolism of these cofactors. Heme and vitamin B12 synthesis share common pathways in bacteria (31). The change in expression of these genes may be a response to the strong downregulation of TonB-dependent receptor, LA2641, which may have a role in cofactor uptake. Alternatively, general disruption of the OM proteome may otherwise prevent transport of B12 across the OM. The possibility cannot be excluded that secondary suppressor mutations may mask obvious phenotypes in this strain and lead to unexpected transcriptional changes.
Since our findings indicate that LipL32 is not required for normal in vitro growth or pathogenesis in different infection models, the role of this protein remains enigmatic. The aspects of the environmental phase of transmission of Leptospira are not well characterized. Although the LipL32 mutant did not show decreased ability to survive in water, it is possible that LipL32 may be important in the normal transmission or environmental phase of the life cycle. We recently showed that an ortholog of LipL32 is found in Pseudoalteromonas tunicata, a marine bacterium that is associated with tunicates (19). However, LipL32 is retained in Leptospira borgpetersenii, which does not persist well outside the host and has significantly reduced its genome to reflect this (7). Furthermore, LipL32 is absent in the saprophytic species Leptospira biflexa, which has only an environmental existence (28). L. interrogans has been shown to form biofilms (30), but the role of LipL32 in this process is unknown.
In conclusion, it is intriguing that L. interrogans produces LipL32 at very high levels at great metabolic cost when it does not appear to be essential for survival or virulence. Considering the many aspects of LipL32 expression and conservation that indicate it is an important protein, LipL32 remains a paradox.
Supplementary Material
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC) Australia, Australian Research Council, the Brazilian National Research Council (Instituto Milênio 420067/2005), and the National Institutes of Health (R01 AI052473, R01 AI034431, and D43 TW00919). G.L.M. is supported by an NHMRC Peter Doherty Fellowship.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 22 December 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Athanazio, D. A., E. F. Silva, C. S. Santos, G. M. Rocha, M. A. Vannier-Santos, A. J. McBride, A. I. Ko, and M. G. Reis. 2008. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta Tropica 105176-180. [DOI] [PubMed] [Google Scholar]
- 2.Atzingen, M. V., A. S. Barbosa, T. De Brito, S. A. Vasconcellos, Z. M. de Morais, D. M. Lima, P. A. Abreu, and A. L. Nascimento. 2008. Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiol. 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa, A. S., P. A. Abreu, F. O. Neves, M. V. Atzingen, M. M. Watanabe, M. L. Vieira, Z. M. Morais, S. A. Vasconcellos, and A. L. Nascimento. 2006. A newly identified leptospiral adhesin mediates attachment to laminin. Infect. Immun. 746356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourhy, P., H. Louvel, I. Saint Girons, and M. Picardeau. 2005. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 1873255-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branger, C., B. Chatrenet, A. Gauvrit, F. Aviat, A. Aubert, J. M. Bach, and G. Andre-Fontaine. 2005. Protection against Leptospira interrogans sensu lato challenge by DNA immunization with the gene encoding hemolysin-associated protein 1. Infect. Immun. 734062-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branger, C., C. Sonrier, B. Chatrenet, B. Klonjkowski, N. Ruvoen-Clouet, A. Aubert, G. Andre-Fontaine, and M. Eloit. 2001. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect. Immun. 696831-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulach, D. M., R. L. Zuerner, P. Wilson, T. Seemann, A. McGrath, P. A. Cullen, J. Davis, M. Johnson, E. Kuczek, D. P. Alt, B. Peterson-Burch, R. L. Coppel, J. I. Rood, J. K. Davies, and B. Adler. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA 10314560-14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, S. L., and E. J. Rubin. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296179-185. [DOI] [PubMed] [Google Scholar]
- 9.Choy, H. A., M. M. Kelley, T. L. Chen, A. K. Moller, J. Matsunaga, and D. A. Haake. 2007. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 752441-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croda, J., C. P. Figueira, E. A. Wunder, Jr., C. S. Santos, M. G. Reis, A. I. Ko, and M. Picardeau. 2008. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect. Immun. 765826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen, P. A., S. J. Cordwell, D. M. Bulach, D. A. Haake, and B. Adler. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 702311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, P. A., X. Xu, J. Matsunaga, Y. Sanchez, A. I. Ko, D. A. Haake, and B. Adler. 2005. Surfaceome of Leptospira spp. Infect. Immun. 734853-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis. MediSci, Melbourne, Australia.
- 14.Flannery, B., D. Costa, F. P. Carvalho, H. Guerreiro, J. Matsunaga, E. D. Da Silva, A. G. Ferreira, L. W. Riley, M. G. Reis, D. A. Haake, and A. I. Ko. 2001. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J. Clin. Microbiol. 393303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerreiro, H., J. Croda, B. Flannery, M. Mazel, J. Matsunaga, M. Galvao Reis, P. N. Levett, A. I. Ko, and D. A. Haake. 2001. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 694958-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 682276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haake, D. A., M. A. Suchard, M. M. Kelley, M. Dundoo, D. P. Alt, and R. L. Zuerner. 2004. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 1862818-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauk, P., F. Macedo, E. C. Romero, S. A. Vasconcellos, Z. M. de Morais, A. S. Barbosa, and P. L. Ho. 2008. LipL32, the major leptospiral lipoprotein: the C terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infect. Immun. 762642-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoke, D. E., S. Egan, P. A. Cullen, and B. Adler. 2008. LipL32 is an extracellular matrix-interacting protein of Leptospira spp. and Pseudoalteromonas tunicata. Infect. Immun. 762063-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, S. H., K. A. Kim, Y. G. Park, I. W. Seong, M. J. Kim, and Y. J. Lee. 2000. Identification and partial characterization of a novel hemolysin from Leptospira interrogans serovar Lai. Gene 25419-28. [DOI] [PubMed] [Google Scholar]
- 21.Levett, P. N., R. E. Morey, R. L. Galloway, D. E. Turner, A. G. Steigerwalt, and L. W. Mayer. 2005. Detection of pathogenic leptospires by real-time quantitative PCR. J. Med. Microbiol. 5445-49. [DOI] [PubMed] [Google Scholar]
- 22.Lo, M., D. M. Bulach, D. R. Powell, D. A. Haake, J. Matsunaga, M. L. Paustian, R. L. Zuerner, and B. Adler. 2006. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 745848-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 672729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray, G. L., K. M. Ellis, M. Lo, and B. Adler. 2008. Leptospira interrogans requires a functional heme oxygenase to scavenge iron from hemoglobin. Microbes Infect. 10791-797. [DOI] [PubMed] [Google Scholar]
- 25.Murray, G. L., V. Morel, G. M. Cerqueira, J. Croda, A. Srikram, R. Henry, A. I. Ko, O. A. Dellagostin, D. M. Bulach, R. Sermswan, B. Adler, and M. Picardeau. 2009. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect. Immun. 77810-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray, G. L., A. Srikram, R. Henry, A. Puapairoj, R. W. Sermswan, and B. Adler. 16 December 2008. Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. doi. 10.1016/j.micinf.2008.11.014. [Epub ahead of print.] [DOI] [PubMed]
- 27.Nally, J. E., J. P. Whitelegge, S. Bassilian, D. R. Blanco, and M. A. Lovett. 2007. Characterization of the outer membrane proteome of Leptospira interrogans expressed during acute lethal infection. Infect. Immun. 75766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picardeau, M., D. M. Bulach, C. Bouchier, R. L. Zuerner, N. Zidane, P. J. Wilson, S. Creno, E. S. Kuczek, S. Bommezzadri, J. C. Davis, A. McGrath, M. J. Johnson, C. Boursaux-Eude, T. Seemann, Z. Rouy, R. L. Coppel, J. I. Rood, A. Lajus, J. K. Davies, C. Medigue, and B. Adler. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS ONE 3e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ristow, P., P. Bourhy, F. W. da Cruz McBride, C. P. Figueira, M. Huerre, P. Ave, I. S. Girons, A. I. Ko, and M. Picardeau. 2007. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ristow, P., P. Bourhy, S. Kerneis, C. Schmitt, M. C. Prevost, W. Lilenbaum, and M. Picardeau. 2008. Biofilm formation by saprophytic and pathogenic leptospires. Microbiology 1541309-1317. [DOI] [PubMed] [Google Scholar]
- 31.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 27841148-41159. [DOI] [PubMed] [Google Scholar]
- 32.Stevenson, B., H. A. Choy, M. Pinne, M. L. Rotondi, M. C. Miller, E. Demoll, P. Kraiczy, A. E. Cooley, T. P. Creamer, M. A. Suchard, C. A. Brissette, A. Verma, and D. A. Haake. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS ONE 2e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, Q., K. McShan, and F. T. Liang. 2008. Essential protective role attributed to the surface lipoproteins of Borrelia burgdorferi against innate defences. Mol. Microbiol. 6915-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.