Abstract
We identified the mutated gene locus in a pigment-overproducing Vibrio cholerae mutant of strain A1552. The deduced gene product is suggested to be an oxidoreductase based on partial homology to putative homogentisate 1,2-dioxygenase in Pseudomonas aeruginosa and Mesorhizobium loti, and we propose that the gene VC1345 in the V. cholerae genome be denoted hmgA in accordance with the nomenclature for other species. The hmgA::mini-Tn5 mutant showed a nonpigmented phenotype after complementation with a plasmid clone carrying the WT hmgA+ locus. Microarray transcription analysis revealed that expression of hmgA and the neighboring genes encoding a postulated two-component sensor system was growth phase dependent. Results from quantitative reverse transcription-PCR analysis showed that hmgA operon expression was reduced in the rpoS mutant, but pigment production by the WT V. cholerae or the hmgA mutant was not detectably influenced by the stationary-phase regulator RpoS. The pigmented mutant showed increased UV resistance in comparison with the WT strain. Interestingly, the pigment-producing mutant expressed more toxin-coregulated pilus and cholera toxin than WT V. cholerae. Moreover, the hmgA mutant showed a fivefold increase in the ability to colonize the intestines of infant mice. A possible mechanism by which pigment production might cause induction of the ToxR regulon due to generation of hydrogen peroxide was supported by results from tests showing that externally supplied H2O2 led to higher TcpA levels. Taken together, our findings suggest that melanin pigment formation may play a role in V. cholerae virulence factor expression.
The bacterium Vibrio cholerae causes the severe diarrheal disease cholera, and the primary virulence factor is the secreted cholera toxin (CTX), which is responsible for the clinical aspects of cholera disease (18). The other major virulence factor produced by V. cholerae is the toxin-coregulated pilus, a type IV bundle-forming pilus required for intestinal colonization by V. cholerae (1, 9, 29, 30). Expression of the V. cholerae virulence genes is regulated by a cascade of transcription factors.
Melanization has been considered to be important in microbial pathogenesis because it has been associated with virulence in many microorganisms (7, 24). Melanin appears to contribute to virulence by reducing the susceptibility of melanized microbes to host defense mechanisms. Although melanin pigments are not considered essential for the growth of microorganisms, they can increase the survival of microorganisms by protecting DNA and other molecules from UV light, enhance virulence, inhibit biofilm degradation, protect enzymes from proteases, and protect microorganisms from hydrolytic enzymes, and they may even act as proton and nutrient sinks in biofilm (34, 36). Melanization, and its consequences for mammalian virulence, has been most extensively studied in Cryptococcus neoformans (24). The pathway of tyrosine metabolism, which could lead to pigment formation, involves two branches: melanin formation via the hydroxyphenylalanine intermediate, which requires tyrosinase activity, and tyrosine catabolism involving homogentisic acid (HGA) formation and its further conversion to maleylacetoacetic acid (2). HGA is the main p-diphenolic intermediate of the normal l-tyrosine catabolism pathway in animals and bacteria. HGA is known to auto-oxidize, leading to the formation of reddish-brown pigment called pyomelanin, HG-melanin, or ochronotic pigment. In vitro studies have shown that HGA auto-oxidation is catalyzed by Mn2+ and involves H2O2 formation during the reaction (2, 16, 20). Several bacterial species are able to produce pigment via the HGA intermediate, including Shewanella colwelliana, Hyphomonas species, and Legionella pneumophila (14, 28).
Most of the V. cholerae strains do not form any detectable pigment under the experimental growth conditions normally used, but some pigmented mutants were detected after N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis (4, 7). The mutated gene(s) in such mutants remains unverified. It has been shown that pigment production in V. cholerae can be induced in response to stress, particularly hyperosmotic shock and elevated temperatures (1, 3, 5, 22). The pigment formation is initiated in late-exponential to postexponential growth of the bacteria (1, 5). V. cholerae is a member of a relatively large group of environmental bacteria that produce melanins, including species of Aeromonas, Burkholderia, Caulobacter, Mycobacterium, Proteus, Pseudomonas, Serratia, and Legionella (3). Melanins are also broadly distributed among eukaryotic microorganisms, including fungi and protozoa (24, 25).
The HGA branch is believed to be the only catabolic pigment-producing branch present in V. cholerae (14, 26), although the existence of other types of pigments cannot yet be excluded. However, to date, there is little information on the genes involved in V. cholerae melanization. We have identified the transposon targeted gene in a pigment-overproducing V. cholerae mutant as a putative oxidoreductase.
MATERIALS AND METHODS
Bacterial strains.
The V. cholerae strain used was a smooth variant of A1552 (wild type [WT], El Tor, Inaba; Rifr). Escherichia coli strains DH5α and S17-1 λpir were used for standard DNA manipulations and as donors in mating in transposon mutagenesis, respectively.
Growth media.
LB medium, LB agar plates, and several defined media were used for preliminary studies of growth and pigment production of the mutant and the WT strains of V. cholerae.
Mutagenesis.
Tn5 mutagenesis of V. cholerae O1 El Tor, strain A1552, was performed by conjugation between the donor, E. coli S17-1 λpir containing a plasmid carrying the mini-Tn5 transposon TnKm2, and the recipient, a rifampin-resistant V. cholerae O1 El Tor A1552. Conjugation was performed by mixing equal volumes of the donor and recipient, followed by filtration through a 0.45-μm-pore-size filter (Nalgene). The filters were then placed on LB agar plates, incubated for 6 h at 37°C, and suspended in LB, and the bacteria were recovered by vortexing. The exconjugants were selected by plating the suspension onto LB plates supplemented with 100 μg/ml of rifampin and 50 μg/ml of kanamycin. A total of 30,000 exconjugants were screened on plates to identify mutants exhibiting a pigment production phenotype.
In-frame deletion mutant construction of the rpoS gene of V. cholerae.
Mutagenesis of the V. cholerae strain A1552 in the rpoS locus was performed by making an in-frame deletion that removed most of the codons, using a procedure described previously (27, 32, 38).
Briefly, a 500-bp 5′ flanking sequence of the gene, including several nucleotides of the coding region, was PCR amplified with the primers rpoS-A (5′-CGCTCTAGACTAAACCTTCGGATGAGAAGA-3′) and rpoS-B (5′-CCCATCCACTATAAACTAACACTCTTCTACTTTGGTTACGGT-3′). The primers rpoS-C (5′-TGTTAGTTTATAGTGGATGGGGCGCTGTTTAACGTCGAATAC-3′) and rpoS-D (5′-CGCTCTAGACAAAGAGATTGGTGCCATGAC-3′) were used to amplify several nucleotides of the 3′ region of the gene plus 500 bp of the downstream flanking sequence. The two PCR products were annealed at their overlapping region and amplified by PCR to produce a single DNA fragment, using the outer primers (rpoS-A and rpoS-D). The resulting PCR product, lacking most of the coding sequence of the gene, was digested with XbaI enzyme and ligated into a similarly digested pCVD442 suicide plasmid. pCVD442::ΔrpoS was introduced into E. coli SM10λpir by electroporation. The donor, E. coli S10λpir containing the plasmid pCVD442::ΔrpoS, was used for conjugal transfer to rifampin-resistant V. cholerae O1 El Tor A1552. A mixture containing equal volumes of the donor and recipient in LB was incubated for 6 h at 30°C. The exconjugants were selected by plating the suspension onto LB plates supplemented with 100 μg/ml of rifampin and 100 μg/ml of carbenicillin (Cb) at 30°C. After selection of the desired transconjugants on rifampin-plus-Cb plates, they were streaked on LB plates with 10% sucrose at 30°C. Several colonies were purified from the plates, tested for Cb sensitivity, and then analyzed for the deletion using colony PCR. The primers VC1348-A (5′-CGCTCTAGACGAGTTTCATCGCCTCAATCC-3′), VC1348-B (5′-CCCATCCACTATAAACTAACAGCAATGGTACCGATTCAACTG-3′), VC1348-C (5′-TGTTAGTTTATAGTGGATGGGGGCGGTTGCCATGTTTAAAGT-3′), and VC1348-D (5′-CGCTCTAGACCAATTGCAGCCATGATTTTG-3′) were used for the construction of a ΔVC1348 mutant. The primers VC1349-A (5′-CGCTCTAGACGCAGCACTTCGGTTTCCAGA-3′), VC1349-B (5′-CCCATCCACTATAAACTAACACGCCATCAAGCAAAACAGTAG-3′), VC1349-C (5′-TGTTAGTTTATAGTGGATGGGGGTGAGCGTTTAAACCAGTAG-3′), and VC1349-D (5′-CGCTCTAGACGAAAGAGTGAACATGCGCAT-3′) were used for the construction of a ΔVC1349 mutant. The procedure for the mutant construction was the same as described above for the ΔrpoS mutant construction. The ΔrpoS hmgA::km double mutant was obtained by phage-mediated transduction using the CP-T1ts phage (8). The CP-T1ts stock was obtained from Andrew Camilli. Bacteriophage CP-T1ts propagated on strain SNW29 was prepared by a plate method. V. cholerae cells were grown to late exponential phase (an optical density at 600 nm [OD600] of ∼0.7) and infected at a multiplicity of infection of 10−5 with CP-T1ts. Bacteriophage was allowed to adsorb for 10 min at room temperature and was then mixed with 5 ml of 45°C LB soft agar (0.5%) and poured onto LB agar plates. The plates were incubated at 30°C. Bacteriophage was recovered from the soft agar layer of the plates by resuspending the top agar in 5 ml of LB broth and was incubated at 4°C to allow bacteriophage diffusion. Agar and bacteria were removed by two successive centrifugations at 15,000 × g for 1 min. Bacteriophage was concentrated by centrifugation at 16,000 × g for 2 h at 4°C. The pellet containing the bacteriophage was suspended in 500 μl of LB broth and used for transduction. Phage transduction was done as follows. The recipient strain (SNW30) was grown in 10 ml LB to late exponential phase (OD600, ∼0.7), spun down, and resuspended in 1 ml LB broth; 0.5 ml of the bacterial suspension, 0.5 ml of phage suspension, and 0.5 ml of adsorption buffer (0.015 M CaCl2+, 0.03 M MgCl2) were mixed and incubated at 30°C for 20 min. The cells were washed once in 0.9% NaCl, spun down, and resuspended in 0.3 ml of NaCl, and 0.1-ml samples of the suspension were spread out on plates containing 30 μg/ml kanamycin (Sigma). Transductant colonies were purified and used for further experiments.
Cloning and analysis of the transposon insertion site.
Chromosomal DNA was isolated from the V. cholerae transposon insertion mutants using the Marmur procedures (19). After digestion of chromosomal DNA with EcoRI restriction enzyme (Fermentas Life Science) according to the manufacturers' instructions, the DNA fragments were separated on a 0.7% agarose (Sigma) gel by electrophoresis at a voltage of 5 V/cm. The DNA fragments between 3 and 8 kb in size were extracted from the agarose gel using a MiniElute Gel Extraction kit according to the manufacturers' instructions (Qiagen) and subsequently ligated using ligase enzyme (Takara Bio Inc.) into the cloning vector pUC18 that had been digested with the EcoRI enzyme. The pool of plasmids was introduced by transformation into E. coli strain DH5α by electroporation, followed by the selection of resistant transformants on plates containing 30 μg/ml kanamycin (Sigma)- and 100 μg/ml Cb (Sigma)-resistant transformants.
PCR primers and complementation.
The PCR primers used for obtaining a plasmid clone with a WT locus allowing complementation tests of the pigment-overproducing mutant were 5′-ATCAACGAGTCCTCTGACGATAA-3′ and 5′-CATGGTTAACGTAAGCGGAACCA-3′. The DNA fragment containing the oxidoreductase gene was obtained after amplification by PCR using WT V. cholerae chromosomal DNA as a template. The PCR product was purified from an agarose gel and ligated into the pCR2.1 vector plasmid (Invitrogen). After amplification in the TOP10F′ E. coli strain, the plasmid was isolated with a Qiaprep Spin Miniprep kit (Qiagen). The hmgA+ plasmid clone (pSNW501) was electroporated into WT and SNW29 mutant V. cholerae (see Results). As a negative control, the vector pCR2.1 without an insert was similarly introduced into the WT and mutant strains. Furthermore, the homogentisate 1,2-dioxygenase was cloned into another vector, pUC18 (36), background at an EcoRI restriction site, resulting in plasmid pSNW502 (denoted phmgA). In both pSNW501 and pSNW502, expression depended on the lac promoter in the vector.
RNA extraction and qRT-PCR.
RNA was prepared using Trizol according to the manufacturer's instructions (Invitrogen), and the concentration was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). cDNA was prepared from 200 ng RNA from each sample using the QuantiTect cDNA kit (Qiagen), which includes a DNase treatment step. Real-time reverse transcription (RT)-PCR was performed using the primers specific for VC1344 (VC1344-F, 5′-CCAAGGAAACGATGATGATGG-3′, and VC1344-R, 5′-GAACGGTGTTTAGCAATTTCG-3′) and VC1345 (VC1345-F, 5′-CATCGTGAAGGCACTTGTTC-3′, and VC1345-R, 5′-GCAGTTCGCCTTCCCACTC-3′). Quantitative RT-PCR (qRT-PCR) experiments were done using Power SYBR Green qPCR MasterMix (Applied Biosystems). Each reaction mixture contained 300 nM primers, 100 ng template, and reference dye. Three independent samples were tested in triplicate. For each sample, the mean cycle threshold of the test transcript was normalized to that of tmRNA.
UV sensitivity test.
Bacterial strains were grown in LB medium with aeration by shaking at 37°C for 24 h. The bacterial suspensions were transferred into a petri dish and irradiated with UV doses of 0, 100, 200, and 300 μJ/m2. All experiments were repeated three to five times, and an average was calculated from the analyses of surviving bacterial cells after serial dilutions of bacterial suspensions were plated on LB agar incubated in the dark at 37°C overnight.
SDS-PAGE and Western blotting.
For TcpA and CTX detection, V. cholerae strains were grown as described previously (12). Bacteria were harvested by centrifugation at 10,000 × g for 10 min at 4°C. The resulting pellet was resuspended in 0.20 volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing 2-mercaptoethanol and used for the detection of TcpA. The culture supernatant fluid was precipitated with 10% trichloroacetic acid (TCA). Briefly, 1 volume (250 μl) of 50% TCA stock was added to 4 volumes (1 ml) of protein sample. The protein-TCA mixture was kept on ice for 15 min, and subsequently, the tube was centrifuged at 15,000 × g for 5 min. The supernatant was removed, the protein pellet was washed with 200 μl of cold acetone, and the tube was centrifuged at 15,000 × g for 5 min. A total of two acetone washes were done. The protein pellet was dried by placing the tube in a 95°C heat block for 5 to 10 min to drive off the acetone. The resulting pellets were dissolved in sample buffer containing 10% glycerol, 0.05% bromophenol blue, 2% SDS, 5% 2-mercaptoethanol, and 10 mM Tris-HCl, pH 6.8, and resolved by 12.5% SDS-PAGE with a discontinuous buffer system at a constant voltage of 60 V for the stacking gel and 120 V for the resolving gel (17). Proteins with known molecular masses (Fermentas) were used as molecular mass markers. The gels were fixed with methanol and glacial acetic acid. The proteins were visualized by Coomassie blue staining. Western blot analyses were performed as described previously (31), and detection was done by using the ECL+ chemiluminescence system (GE Healthcare). Anti-TcpA polyclonal antiserum (a gift from R. Taylor [13]) and anti-CTX A and B subunit antiserum (Sigma) were used for detection of TcpA in whole-cell extracts and CTX in culture supernatants, respectively. The anti-CRP polyclonal antiserum against E. coli cyclic AMP receptor protein (CRP) (15, 35) was used for the internal control of cytoplasmic protein when the TcpA immunoblot analysis was performed. The enhanced-chemiluminescence immunodetection method was applied to detect the reaction bands according to the manufacturer's (GE Healthcare) instructions.
TcpA stability assay.
To determine the intracellular and surface-bound TcpA stability, we used a technique described previously (13). The protein stability was monitored after protein synthesis had been inhibited by the addition of 25 μg/ml chloramphenicol to bacterial cultures grown to 40 Klett units in LB medium at 37°C. Samples to be analyzed by Western blotting were removed at the indicated time points: 0, 5, 10, 15, 30, 45, and 60 min after the addition of chloramphenicol.
Measurement of pigment production.
Bacterial strains were grown in LB medium at 37°C with shaking conditions overnight. One hundred microliters of overnight bacterial culture was inoculated into 20 ml LB medium in the presence or absence of 5 mM l-tyrosine (Sigma). Cultures were grown at 37°C under shaking conditions overnight. The culture supernatants were removed by centrifugation at 15,000 × g for 10 min and filtered through a 0.2-μm Millipore filter (Millipore AB). The pigmentation of the culture supernatants was measured by the OD400. The data represent the results of three independently performed experiments.
Genome database searches.
Genome database accession number searches were done for the complete whole-genome sequence of V. cholerae O1 biovar El Tor strain N16961 chromosomes I and II at the NCBI website (http://www.ncbi.nlm.nih.gov).
Infant mouse competition assay.
Approximately 105 WT A1552 and SNW29 cells were inoculated intragastrically into 6-day-old CD-1 mice (Charles River Laboratories). The mice were euthanized after 20 h, and the bacteria colonizing the intestines were quantified as described previously (6).
Microarray experiments.
Bacteria were cultured statically for 4 h in AKI medium (12) at 37°C and then shifted to aerobic growth for 6 h using shaken culture flasks (12). Triplicate samples were taken every hour for the duration of the experiment (10 h). Bacteria were collected on a 0.2-μm bacterial filter (diameter, 47 mm) by quick vacuum filtration (2 to 5 s) followed by immediate lysis with Trizol reagent (Invitrogen), frozen on dry ice, and stored at −80°C until RNA isolation was performed. RNA was isolated, treated with DNase I, and cleaned with an RNeasy kit (Qiagen). Labeling of cDNA and microarray hybridizations were performed as described previously (13). RNA from bacteria exponentially grown in LB (OD600, ∼0.3) was used as a reference. Microarray data analysis was done as described previously (8).
RESULTS AND DISCUSSION
Analysis of a pigment-producing V. cholerae mutant.
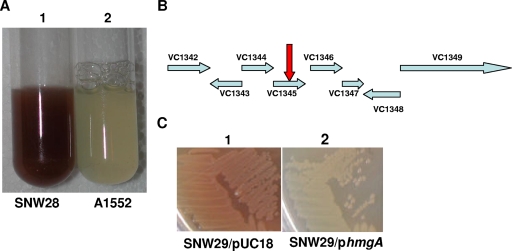
A transposon mutant library for the V. cholerae O1 El Tor strain was obtained by using the mini-Tn5 transposon as described in Materials and Methods. Among the 30,000 transposon mutants, we found two pigmented colonies in a screening on solid medium. One mutant isolate, denoted SNW28, was chosen for further analysis. Upon growth in liquid medium, the culture of the mutant turned distinctly brown (Fig. 1A). In order to map the mutated locus in the SNW28 isolate, we extracted total chromosomal DNA from the strain and carried out a cloning experiment using EcoRI-digested DNA and the vector pUC18. By selecting for kanamycin resistance, we obtained clones carrying the whole transposon and some flanking DNA of the chromosome. The cloned DNA was sequenced, and the exact position of transposon insertion was revealed. As shown in Fig. 1B, we found that the insertion had occurred after nucleotide 681 in an open reading frame corresponding to the locus denoted VC1345 (accession number, AE003852.1; GeneID, 2614799) in the V. cholerae genome. The second brown-pigmented mutant was found to have a transposon insertion in the VC1345 gene. This gene locus is located on chromosome 1 in V. cholerae, and the search of the genome databases revealed that the gene sequence was suggested to encode the enzyme oxidoreductase with the proposed designation HmgA. The putative 378-amino-acid-long V. cholerae HmgA oxidoreductase protein sequence was used in a BLAST search for homologous proteins in other organisms. Several similar sequences were indicated in the search results: for example, the V. cholerae protein showed 25% identity and 42% similarity to homogentisate 1,2-dioxygenase from Pseudomonas aeruginosa, 25% identity and 44% similarity to that from Mesorhizobium loti, 27% identity and 42% similarity to that from Caulobacter crescentus, and 29% identity and 48% similarity to homogentisate oxygenase-related protein from Sulfolobus solfataricus. The V. cholerae hmgA gene is located downstream of a putative 4-hydroxyphenylpyruvate dioxygenase gene and upstream of a conserved hypothetical protein, which is homologous to fumarylacetoacetate hydrolase family proteins from C. crescentus and Mycobacterium tuberculosis and to 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase-related proteins in S. solfataricus, Archaeoglobus fulgidus, and Thermoplasma acidophilum.
FIG. 1.
(A) Pigment production after bacterial growth in LB liquid medium at 37°C with shaking. (B) Genetic map of the mini-Tn5-Km2 cassette insertion in the brown-pigment-overproducing V. cholerae mutant at chromosome position VC1345 (accession number, AE003852.1; GeneID, 2614799) (arrow). (C) Complementation analysis of the brown-pigment-producing mutant.
Genetic transfer by phage-mediated transduction of the transposon-targeted gene from SNW28 to the original WT A1552 strain background confirmed that the hmgA::mini-Tn5 insertion mutation per se was the reason behind the pigmented phenotype, and the transductant derivative SNW29 was used for further studies.
To verify that the pigment production really was caused by the insertion mutation in the VC1345 open reading frame, we cloned the WT hmgA allele under the control of a vector plasmid promoter and carried out a trans-complementation test. We used PCR primers and amplified a fragment carrying the WT locus as described in Materials and Methods. As shown in Fig. 1C, the hmgA+ clone complemented the mutation, and in its presence, the pigment production was lost.
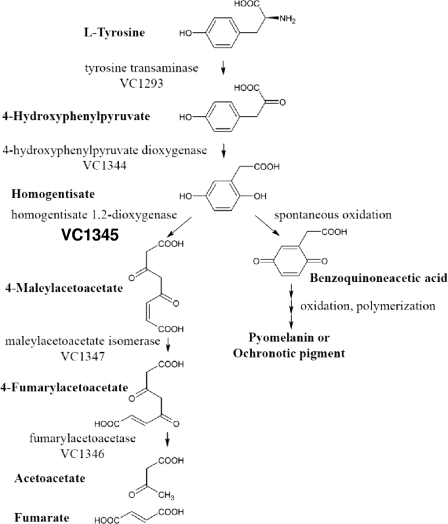
HGA is the main p-diphenolic intermediate of the normal l-tyrosine catabolic pathway in animals and bacteria. HGA is known to auto-oxidize, leading to formation of reddish-brown pigment called pyomelanin, HG-melanin, or ochronotic pigment. The putative involvement of the V. cholerae hmgA gene product in a catabolic scheme is illustrated in Fig. 2.
FIG. 2.
Catabolic pathway of tyrosine metabolism and representative genetic loci. Predicted enzymes encoded by the respective genetic loci in V. cholerae are as follows: VC1344, 4-hydroxyphenylpyruvate dioxygenase; VC1345, homogentisate 1,2-dioxygenase; VC1347, maleylacetoacetate isomerase; VC1346, fumarylacetoacetase. The proposed pathway is based in part on the pathway described for Streptomyces avermitilis (5).
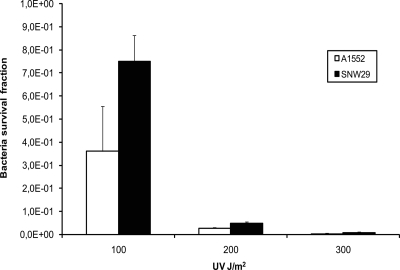
We also tested whether the pigment-producing mutant SNW29 would be altered with respect to sensitivity to UV irradiation in comparison with the WT strain, A1552. We observed that the melanin pigment-producing mutant strain SNW29 was more resistant to UV irradiation than the WT V. cholerae strain (Fig. 3).
FIG. 3.
UV sensitivity test of V. cholerae A1552 (WT) and SNW29. Bacterial growth, UV irradiation, and plating to determine bacterial survival were performed as described in Materials and Methods.
Growth phase-dependent expression of the hmgA gene.
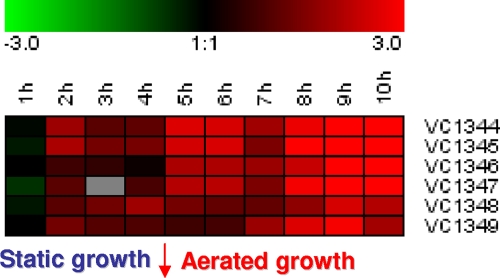
We analyzed the expression of the hmgA operon in different growth phases of the WT V. cholerae strain A1552 cultured at 37°C. Microarray expression profiling was used to analyze the expression of the hgmA gene and the neighboring genes as shown in Fig. 2, e.g., VC1344 and VC1346 to VC1349, at the transcriptional level. For this experiment, total bacterial RNA was obtained from the different growth phases after growth in AKI medium (see Materials and Methods). In brief, cDNA derived from RNA prepared from cells grown under these conditions was hybridized to microarrays in the presence of the same differentially labeled reference cDNA resulting from RNA isolated from exponentially grown bacteria in LB. We observed that the expression of the VC1344-to-VC1349 gene cluster was influenced by the growth phase, as revealed by the microarray expression profiling data (Fig. 4) when the cells had reached the stationary phase (after 10 h, the OD600 was 3.86 for A1552 and 3.7 for SNW30 and did not increase any further) (Fig. 4). Two of the neighboring genes, VC1348 and VC1349, annotated to encode a putative response regulator and a sensor kinase, respectively, were observed to have the highest expression levels in the stationary phase (Fig. 4). We speculate that this putative two-component system might be involved in sensing some environmental signal(s) and thus influences colonization ability.
FIG. 4.
Expression of the VC1344-VC1349 gene cluster during 10 h of bacterial growth under AKI conditions. Gene expression of WT A1552 was analyzed using RNA from exponentially grown bacteria in LB culture as a reference. The expression ratios are represented by shades of color according to the log2 (induction) scale shown.
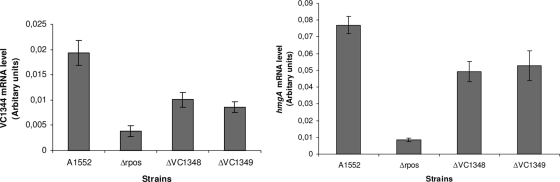
To test if VC1348 and/or VC1349 mutants would affect expression of the VC1344-VC1345 operon, we introduced deletion mutations into each gene and performed qRT-PCR analyses as summarized in Fig. 5. Mutation in either VC1348 or VC1349 caused reduced expression levels (to 50 to 70% of the WT level) of both VC1344 and VC1345. Determining how the putative regulatory loci might be involved in the control of pigment production will require further study of the VC1348-VC1349 genes and gene products.
FIG. 5.
Analyses of VC1344 and VC1345 (hmgA) gene expression by qRT-PCR as described in Materials and Methods. The data represent the results of three independently performed experiments. The error bars indicate the standard deviations from three experiments.
The finding that the expression level of the VC1344-to-VC1349 gene cluster was increasing with increasing cell density prompted us to determine if the sigma factor RpoS might be required for expression of the tyrosine metabolic pathway and brown-pigment production in V. cholerae.
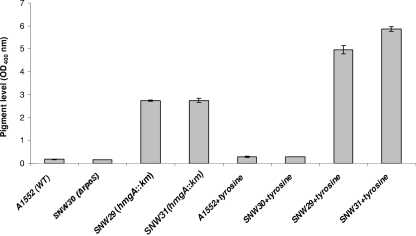
We compared the levels of transcription of the VC1344 and hmgA genes in WT V. cholerae strain A1552 and in SNW30 by qRT-PCR. We found that the expression of both the hmgA and VC1344 genes was ∼8-fold reduced in the ΔrpoS mutant of V. cholerae strain A1552 (Fig. 5). We also tested the levels of pigment production in WT V. cholerae, the hmgA mutant (SNW29), the rpoS mutant (SNW30), and the hmgA rpoS double mutant (SNW31) in the absence or presence of exogenously added tyrosine in the culture medium. As shown in Fig. 6, a slight increase in the production of pigment was observed in the hmgA rpoS double mutant (SNW31) grown in the presence of tyrosine compared with the hmgA single mutant (SNW29). However, there were no differences observed in pigment production between the WT and the rpoS single mutant (SNW30) strain when grown either in the absence or in the presence of tyrosine.
FIG. 6.
Levels of pigment production in the WT V. cholerae strain A1552, the rpoS mutant (SNW30), the hgmA mutant (SNW29), and the hgmA rpoS mutant (SNW31) with and without tyrosine supplement in growth medium. The error bars show the standard variations of three experiments.
Effect on V. cholerae virulence factor expression of the hmgA::mini-Tn5 mutation.
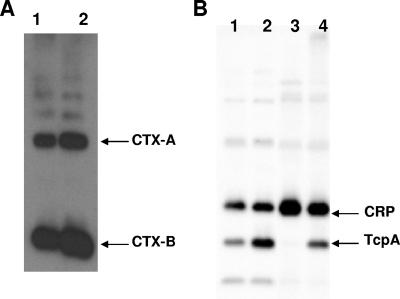
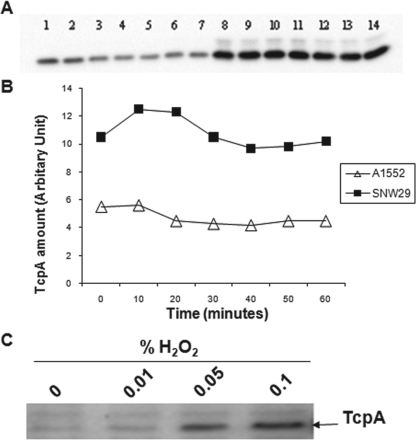
Earlier studies with mutant V. cholerae suggested that there might be some connection between melanin pigment production and expression of CTX. Coyne and al-Harthi (4) observed that WT V. cholerae 569B is not normally melanogenic in culture while its hypertoxic mutant strain, HTX-3, synthesized a pyomelanin. Furthermore, pyomelanin-producing mutants of V. cholerae were suggested to be substantially more virulent than their nonmelanogenic parental strain (10, 14). However, the genetic alteration(s) in those hypertoxigenic or hypervirulent strains was not revealed. Interestingly, we observed an elevated level of secreted CTX in the hmgA::mini-Tn5 mutant in comparison with the WT strain, A1552 (Fig. 7A). In order to further investigate the putative link between pigment formation and virulence factor expression, we analyzed the level of the toxin-coregulated pilus, Tcp, and as an internal control, the CRP transcriptional factor, in strains A1552 and SNW29. As shown in Fig. 7B, there was a clear increase in the level of TcpA protein, the main Tcp pilus subunit, as a result of the hmgA gene mutation. Two possible reasons for the elevated levels of TcpA in the hmgA mutant derivatives could be that the genetic expression of the tcp locus was increased or that the stability and turnover of the TcpA protein were altered. An experiment designed to monitor the relative stability of TcpA in the mutant and parental strains did not reveal much difference in TcpA stability (Fig. 8A and B). The fact that the hmgA mutant also secreted a higher level of CTX would presumably favor the hypothesis that the mutant derivative is altered in its genetic expression of the virulence factors. A number of parameters, like temperature, pH, osmolarity, amino acids, oxidative stress, and bile, are known to modulate the expression of the ToxR regulon, and such environmental signals exert their effects at different levels of the regulatory cascade (22). Martin and Batkoff (20) reported that the HGA auto-oxidation at physiological pH can generate superoxide radicals and hydrogen peroxide (H2O2) in the metabolic disorder alkaptonuria in eukaryotic cells. We considered that the SNW29 strain perhaps produced more CTX and TcpA as a result of some stress-induced effect on the ToxR regulon. To test if, for example, H2O2 per se could induce higher expression of CTX and the toxin-coregulated pilus, we monitored the level of TcpA after incubation of V. cholerae strain A1552 in the presence of different H2O2 concentrations. As shown in Fig. 8C, the TcpA level was increased by increasing concentration of H2O2.
FIG. 7.
(A) Immunoblot analyses of CTX levels in V. cholerae WT A1552 (lane 1) and SNW29, a hyperpigmented strain (lane 2), with anti-CTX polyclonal antiserum. (B) TcpA production in A1552 (WT) and SNW29 (hyperpigmented) strains. The samples in lane 1 (A1552) and lane 2 (SNW29) were from bacteria grown in yeast extract-peptone (YEP) medium at 37°C under static culture conditions. The samples in lane 3 (A1552) and lane 4 (SNW29) were from bacteria grown in YEP medium at 37°C with shaking. The CRP protein was detected as an internal control for cytoplasmic protein. The arrows show the positions of the TcpA and CRP proteins. A total of 3 μg of protein was loaded in each lane.
FIG. 8.
Comparison of TcpA protein stabilitiesy in WT strain A1552 and the mutant SNW29. (A) Immunoblot analysis using anti-TcpA polyclonal antiserum. Lanes 1 to 7, A1552; lanes 8 to 14, SNW29. The times after addition of chloramphenicol were as follows: lanes 1 and 8, 0 min; lanes 2 and 9, 5 min; lanes 3 and 10, 10 min; lanes 4 and 11, 15 min; lanes 5 and 12, 30 min; lanes 6 and 13, 45 min; lanes 7 and 14, 60 min. (B) Quantitative analysis of TcpA levels using the Quantity One program (Bio-Rad). (C) Effect of H2O2 on TcpA expression in V. cholerae strain A1552. Shown is immunoblot analysis using anti-TcpA antiserum. The bacteria were grown in the presence of different concentrations of H2O2.
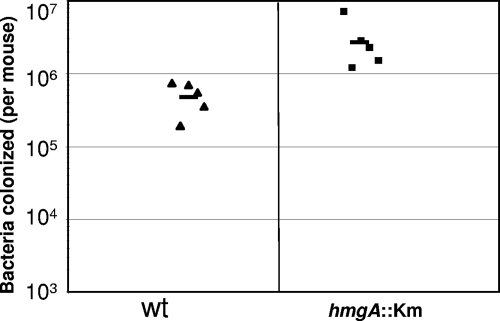
The toxin-coregulated pilus is known to be essential for colonization of the intestinal epithelium by V. cholerae (11, 21). Using colonization of the infant mouse intestine as an infection model, the hmgA::mini-Tn5 mutant was examined, and its colonization efficiency was compared to that of the parental strain, A1552. Colonization by the mutant strain was about fivefold higher than that by the WT strain (Fig. 9). As colonization efficiency is also dependent on bacterial growth, the growth rates of strains SNW29 and A1552 in LB were determined, and we observed no growth rate difference between the WT and the mutant (data not shown). Taken together, the results suggest that the increased colonization of strain SNW29 in the infant mouse model might be due to the increased expression of TcpA.
FIG. 9.
Infant mouse colonization assay using strains A1552 (wt) and SNW29 (hgmA::Km) as described in Materials and Methods.
In summary, we identified the mutant locus in a pigment-overproducing V. cholerae mutant. Our characterization of the mutant has revealed that melanin pigment formation can play a role in virulence factor expression of the bacteria and thereby influence the colonization ability.
Acknowledgments
We thank Bernt Eric Uhlin for valuable suggestions and support.
This work was supported by grants from the Swedish Research Council, the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), and the Faculty of Medicine at Umeå University, and it was performed within the Umeå Centre for Microbial Research (UCMR). The work was carried out in the frame of the European Virtual Institute for Functional Genomics of Bacterial Pathogens (CEE LSHB-CT-2005-512061) and with affiliation to the ERA-NET PathoGenoMics program.
Editor: A. Camilli
Footnotes
Published ahead of print on 22 December 2008.
REFERENCES
- 1.Attridge, S. R., P. A. Manning, J. Holmgren, and G. Jonson. 1996. Relative significance of mannose-sensitive hemagglutinin and toxin-coregulated pili in colonization of infant mice by Vibrio cholerae El Tor. Infect. Immun. 643369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carreira, A., L. M. Ferreira, and V. Loureiro. 2001. Brown pigments produced by Yarrowia lipolytica result from extracellular accumulation of homogentisic acid. Appl. Environ. Microbiol. 673463-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatfield, C. H., and N. P. Cianciotto. 2007. The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect. Immun. 754062-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coyne, V. E., and L. al-Harthi. 1992. Induction of melanin biosynthesis in Vibrio cholerae. Appl. Environ. Microbiol. 582861-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denoya, C. D., D. D. Skinner, and M. R. Morgenstern. 1994. A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli. J. Bacteriol. 1765312-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 642246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez, B. L., and J. D. Nosanchuk. 2003. Melanin and fungi. Curr. Opin. Infect. Dis. 1691-96. [DOI] [PubMed] [Google Scholar]
- 8.Hava, D. L., and A. Camilli. 2001. Isolation and characterization of a temperature-sensitive generalized transducing bacteriophage for Vibrio cholerae. J. Microbiol. Methods 46217-225. [DOI] [PubMed] [Google Scholar]
- 9.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 1681487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivins, B. E., and R. K. Holmes. 1980. Isolation and characterization of melanin-producing (mel) mutants of Vibrio cholerae. Infect. Immun. 27721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivins, B. E., and R. K. Holmes. 1981. Factors affecting phaeomelanin production by a melanin-producing (mel) mutant of Vibrio cholerae. Infect. Immun. 34895-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwanaga, M., and K. Yamamoto. 1985. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J. Clin. Microbiol. 22405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirn, T. J., M. J. Lafferty, C. M. Sandoe, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35896-910. [DOI] [PubMed] [Google Scholar]
- 14.Kotob, S. I., S. L. Coon, E. J. Quintero, and R. M. Weiner. 1995. Homogentisic acid is the primary precursor of melanin synthesis in Vibrio cholerae, a Hyphomonas strain, and Shewanella colwelliana. Appl. Environ. Microbiol. 611620-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouokam, J. C., S. N. Wai, M. Fällman, U. Dobrindt, J. Hacker, and B. E. Uhlin. 2006. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect. Immun. 742022-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Du, B. N. Jr. 1991. Alcaptonuria and ochronotic arthritis. Mol. Biol. Med. 831-38. [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 18.Levin, B. R., and R. V. Tauxe. 1996. Cholera: nice bacteria and bad viruses. Curr. Biol. 61389-1391. [DOI] [PubMed] [Google Scholar]
- 19.Marmur, J. 1961. A procedure for the isolation deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3208-218. [Google Scholar]
- 20.Martin J. P., Jr., and B. Batkoff. 1987. Homogentistic acid autooxidation and oxygen radical generation: implications for the etiology of alkaptouric arthritis. Free Rad. Biol. Med. 3241-250. [DOI] [PubMed] [Google Scholar]
- 21.Mekalanos, J. J., R. J. Collier, and W. R. Romig. 1978. Purification of cholera toxin and its subunits: new methods of preparation and the use of hypertoxinogenic mutants. Infect. Immun. 20552-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nag, S., S. Das, and K. Chaudhuri. 2005. In vivo induced clpB1 gene of Vibrio cholerae is involved in different stress responses and affects in vivo cholera toxin production. Biochem. Biophys. Res. Commun. 3311365-1373. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nosanchuk, J. D., and A. Casadevall. 2003. The contribution of melanin to microbial pathogenesis. Cell Microbiol. 5203-223. [DOI] [PubMed] [Google Scholar]
- 25.Nosanchuk, J. D., and A. Casadevall. 2006. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 503519-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruzafa, C., A. Sanchez-Amat, and F. Solano. 1995. Characterization of the melanogenic system in Vibrio cholerae, ATCC 14035. Pigment Cell Res. 8147-152. [DOI] [PubMed] [Google Scholar]
- 27.Skorupski, K. and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 16947-52. [DOI] [PubMed] [Google Scholar]
- 28.Steinert, M., M. Flugel, M. Schuppler, J. H. Helbig, A. Supriyono, P. Proksch, and P. C. Luck. 2001. The Lly protein is essential for p-hydroxyphenylpyruvate dioxygenase activity in Legionella pneumophila. FEMS Microbiol. Lett. 20341-47. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 842833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 642853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaitkevicius, K., B. Lindmark, G. Ou, T. Song, C. Toma, M. Iwanaga, J. Zhu, A. Andersson, M. L. Hammarström, S. Tuck, and S. N. Wai. 2006. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. USA 1039280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Weiner, R. M. 1997. Biopolymers from marine prokaryotes. Trends Biotechnol. 15390-394. [DOI] [PubMed] [Google Scholar]
- 35.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 1826347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler, M. H., and A. A. Bell. 1988. Melanins and their importance in pathogenic fungi. Curr. Top. Med. Mycol. 2338-387. [DOI] [PubMed] [Google Scholar]
- 37.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 993129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]