Abstract
We previously reported that infection by Fusobacterium nucleatum strongly induced the expression of both human beta-defensin 2 (HBD-2) and HBD-3 by gingival epithelial cells. The aim of this study was to characterize the pattern recognition receptors (PRRs) and regulatory mechanisms involved in the induction of HBD-2 and -3 expression by F. nucleatum in gingival epithelial cells. Immortalized human gingival epithelial HOK-16B cells were infected with live or heat-killed F. nucleatum, and the expression of HBDs and interleukin-8 (IL-8) was examined by real-time reverse transcription-PCR and enzyme-linked immunosorbent assay, respectively. Live, but not heat-killed, F. nucleatum invaded HOK-16B cells, as seen by confocal microscopy and flow cytometry. Live F. nucleatum induced both HBD-2 and -3 efficiently, whereas heat-killed bacteria induced only HBD-3 at a reduced level. Knockdown of NACHT-LRR- and pyrin domain-containing protein 2 (NALP2), the most abundant intracellular PRR in HOK-16B cells, by RNA interference (RNAi) significantly reduced the induction of HBD-3 but not HBD-2 and IL-8. In addition, knockdown of Toll-like receptor 2 (TLR2) by RNAi reduced the upregulation of HBD-2 and -3 but not IL-8. Heat-killed F. nucleatum was hindered in its ability to activate TLR2 and JNK signaling pathways. Theses data show that TLR2 and NALP2 mediate the induction of HBDs by F. nucleatum in gingival epithelial cells, but thresholds for the two HBD genes are different.
Gingival epithelium is the first line of defense against subgingival plaque-associated bacteria and provides not only a physical but also a chemical barrier against bacterial infection. The barrier function of the epithelium is attributed to its unique architectural integrity and the production of antimicrobial peptides, such as human beta-defensins (HBDs) (5). The epithelia of most body sites express HBD-1 constitutively, but HBD-2 and -3 are expressed only during infection or inflammation (32). However, gingival epithelium expresses HBD-2 in the absence of inflammation, presumably due to the constant exposure to oral bacteria (6, 32). The influence of oral bacteria on the expression of HBDs by gingival epithelial cells has been studied extensively in vitro. Components from several species of oral bacteria, including Streptococcus gordonii, Fusobacterium nucleatum, and Porphyromonas gingivalis, have been shown to induce HBD-2 expression (2, 20, 28, 31); in addition, infection of oral epithelial cells with live Aggregatibacter actinomycetemcomitans induces HBD-3 (9). Previously, we reported that infection by F. nucleatum or Prevotella intermedia strongly induced the expression of both HBD-2 and -3 by gingival epithelial cells (13).
Signaling pathways involved in the expression of HBDs also have been studied. The promoter region of HBD-2 contains numerous regulatory elements, including the binding sites for NF-κB, AP-1, AP-2, and NF-IL-6, whereas the promoter of HBD-3 contains no discernible NF-κB binding elements (14). Chung and Dale reported that various species of bacteria utilized different signaling pathways in the induction of HBD-2. In contrast to S. gordonii that used JNK and p38 mitogen-activated protein kinases (MAPKs) but not NF-κB to induce HBD-2 from gingival epithelial cells, A. actinomycetemcomitans and P. gingivalis used both MAPKs and NF-κB (2). HBD-3 induction by Staphylococcus aureus in skin keratinocytes was shown to involve p38 and AP-1 (21).
The regulation of HBD expression by bacteria, which is a part of the innate immune response, must be initiated by the recognition of unique microbial molecular patterns by pattern recognition receptors (PRRs). An array of PRRs is distributed at the surface, cytoplasm, and endosomal compartments of host cells. Toll-like receptors (TLRs) are expressed either on the cell surface (TLR1, -2, -4, -5, -6, and -10) or on the endosomal compartments (TLR3, -7, -8, and -9) (17). In contrast, a new family of PRRs, the nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), has been characterized as the cytoplasmic sensors of microbes (33). Ligands for human TLRs are known except TLR10, and each TLR recognizes a specific microbial component, such as lipoproteins, lipopolysaccharide (LPS), flagellin, single-stranded RNA, and CpG DNA (17). NOD1 and NOD2 recognize peptidoglycan. Neuronal apoptosis inhibitory protein 5 is activated by flagellin, and NACHT-LRR- and pyrin domain-containing protein 3 (NALP3) is also known to be activated by bacterial pore-forming toxins, bacterial mRNA, and gout-associated crystals (33). However, ligands for many other NLRs are not known. Although it has been established that P. gingivalis uses, in part, protease-activated receptor 2 during HBD-2 induction, which PRRs mediate HBD induction by other oral bacteria in gingival epithelial cells is not known (3).
F. nucleatum is a gram-negative, anaerobic, non-spore-forming, spindle-shaped or fusiform rod. F. nucleatum is considered an important bridging organism between early and late colonizers of plaque biofilm and is carried by 80% of adults regardless of periodontal health (24, 26). Although F. nucleatum can contribute to the growth of plaque biofilm and the development of periodontitis, its role in periodontal health and disease is controversial (8). The purpose of the present study was to characterize the PRRs and regulatory mechanisms involved in F. nucleatum-induced expression of HBD-2 and -3 by gingival epithelial cells.
MATERIALS AND METHODS
Human materials.
The use of human materials was approved by Institutional Review Board at Seoul National University Dental Hospital. Normal human gingival tissue specimens were obtained from healthy volunteers (age range, 20 to 30 years) undergoing oral surgery under the informed consent, and peripheral blood mononuclear cells (PBMC) were isolated from peripheral blood provided by the Red Cross (Seoul, South Korea).
Bacteria culture.
Culture, counting, and staining of F. nucleatum ATCC 25586 (American Type Culture Collection, Bethesda, MD) was cultured in brain heart infusion (BHI) broth (Difco) supplemented with 5 μg of hemin (Sigma, St. Louis, MO)/ml plus 10 μg of vitamin K under an anaerobic atmosphere (5% H2, 10% CO2, and 85% N2)/ml. The bacteria were enumerated by flow cytometry and sometimes stained with 5 μM 5 (and 6-)-carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR) before use. Heat-killed bacteria were prepared by heating the bacteria at 95°C for 1 h.
Cell culture.
Normal human gingival epithelial cells were isolated from excised retromolar gingival tissue and primary cultures were established in keratinocyte growth medium containing 0.15 mM calcium and a supplementary growth-factor bullet kit (KGM; Clonetics, San Diego, CA) as previously reported (23). Cells were subcultured at every 70% confluence, and the second passage cells were used for experiments. HOK-16B, kindly provided by N.-H. Park (University of California at Los Angeles) is an immortalized cell line established by the transfection of human papillomavirus type 16 DNA into primary human oral keratinocytes cultured from excised gingival tissue (27). HOK-16B cells were maintained in KGM with supplements. The reporter CHO cell line that expresses a human CD14 and a human TLR2 and drives the expression of surface CD25 as a result of TLR2-mediated NF-κB translocation was kindly provided by D. Golenbock (University of Massachusetts Medical School, Worcester, MA). The CHO/CD14/TLR2 cells were maintained in Ham F-12 medium (Gibco, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 1 mg of G418 (Invitrogen, Carlsbad, CA)/ml, 400 U of hygromycin (Amresco, San Francisco, CA)/ml, and 1× penicillin-streptomycin (Gibco).
Infection of epithelial cells with bacteria.
HOK-16B cells were plated at 6 × 104 cells/500 μl/well in triplicate into 24-well plates 1 day before infection. At 80% confluence, cells were infected with live or heat-killed bacteria at the multiplicities of infection (MOI) of 1,000 in KGM containing 2% heat-inactivated human sera (Sigma) and cultured at 37°C in a water-saturated atmosphere of 95% air and 5% CO2 for 24 h. The antibiotics in KGM were removed to prolong the survival of bacteria. In this culture condition, F. nucleatum, an absolute anaerobe did not outgrow but survived long enough to invade into cells. In addition, the viability of HOK-16B cells was not affected. Sometimes, HOK-16B cells were stimulated with 10 μg of Pam3CSK4 (InvivoGen, San Diego, CA)/ml for 24 h. The culture supernatant collected from each well was saved at -80°C for enzyme-linked immunosorbent assay (ELISA), and the total RNA was extracted from combined cells of triplicate wells by using TRIzol (Invitrogen). Experiments were repeated three times.
Examination of bacterial invasion into HOK-16B cells.
After infection with CFSE-labeled F. nucleatum, HOK-16B cells were stained with rhodamine-phalloidin (Sigma) and Hoechst 33342 (Molecular Probes) and then analyzed by confocal microscopy, as described previously (13).
Alternatively, bacterial invasion was examined by modified flow cytometric phagocytosis assay described previously (12). Briefly, HOK-16B cells were infected with CFSE-labeled bacteria at an MOI of 1,000 for 24 h and detached with trypsin-EDTA (Gibco). After quenching the fluorescence of the bacteria bound on the surface with 500 μl of trypan blue (400 mg/ml prepared in 0.85% saline solution), cells were analyzed by flow cytometry. To examine bacterial adhesion, HOK-16B cells were incubated with CFSE-labeled bacteria by centrifugation at 900 × g for 10 min and analyzed by flow cytometry without quenching.
Real-time reverse transcription PCR (RT-PCR).
Total RNA (2 μg) from HOK-16B, normal gingival epithelial cells, or PBMC was subjected to reverse transcription with (dT)18 and Superscript II enzyme (Invitrogen) in a 25-μl reaction mix at 42°C for 1 h. Real time-PCR was performed in a 20-μl reaction mix containing 1 μl of template cDNA, SYBR Premix Ex Taq, ROX Reference Dye (Takara Bio, Otsu, Japan), and each primer (0.2 μM). Primer sequences used are listed in Table 1. All primers were designed to amplify at least two exons in order to prevent the amplification of contaminating gDNA. Amplification was performed in a fluorescence thermocycler (7500 real-time PCR; Applied Biosystems, Foster City, CA) under the following conditions: initial denaturation at 94°C for 1 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s, and elongation at 72°C for 33 s. The specificity of the PCR product was verified by melting curve analysis and examination on a 3% agarose gel. The housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was amplified in parallel with the gene of interest. Relative copy numbers compared to GAPDH were calculated by using 2−ΔCT. Real-time PCR was performed in triplicate for each RNA sample.
TABLE 1.
Primer sequences used for real-time PCR
| Primer | Orientation | Sequence (5′-3′) |
|---|---|---|
| GAPDH | Forward | CAGCCTCAAGATCATCAGCA |
| Reverse | CCATCCACAGTCTTCTGGGT | |
| HBD-2 | Forward | ATCAGCCATGAGGGTCTTGT |
| Reverse | GGATCGCCTATACCACCAAA | |
| HBD-3 | Forward | TGTTTGCTTTGCTCTTCCTG |
| Reverse | CTTTCTTCGGCAGCATTTC | |
| TLR2 | Forward | ATTGTGCCCATTGCTCTTTC |
| Reverse | ACCCACACCATCCACAAAGT | |
| TLR3 | Forward | CGACGAAATGTCTGGATTTG |
| Reverse | GATGCACACAGCATCCCA | |
| TLR4 | Forward | AACCAAGAACCTGGACCTGA |
| Reverse | GAGAGGTGGCTTAGGCTCTG | |
| TLR7 | Forward | TGCTCTGCTCTCTTCAACCA |
| Reverse | CCAAGGAGTTTGGAAATTAGGA | |
| TLR8 | Forward | TGCTGCAAGTTACGGAATGA |
| Reverse | CGCATAACTCACAGGAACCA | |
| TLR9 | Forward | TCCTTCCCTGTAGCTGCTGT |
| Reverse | GTAGGAAGGCAGGCAAGGTA | |
| NOD1 | Forward | TGGTGGCCAAGTGATTGTAA |
| Reverse | CAATCTCCACCTCTTGCCAT | |
| NOD2 | Forward | GGT GTCTGCAAGGCTCTGTA |
| Reverse | TCAATGCCAAGAAGTTCTGC | |
| NALP2 | Forward | GAAATGTCATCTGCCTGGGT |
| Reverse | ATCTCCCGGAACTTCGTCTT | |
| NALP3 | Forward | TGAAGAGGAGTGGATGGGTT |
| Reverse | TTCAATGCACTGGAATCTGC | |
| NALP6 | Forward | GCGCAGGAGAAGAAGAAGAA |
| Reverse | CACAGTGGGTCAGTCATTGC | |
| IPAF | Forward | CAGAACCCTGCTGACTGAGA |
| Reverse | CCAAATGGAAAGGTCAAAGG |
ELISA.
The amounts of interleukin-8 (IL-8) secreted into medium during coculture with bacteria were measured by using ELISA kits (R&D Systems, Minneapolis, MN), according to the manufacturer's instructions. Fresh KGM was used as a negative control, and a recombinant IL-8 provided within the kit was used as a positive control.
RNA interference (RNAi) in HOK-16B cells and bacterial infection.
Stealth small interfering RNA (siRNA) duplex oligoribonucleotides against Homo sapiens TLR2 (GenBank no. NM_003264) and Homo sapiens NALP2 (GenBank no. NM_017852) were designed by using the BLOCK-iT RNAi Designer program (https://rnaidesigner.invitrogen.com/rnaiexpress/) and synthesized by Invitrogen. The oligonucleotide sequences used are as follow: sense (5′-UGAAGCAUCAAUCUCAAGUUCCUCA-3′) and antisense (5′-UGAGGAACUUGAGAUUGAUGCUUCA-3′) for TLR2 and sense (5′-UGCUCUCUCAGACAGAUCCAUUCGG-3′) and antisense (5′-CCGAAUGGAUCUGUCUGAGAGAGCA-3′) for NALP2. The BLOIT-iT (Invitrogen) fluorescent oligonucleotide that is not homologous to any known genes was used as transfection efficiency detector and a negative control. HOK-16B or normal gingival epithelial cells were plated at 4 × 104 cells/well into a 24-well plate. After overnight incubation, cells were transfected with siRNAs by using a BLOIT-iT transfection kit (Invitrogen). After culture for 24 h, the cells were infected with live F. nucleatum at MOI of 1,000 for 24 h. In case of TLR2, cells were retransfected with siRNA upon infection with F. nucleatum to prevent rebound of TLR2 transcription. Experiments were repeated two to four times. The F. nucleatum-induced upregulation of HBDs were calculated by subtracting the expression levels in uninfected from those in infected cells.
Western blot.
HOK-16B cells were lysed with radioimmunoprecipitation assay buffer (50 nM Tris-HCl, 1% NP-40, 150 nM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM NaF, 1 mM Na3VO4, and protease inhibitors). Proteins (30 μg/lane) were separated through an 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, transferred to a polyvinylidene difluoride membrane, and probed with anti-human NALP2 polyclonal antibody (Abcam, Cambridge, MA).
Flow cytometry.
HOK-16B cells were stained with fluorescein isothiocyanate (FITC)-conjugated mouse anti-human TLR2 monoclonal antibody clone TL2.1 (Imgenex, San Diego, CA) and analyzed with FACSCalibur (BD Biosciences).
The CHO cells were plated at 2 × 105 cells/well into a 24-well plate. After overnight incubation, cells were stimulated with live or heat-killed F. nucleatum, Pam3CSK4, or heat-treated (1 h at 95°C) Pam3CSK4 for 16 h. After being stained with FITC-conjugated mouse anti-human CD25 MAb, clone 2A3 (BD Biosciences, San Diego, CA), cells were analyzed with FACSCalibur (BD Biosciences).
Examination of signaling pathways.
HOK-16B cells (1.2 × 106) plated in a 100-mm dish were stimulated with live or heat-killed F. nucleatum at MOI of 1,000 for 15 min. After a wash with phosphate-buffered saline, protein lysates were prepared with lysis buffer included in a PathScan Inflammation Multi-Target ELISA kit (Cell Signaling, Danvers, MA) and subjected to ELISA analyses of NF-κB p65 and phosphorylated forms of NF-κB p65, IκB, p38, JNK, and STAT3 in duplicates, using the kit. Experiments were repeated twice.
Statistics.
The differences between the two groups were analyzed with the two-tailed nonpaired Student t test. The data were considered statistically significant at a P value of <0.05.
RESULTS
Invasion of F. nucleatum and induction of HBDs.
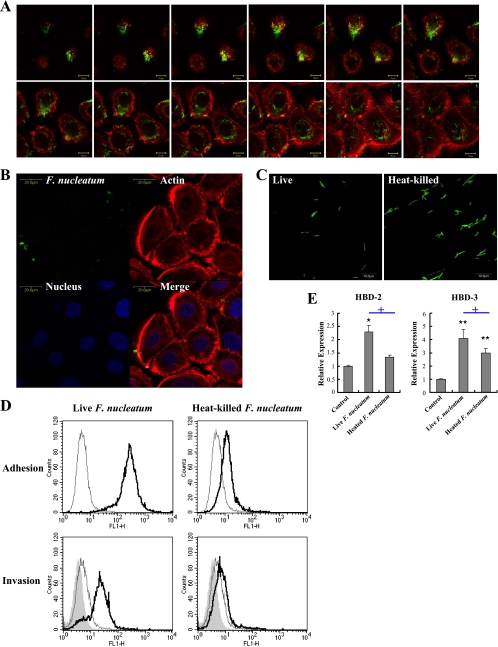
We previously examined that F. nucleatum is highly invasive (13), suggesting that this bacterium might stimulate intracellular PRRs. Since invasion by F. nucleatum requires bacterial protein synthesis (10), we studied the role of bacterial invasion in inducing HBD expression in gingival epithelial cells using heat-killed F. nucleatum. Immortalized human gingival epithelial (HOK-16B) cells were cocultured with live or heat-killed F. nucleatum at an MOI of 1,000 in 95% air and 5% CO2 for 24 h. Upon infection with live F. nucleatum, internalized bacteria were observed in the cytoplasm around nucleus, infecting most cells (Fig. 1A), whereas heat-killed bacteria hardly adhered to and invaded the cells, despite the intact morphology of bacteria (Fig. 1B to D). In contrast to live F. nucleatum that upregulated both HBD-2 and -3, heat-killed bacteria upregulated only HBD-3 at a reduced level (Fig. 1E). Collectively, these results indicate that bacterial adhesion and/or invasion might be important for the induction of HBD-2 and -3 expression in gingival epithelial cells.
FIG. 1.
Bacterial invasion and the induction of HBDs by F. nucleatum in HOK-16B cells. (A) HOK-16B cells were cocultured with live CFSE-labeled F. nucleatum at an MOI of 1,000 for 24 h. After fixation and permeabilization, cells were stained with rhodamine-phalloidin and then examined under a confocal microscope with serial z-section. (B) HOK-16B cells cocultured with heat-killed CFSE-labeled F. nucleatum at an MOI of 1,000 for 24 h were stained for actin (red) and nuclei (blue) and examined under a confocal microscope. (C) The morphology of live and heat-killed bacteria was examined under a confocal microscope. (D) Adhesion and invasion of live or heat-killed CFSE-labeled F. nucleatum to HOK-16B cells were analyzed by flow cytometry as described in Materials and Methods. The thin empty line represents live HOK-16B cells alone, the thick empty line represents live HOK-16B cells with the bacteria, and the shaded curve represents fixed HOK-16B with the bacteria. (E) HOK-16B cells were cocultured with live or heat-killed F. nucleatum at an MOI of 1,000 for 24 h. Expression of HBD-2 and -3 was evaluated by real-time RT-PCR. The means ± the standard errors of the mean (SEM) of the relative copy numbers were expressed as the fold induction compared to those of the control culture without bacteria. *, P < 0.05, and **, P < 0.005 versus control; +, P < 0.05 versus live F. nucleatum.
PRRs expressed by HOK-16B cells.
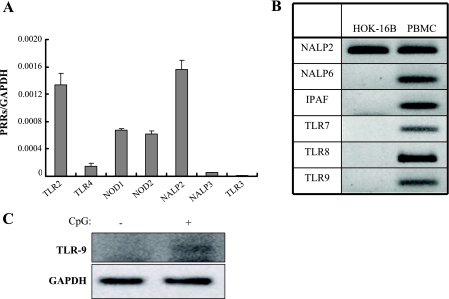
In order to determine which PRRs might mediate the induction of HBDs by F. nucleatum, the expression of various PRRs in HOK-16B cells was examined by real-time RT-PCR. Among the genes for which expression was detected, NALP2 and TLR3 showed the highest and lowest levels, respectively (Fig. 2A). TLR7, TLR8, TLR9, IPAF, and NALP6 transcripts were not detected (Fig. 2B). PBMC expressed all of these receptors at high levels, confirming the validity of primers used (Fig. 2B). In addition, real-time RT-PCR using RNA from primary cells confirmed the absence of TLR7, TLR8, TLR9, IPAF, and NALP6 transcripts in gingival epithelial cells. Stimulation of HOK-16B cells with CpG DNA, a synthetic ligand to TLR9, induced its expression at low level (Fig. 2C). These results indicate that unstimulated gingival epithelial cells express a limited repertoire of PRRs compared to PBMC.
FIG. 2.
PRRs expressed in HOK-16B cells. Expression of TLR2, TLR3, TLR4, TLR7, TLR8, TLR9, NOD1, NOD2, IPAF, NALP2, NALP3, and NALP6 in HOK-16B cells was evaluated by real-time RT-PCR. (A) The levels of gene expression were presented as relative copy numbers compared to GAPDH expression. (B) PCR products of the genes for which expression was not detected were visualized on agarose gels. PCR products using RNA from PBMC served positive controls. (C) HOK-16B cells were cultured in the absence or presence of 5 μM CpG DNA for 24 h, and the expression of TLR9 was examined by RT-PCR.
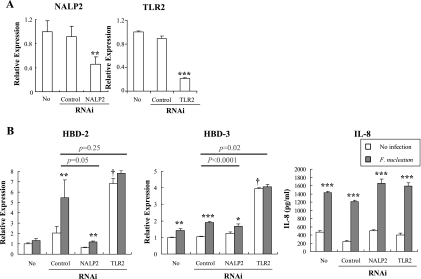
Role of NALP2 and TLR2 in the induction of HBDs by F. nucleatum.
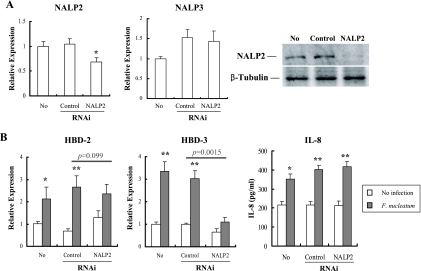
Although the ultimate destiny of internalized F. nucleatum is not known, some bacterial components may be delivered to the cytosol. Hence, the role of NALP2, the most abundant intracellular PRR in HOK-16B cells, in the induction of HBDs by F. nucleatum was evaluated by using RNAi. As shown in Fig. 3A, NALP2-specific siRNA suppressed the expression of its mRNA and protein but not that of NALP3 transcripts. In addition, transfection with control or NALP2 siRNA had no significant effects upon the basal levels of HBD expression or IL-8 secretion. The knockdown of NALP2 significantly reduced the F. nucleatum-induced upregulation of HBD-3 by ca. 78% (P = 0.0015). The upregulation of HBD-2 also showed tendency to decrease by ca. 46% (P = 0.099), but the induction of IL-8 was not affected (P = 0.54) (Fig. 3B).
FIG. 3.
Role of NALP2 in induction of HBD-2, HBD-3, and IL-8 by F. nucleatum in HOK-16B cells. HOK-16B cells were transfected with control nonsilencing or NALP2-specific siRNA. After 24 h of incubation, cells were infected with live F. nucleatum at MOI 1000 for 24 h. (A) Gene-silencing was assessed by real-time RT-PCR and Western blotting 24 h after transfection. (B) The expression of HBDs and secretion of IL-8 by F. nucleatum was evaluated by real-time RT-PCR and ELISA, respectively. The mean ± the SEM values of the relative copy numbers from nine real-time RT-PCR assays were expressed as relative expression compared to that of control cultures without any treatment. *, P < 0.05; and **, P < 0.001 versus no infection. P values indicate differences in the F. nucleatum-induced upregulation of HBDs between control and NALP2-specific RNAi.
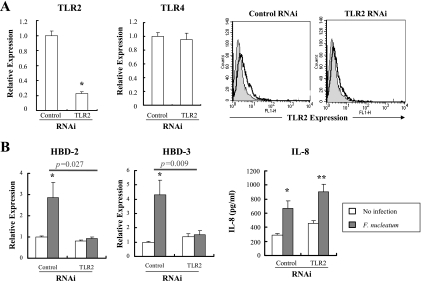
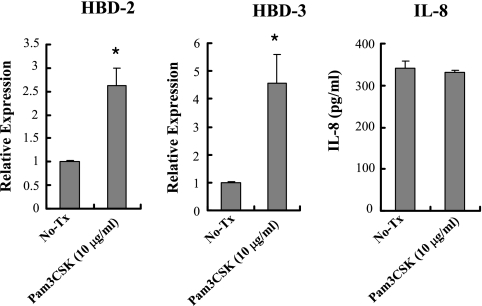
We investigated whether other PRRs expressed on the cell surface contributed to HBD induction. Like other gingival epithelial cells, HOK-16B cells expressed TLR2 and TLR4 on the cell surface (29). LPS from F. nucleatum, a ligand to TLR4, is known as a poor inducer of HBD-2 from cultured human oral keratinocytes, even in the presence of human serum as a source of LPS binding protein or soluble CD14 (20). Therefore, we studied the role of TLR2 in inducing HBDs by RNAi assay. Transfection with TLR2-specific siRNA decreased the level of TLR2 transcripts and proteins but had no effect on TLR4 expression (Fig. 4A). Knockdown of TLR2 RNA did not affect the basal levels of HBDs and reduced the F. nucleatum-induced upregulation of HBD-2 and -3 by ca. 93 and 96%, respectively (P = 0.027 and P = 0.009). The induction of IL-8 was not affected (P = 0.54) (Fig. 4B). The role of TLR2 in inducing HBDs and IL-8 was confirmed using a synthetic ligand to TLR2, Pam3CSK4. Stimulation of HOK-16B cells with 10 μg of Pam3CSK4/ml upregulated HBD-2 and -3 but not IL-8 (Fig. 5), a finding consistent with the RNAi assay results.
FIG. 4.
Role of TLR2 in the induction of HBD-2, HBD-3, and IL-8 by F. nucleatum in HOK-16B cells. HOK-16B cells were transfected with control nonsilencing or TLR2-specific siRNA. After 24 h of incubation, the cells were infected with live F. nucleatum at an MOI of 1,000 for 24 h. (A) Gene silencing was assessed by real-time RT-PCR and flow cytometry 24 h after transfection. (B) The expression of HBDs and secretion of IL-8 by F. nucleatum were evaluated by real-time RT-PCR and ELISA, respectively. The mean ± the SEM values of the relative copy numbers from 12 real-time RT-PCR assays were expressed as relative expression compared to that of control cultures without any treatment. *, P < 0.05; and **, P < 0.001 versus no infection. P values indicate differences in the F. nucleatum-induced upregulation of HBDs between control and TLR2-specific RNAi.
FIG. 5.
TLR2-mediated induction of HBD-2 and -3 in HOK-16B cells. HOK-16B cells were stimulated with a synthetic TLR2 ligand, Pam3CSK4 (10 μg/ml), for 24 h. The expression of HBDs and the secretion of IL-8 were evaluated by real-time RT-PCR and ELISA. The mean ± the SEM values of the relative copy numbers from six real-time RT-PCR assays were expressed as a ratio compared to that of control cultures. *, P < 0.01.
The role of NALP2 and TLR2 in the induction of HBDs by F. nucleatum was further addressed in normal gingival epithelial cells. Similar reduction in the level of NALP2 and TLR2 transcripts was observed by NALP2- and TLR2-specific RNAi, respectively (Fig. 6A). However, unlike the situation observed in HOK-16B cells, transfection with TLR2 siRNA significantly upregulated the basal levels of HBD-2 and -3 in normal gingival epithelial cells. NALP2-specific and TLR2-specific RNAi reduced the F. nucleatum-induced upregulation of HBD-3 by ca. 52 and 85%, respectively, compared to the control RNAi experiment (P < 0.0001 and P = 0.02). Similarly, the upregulated level of HBD-2 by F. nucleatum infection was reduced by ca. 84 and 71%, respectively, although significance was not achieved (P = 0.05 and P = 0.24). In contrast, upregulation of IL-8 was not reduced at all (Fig. 6B). These results indicate that NALP2 and TLR2 have roles in the induction of HBDs by F. nucleatum in gingival epithelial cells.
FIG. 6.
Role of NALP2 and TLR2 in the induction of HBD-2, HBD-3, and IL-8 by F. nucleatum in normal gingival epithelial cells. Normal gingival epithelial cells were transfected with control nonsilencing, NALP2-specific, or TLR2-specific siRNA. After 24 h of incubation, cells were infected with live F. nucleatum at an MOI of 1,000 for 24 h. (A) Gene silencing was assessed by real-time RT-PCR. **, P < 0.01; and ***, P < 0.0001 versus control RNAi. (B) The expression of HBDs and secretion of IL-8 by F. nucleatum were evaluated by real-time RT-PCR and ELISA, respectively. The mean ± the SEM values of the relative copy numbers from six real-time RT-PCR assays were expressed as relative expression compared to that of control cultures without any treatment. *, P < 0.05; **, P < 0.01; and ***, P < 0.0001 versus no infection. †, P < 0.05 versus control RNAi. P values indicate difference in the F. nucleatum-induced upregulation of HBDs between control and NALP2- or TLR2-specific RNAi.
Impaired activation of TLR2 and signaling pathways by heat-killed F. nucleatum.
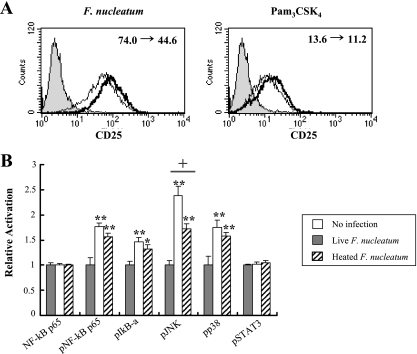
Because TLR2 was found to be involved in inducing both HBD-2 and -3 in HOK-16B cells, we wondered why heat-killed F. nucleatum induced expression of HBD-3 but not HBD-2. The ability of live and heat-killed F. nucleatum to activate TLR2 was examined using a CHO reporter cell line (CHO/CD14/TLR2) that expresses functional TLR2 and surface CD25 in an NF-κB-dependent manner (34). Heat-killed F. nucleatum slightly decreased the TLR2-mediated expression of CD25. The ability of heat-treated Pam3CSK4 to activate TLR2 was similarly reduced (Fig. 7A).
FIG. 7.
Impaired activation of TLR2 and signaling pathways. (A) After coculture of the CHO/CD14/TLR2 cells with live F. nucleatum at an MOI of 2.5 (thick line at left panel) or 10 μg of Pam3CSK4/ml (thick line at right panel) for 16 h, the cells were stained for surface CD25 expression and analyzed by flow cytometry. The expression of CD25 by coculture of the CHO/CD14/TLR2 cells with heat-treated F. nucleatum or Pam3CSK4 was overlaid in thin line. Cells cultured in medium alone served a negative control (filled). The change of mean fluorescence intensity by heat treatment is shown in the upper right corner. The data shown are representative of three similar results. (B) HOK-16B cells plated in a 100-mm dish were stimulated with live or heat-killed F. nucleatum at an MOI of 1,000 for 15 min. After being washed with phosphate-buffered saline, protein lysates were prepared and subjected to ELISA analyses of NF-κB p65 and phosphorylated forms of NF-κB p65, IκB, p38, JNK, and STAT3. The mean ± the SEM values of optical density from four assays were expressed as the relative activation compared to control cultures without bacteria. *, P < 0.05; and **, P < 0.01 versus no infection. †, P < 0.05 live versus heated.
Next, we examined the activation of signaling pathways involved in inflammation. In preliminary Western blotting analysis, the maximum level of p38 and ERK activation by F. nucleatum was observed 15 min after infection. Live F. nucleatum significantly increased the phosphorylated forms of NF-κBp65, IκBα, JNK, and p38 but not of phosphorylated STAT3 or nonphosphorylated NF-κBp65; moreover, a significant reduction in JNK activation was observed when heat-killed F. nucleatum were used (Fig. 7B). These results indicate that activation of TLR2 and downstream signaling pathways is impaired when heat-killed F. nucleatum were used for induction.
DISCUSSION
We investigated the molecular mechanisms involved in the induction of HBDs in gingival epithelial cells by F. nucleatum and showed that NALP2 and TLR2 mediate the induction of HBDs, especially HBD-3.
Since NALP2 is known as a negative regulator of NF-κB (1), we initially hypothesized that gingival epithelial cells might express enough NALP2 to cope with the persistent stimuli of commensal bacteria. However, knockdown of NALP2 RNA in HOK-16B or normal gingival epithelial cells did not increase the basal levels of HBD-2, HBD-3, or IL-8, but rather significantly decreased F. nucleatum-induced expression of HBD-3 (Fig. 3B and 6B). Although significance was not achieved, F. nucleatum-induced expression of HBD-2 was also reduced. Complete knockout of NALP2 may be helpful for clear conclusion. The role of NALP2 as a negative regulator of NF-κB has been shown by overexpressing NALP2 in HEK293T cells or by knocking down endogenous NALP2 in THP-1 cells; however, the function of NALP2 in other cell types is not known. Whether NALP2 has unique physiologic functions in nonhematopoietic and hematopoietic cells warrants further investigation.
Through interaction with adaptor molecules MAL and MyD88, TLR2 induces activation of NF-κB, JNK, and p38, the signaling pathways required for the expression of HBD-2 and -3 (25). Knockdown of TLR2 by RNAi suppressed the F. nucleatum-induced upregulation of HBD-2 and -3 in HOK-16B cells but HBD-3 alone in normal gingival epithelial cells (Fig. 4B and 6B). The discrepancy may be attributed to interindividual variation of normal gingival epithelial cells used. Furthermore, TLR2 RNAi significantly increased the basal levels of HBDs in normal gingival epithelial cells (Fig. 6A), the reason for which is not clear at this moment. However, Pam3CSK4, a synthetic ligand to TLR2 upregulated HBD-2 and -3 in both cell types (Fig. 5 and data not shown), corroborating that TLR2 indeed mediates the induction of HBD-2 and -3 by F. nucleatum.
The fact that heat-killed F. nucleatum induced HBD-3 to some degree but not HBD-2 suggests divergent regulation for HBD-2 and -3 expression (Fig. 1E). Although heat-killed bacteria activated both NF-κB and MAPKs, the activation of JNK was significantly reduced compared to live bacteria (Fig. 7B). It has been reported that p38 and JNK, but not NF-κB, are involved in the induction of HBD-2 by F. nucleatum (19). Therefore, reduced JNK activation may be responsible for the inability of heat-killed F. nucleatum to induce HBD-2, suggesting a higher threshold of JNK activation for HBD-2 induction than for HBD-3. Accordingly, in our previous study only two out of eight bacterial species induced HBD-2, whereas five species induced HBD-3 (13).
The inability of heat-killed F. nucleatum to induce HBD-2 might be attributed to its reduced ability to activate TLR2. TLR2, in complex with TLR1 or TLR6, recognizes diverse structures, including lipopeptides, lipoteichoic acid, lipoarabinomannan, zymosan, and porin (17). Although heat-killed F. nucleatum hardly adhered to the CHO/CD14/TLR2 cells (data not shown), its ability to activate TLR2 was just slightly decreased compared to live bacteria (Fig. 7A). Thus, the relationship between bacterial adhesion and TLR2 activation is not clear. A similar reduction in TLR2 activation was observed when heat-treated Pam3CSK4, a typical lipopeptide, was used as a ligand (Fig. 7A), probably due to the altered structure of cysteine and serine residues that fit tightly into the ligand-binding site of TLR2 (15). In addition, heat-labile ligands might also contribute to the expression of HBD-2 through other PRR(s) such as protease-activated receptor 2 (3).
The reduced HBD-3 expression by heat-killed F. nucleatum might be explained in two ways: the reduced TLR2 activation and inaccessibility to NALP2. Unlike live bacteria, heat-killed F. nucleatum hardly invades epithelial cells (Fig. 1B and D). According to the current model, bacterial ligands are delivered to the cytosol via type III or IV secretion systems, pore-forming toxins, or host cell channels present at either cytoplasmic or phagosomal/endosomal membranes, such as the pannexin 1 pore (16). F. nucleatum does not contain the genes to encode pore-forming toxins or type III or IV secretion systems (7, 18); thus, its ligands may be delivered to the cytosol through a channel located in endosomes. Alternatively, internalized F. nucleatum may escape the endosomes into cytosol, similarly to A. actinomycetemcomitans (22). In any case, bacterial invasion may play a role in access to cytoplasmic PRRs and heat-killed F. nucleatum does not seem to activate NALP2.
Although expression of IL-8 is also known to be regulated by NF-κB, JNK, and p38 (11), knockdown of NALP2 or TLR2 did not affect the expression of IL-8, suggesting distinct regulatory mechanisms for HBDs and IL-8. How the same signaling pathways that are activated by different receptors get sorted to different target genes is unclear. It is interesting that TLR2, which often mediates inflammatory responses in macrophages (4, 30), did not mediate IL-8 induction in gingival epithelial cells. Uehara et al. also reported no induction of IL-8, MCP-1, or IL-6 but efficient induction of HBD-2 by a synthetic ligand to TLR2 in two oral epithelial cell lines (29). In their work, tongue, salivary gland, pharyngeal, esophageal, intestinal, cervical, breast, and lung epithelial cells showed similar responses to TLR2 stimulation; only colon adenocarcinoma cell lines induced the inflammatory chemokines and cytokine. Divergent responses of cells from different tissue or body sites to TLR2 stimulation might be dependent on the characteristics of commensal bacteria to which the cells are constantly exposed.
In conclusion, TLR2 and NALP2 mediate the induction of HBD(s), but not IL-8, by F. nucleatum in gingival epithelial cells and HBD-2 and -3 are divergently regulated.
Acknowledgments
This study was supported by BK21 CLS to the School of Dentistry, Seoul National University, and by grant R01-2007-000-10488-0 to Y.C. from the Korea Science and Engineering Foundation.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 22 December 2008.
REFERENCES
- 1.Bruey, J. M., N. Bruey-Sedano, R. Newman, S. Chandler, C. Stehlik, and J. C. Reed. 2004. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-κB and caspase-1 activation in macrophages. J. Biol. Chem. 27951897-51907. [DOI] [PubMed] [Google Scholar]
- 2.Chung, W. O., and B. A. Dale. 2004. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect. Immun. 72352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung, W. O., S. R. Hansen, D. Rao, and B. A. Dale. 2004. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J. Immunol. 1735165-5170. [DOI] [PubMed] [Google Scholar]
- 4.Cole, L. E., K. A. Shirey, E. Barry, A. Santiago, P. Rallabhandi, K. L. Elkins, A. C. Puche, S. M. Michalek, and S. N. Vogel. 2007. Toll-like receptor 2-mediated signaling requirements for Francisella tularensis live vaccine strain infection of murine macrophages. Infect. Immun. 754127-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dale, B. A. 2002. Periodontal epithelium: a newly recognized role in health and disease. Periodontol. 2000 3070-78. [DOI] [PubMed] [Google Scholar]
- 6.Dale, B. A., J. R. Kimball, S. Krisanaprakornkit, F. Roberts, M. Robinovitch, R. O'Nell, E. V. Valore, T. Ganz, G. M. Anderson, and A. Weinberg. 2001. Localized antimicrobial peptide expression in human gingiva. J. Periodontal Res. 36285-294. [DOI] [PubMed] [Google Scholar]
- 7.Desvaux, M., A. Khan, S. A. Beatson, A. Scott-Tucker, and I. R. Henderson. 2005. Protein secretion systems in Fusobacterium nucleatum: genomic identification of type 4 piliation and complete type V pathways brings new insight into mechanisms of pathogenesis. Biochim. Biophys. Acta 171392-112. [DOI] [PubMed] [Google Scholar]
- 8.Feng, Z., and A. Weinberg. 2006. Role of bacteria in health and disease of periodontal tissues. Periodontol 2000 4050-76. [DOI] [PubMed] [Google Scholar]
- 9.Feucht, E. C., C. L. DeSanti, and A. Weinberg. 2003. Selective induction of human beta-defensin mRNAs by Actinobacillus actinomycetemcomitans in primary and immortalized oral epithelial cells. Oral Microbiol. Immunol. 18359-363. [DOI] [PubMed] [Google Scholar]
- 10.Han, Y. W., W. Shi, G. T. Huang, S. K. Haake, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fuobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 683140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann, E., O. Dittrich-Breiholz, H. Holtmann, and M. Kracht. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72847-855. [PubMed] [Google Scholar]
- 12.Ji, S., J. Hyun, E. Park, B. L. Lee, K. K. Kim, and Y. Choi. 2007. Susceptibility of various oral bacteria to antimicrobial peptides and to phagocytosis by neutrophils. J. Periodontal Res. 42410-419. [DOI] [PubMed] [Google Scholar]
- 13.Ji, S., Y. Kim, B.-Y. Min, S. H. Han, and Y. Choi. 2007. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J. Periodontal Res. 42503-510. [DOI] [PubMed] [Google Scholar]
- 14.Jia, H. P., B. C. Schutte, A. Schudy, R. Linzmeier, J. M. Guthmiller, G. K. Johnson, B. F. Tack, J. P. Mitros, A. Rosenthal, T. Ganz, and P. B. McCray, Jr. 2001. Discovery of new human β-defensins using a genomics-based approach. Gene 263211-218. [DOI] [PubMed] [Google Scholar]
- 15.Jin, M. S., S. E. Kim, J. Y. Hoe, M. E. Lee, H. M. Kim, S. Paik, H. Lee, and J. O. Lee. 2007. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 1301071-1082. [DOI] [PubMed] [Google Scholar]
- 16.Kanneganti, T. D., M. Lamkanfi, and G. Núñez. 2007. Intracellular NOD-like receptors in host defense and disease. Immunity 27549-559. [DOI] [PubMed] [Google Scholar]
- 17.Kanzler, H., F. J. Barrat, E. M. Hessel, and R. L. Coffman. 2007. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat. Med. 13552-559. [DOI] [PubMed] [Google Scholar]
- 18.Kapatral, V., N. Ivanova, I. Anderson, G. Reznik, A. Bhattacharyya, W. L. Gardner, N. Mikhailova, A. Lapidus, N. Larsen, M. D'Souza, T. Walunas, R. Haselkorn, R. Overbeek, and N. Kyrpides. 2003. Genome analysis of Fusobacterium nucleatum subsp. vincentii and its comparison with the genome of F. nucleatum ATCC 25586. Genome Res. 131180-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of Human β-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathway, but not the NF-κB transcription factor family. J. Immunol. 168316-324. [DOI] [PubMed] [Google Scholar]
- 20.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 682907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menzies, B. E., and A. Kenoyer. 2006. Signal transduction and nuclear responses in Staphylococcus aureus-induced expression of human β-defensin 3 in skin keratinocytes. Infect. Immun. 746847-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer, D. H., J. E. Lippmann, and P. M. Fives-Taylor. 1996. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect. Immun. 642988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min, B.-M., K. M. Woo, G. Lee, and N.-H. Park. 1999. Terminal differentiation of normal human oral keratinocytes is associated with enhanced cellular TGF-β and phospholipase c-γ1 levels and apoptotic cell death. Exp. Cell Res. 249377-385. [DOI] [PubMed] [Google Scholar]
- 24.Nishihara, T., and T. Koseki. 2004. Microbial etiology of periodontitis. Periodontol. 2000 3614-26. [DOI] [PubMed] [Google Scholar]
- 25.O'Neill, L. A., and A. G. Bowie. 2007. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7353-364. [DOI] [PubMed] [Google Scholar]
- 26.Papapanou, P. N., A. Sellén, J. L. Wennström, and G. Dahlén. 1993. An analysis of the subgingival microflora in randomly selected subjects. Oral Microbiol. Immunol. 824-29. [DOI] [PubMed] [Google Scholar]
- 27.Park, N.-H., B.-M. Min, S. L. Li, M. Z. Huang, H. M. Cherick, and J. Doniger. 1991. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis 121627-1631. [DOI] [PubMed] [Google Scholar]
- 28.Taguchi, Y., and H. Imai. 2006. Expression of beta-defensin-2 in human gingival epithelial cells in response to challenge with Porphyromonas gingivalis in vitro. J. Periodontal Res. 41334-339. [DOI] [PubMed] [Google Scholar]
- 29.Uehara, A., Y. Fujimoto, K. Fukase, and H. Takada. 2007. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Molecular Immunol. 443100-3111. [DOI] [PubMed] [Google Scholar]
- 30.Underhill, D. M., A. Ozinsky, A. M. Hajjar, A. Stevens, C. B. Wilson, M. Basseti, and A. Aderem. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401811-815. [DOI] [PubMed] [Google Scholar]
- 31.Vankeerberghen, A., H. Nuytten, K. Dierickx, M. Quirynen, J. J. Cassiman, and H. Cuppens. 2005. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J. Periodontol. 761293-1303. [DOI] [PubMed] [Google Scholar]
- 32.Weinberg, A., S. Krisanaprakornkit, and B. A. Dale. 1998. Epithelial antimicrobial peptides: review and significance for oral application. Crit. Rev. Oral Biol. Med. 9399-414. [DOI] [PubMed] [Google Scholar]
- 33.Ye, Z., and J. P. Ting. 2008. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr. Opin. Immunol. 203-9. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimura, A., E. Lien, R. R. Ingalls, E. Tuomanen, R. Dziarski, and D. Golenbock. 1999. Cutting edge: recognition of gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 1631-5. [PubMed] [Google Scholar]