Abstract
Continuing threats of devastating outbreaks in poultry and of human infections caused by highly pathogenic avian influenza virus (HPAIV) H5N1 emphasize the need for the further development of rapid and reliable methods of virus detection and characterization. Here we report on the design and comprehensive validation of a low-density microarray as a diagnostic tool for the detection and typing of avian influenza virus (AIV). The array consists of one probe for the conserved matrix gene and 97 probes targeting the HA0 cleavage-site region. Following fragment amplification by a generic PCR approach, the array enables AIV detection, hemagglutinin (HA) subtyping, and pathotyping within a single assay. For validation, a panel of 92 influenza A viruses which included 43 reference strains representing all 16 HA subtypes was used. All reference strains were correctly typed with respect to their HA subtypes and pathotypes, including HPAIV H5N1/Asia, which caused outbreaks in Germany in 2006 and 2007. In addition, differentiation of strains of the Eurasian and North American lineages of the H5 and H7 subtypes was possible. The sensitivity of the microarray for the matrix gene is comparable to that of real-time reverse transcription-PCR (RT-PCR). It is, however, 10- to 100-fold lower than that of real-time RT-PCR with respect to HA subtyping and pathotyping. The specificity of the array was excellent, as no pathogens relevant for differential diagnosis yielded a positive reaction. Validation with field samples included 19 cloacal swab specimens from wild and domestic birds. Influenza A virus was verified in all samples, whereas the HA subtypes could be determined for 14 samples. The results demonstrate that the microarray assay described complements current methods and can accelerate the diagnosis and characterization of AIV.
Influenza A virus, a genus within the family Orthomyxoviridae (11), infects wild birds and poultry as well as humans and other mammals. Its genome consists of a 13.5-kb single-stranded negative-sense RNA organized in eight segments. Influenza A viruses show degrees of high genetic and antigenic variability. A total of 16 hemagglutinin (HA) subtypes and 9 neuraminidase (NA) subtypes have been described (1, 2, 12). Influenza A viruses of all subtypes are found in birds, whereas populations of humans, pigs, and horses are endemically affected by certain species-adapted subtypes. Virus subtyping is routinely accomplished after virus isolation in embryonated chicken eggs and is combined with the hemagglutination inhibition assay or reverse transcription (RT)-PCR, followed by partial or complete sequencing of the HA and NA genes (16, 31, 42).
In the case of avian influenza viruses (AIVs), two pathotypes are differentiated. AIVs of low pathogenicity (LPAIVs) are maintained in aquatic wild birds as reservoir hosts, in which the infection remains localized to the respiratory and intestinal tracts. LPAIVs of subtypes H5 and H7, upon transmission to gallinaceous poultry, can give rise to highly pathogenic AIVs (HPAIVs), which cause severe systemic infections and epidemic disease with high rates of mortality (7). HPAIVs are characterized by intravenous pathogenicity indices of greater than 1.2 (1, 2). Alternatively, they can be pathotyped by sequence analysis of the cleavage site within the hemagglutinin precursor protein HA0 (1, 2, 3, 41). LPAIVs show a monobasic amino acid composition at the HA0 cleavage site, and their HA0 is cleaved extracellularly by tissue-specific, trypsin-like proteases. In contrast, HPAIVs, with very few exceptions (26), have a polybasic HA0 cleavage site, which results in intracellular processibility by ubiquitous subtilisin-like proteases (30, 36).
AIV subtyping and subsequent pathotyping are of utmost significance for diagnostics and for surveillance and epidemiological studies, as well as for the initiation of restrictive measures against HPAIVs. Molecular methods have the advantage of accelerating diagnostics and reducing the risk of handling infectious material. Considering the genetic diversity of influenza A viruses and the constant emergence of new strains (6, 8, 35, 40), the limitations of present RT-PCR-based assays become obvious. Microarray assays, which are capable of detecting multiple targets in parallel, open new possibilities, including the in-depth analysis of virus isolates within a short turnaround time. Recently, microarray assays have been described as valuable methods for the detection, differential diagnosis, and subtyping (of the H1, H2, H3, H5, N1, and N2 subtypes) of influenza A viruses with relevance to human disease (9, 10, 18, 20, 21, 22, 27, 28, 38, 39), as well as for HA and NA subtyping (23, 24, 32, 33).
Here we report on the design of a novel, low-density microarray that facilitates the fast detection of AIV, HA subtyping, and pathotyping. Extensive validation was performed, with emphasis placed on the utilization of the microarray system as a diagnostic tool.
MATERIALS AND METHODS
Viruses, bacteria, and diagnostic samples.
A panel of 92 influenza A virus isolates (Table 1) was obtained as allantoic fluids from the OIE and the German National Reference Laboratory for Avian Influenza. In addition, 19 field samples that consisted of cloacal swab samples from wild and domestic birds were used. Specificity tests were conducted with RNA or DNA from Newcastle disease virus strain La Sota, reovirus strain 1133 (Intervet), infectious bursitis disease virus D78, infectious laryngotracheitis virus A489 (Intervet), Mycoplasma gallinarum, and Mycoplasma gallinaceum.
TABLE 1.
Influenza A viruses used in this study
| No. | Isolatea | Acc. no.b |
|---|---|---|
| 1 | A/swine/Bakum/5/95 (H1N1) | DQ100426 |
| 2 | A/USSR/90/77 (H1N1) | CY010372 |
| 3 | A/Fort Monmoth/1/47 (H1N1) | CY009612 |
| 4 | A/Denver/1/57 (H1N1) | CY008988 |
| 5 | A/Chile/1/83 (H1N1) | |
| 6 | R30/06 (H1N1) | |
| 7 | A/Anser egypticus/Germany/R1419/2006 (H1N1) | AM922139 |
| 8 | A/duck/Potsdam/177/83 (H2N2) | CY005765 |
| 9 | A/bantam/Germany/DZ4/85 (H2N2) | AM922141 |
| 10 | A/Mallard/Germany/Wv1317-21/03 (H2N3) | AM922142 |
| 11 | A/Mallard/Germany/Wv677/04 (H2N3) | AM922143 |
| 12 | A/Mallard/Germany/Wv94345/04 (H2N3) | AM922144 |
| 13 | A/HongKong/1/68 (H3N2) | EF409245 |
| 14 | A/Mallard/439/04 (H3N2) | |
| 15 | A/Mallard/Germany/Wv1303-04/03 (H3N8) | AM087224c |
| 16 | A/Mallard/Germany/Wv64-67/05 (H3N8) | |
| 17 | A/duck/Ukraine/1/63 (H3N8) | AB292668 |
| 18 | A/Mallard/Germany/Wv180 6-09/03 (H4N6) | AM922147 |
| 19 | A/Mallard/Wv1754-57/03 (H4N6) | AM922148 |
| 20 | A/Mallard/Wv1732-34/03 (H4N6) | AM922149 |
| 21 | A/duck/Czechoslowakia/56 (H4N6) | M25283 |
| 22 | A/Mallard/Germany/Wv1027/04 (H4N6) | |
| 23 | A/HongKong/156/97 (H5N1) | AF046088 |
| 24 | A/whooper swan/Germany/R65/06 (H5N1) | DQ464354 |
| 25 | A/dk/Vietnam/TG24-01/05 (H5N1) | AM183677 |
| 26 | A/Teal/Wv632/05 (H5N1) | |
| 27 | A/chicken/Italy/8/98 (H5N2) | AJ305306 |
| 28 | A/Mall/QC/2323-19/2006 (H5N2) | |
| 29 | A/Goose/MB/428/06 (H5N2) | |
| 30 | A/DK/BC/26-6/05 (H5N2) | DQ309440 |
| 31 | A/Mallard/Föhr/Wv1349-51/03 (H5N3) | AM087222 |
| 32 | A/chicken/Italy/22/98 (H5N9) | CY022621 |
| 33 | A/Mal/AB/392/2006 (H5N9) | |
| 34 | A/Mall/BC/544-2005 (H5N9) | |
| 35 | A/Wigeon/Föhr/Wv579/05 (H6) | |
| 36 | A/turkey/Hessen/R04/99 (H6N2) | AJ507207 |
| 37 | A/turkey/Hartzfehn/R26/99 (H6N2) | AJ507208 |
| 38 | A/turkey/LTZ/R63/02 (H6N2) | |
| 39 | A/turkey/Grub/R41/98 (H6N5) | |
| 40 | A/turkey/Grub/R42/98 (H6N5) | |
| 41 | A/turkey/Grub/R43/98 (H6N5) | AJ507206 |
| 42 | A/turkey/Heidemark/R83/99 (H6N7) | |
| 43 | A/broiler/Italy/445/99 (H7N1) | AJ580353 |
| 44 | A/chicken/Brescia/19/02 (H7N1) | AM922154 |
| 45 | A/hen/Italy/444/99 (H7N1) | AJ704810 |
| 46 | A/Mallard/NVP/41/04 (H7N1) | |
| 47 | A/turkey/Italy/472/99 (H7N1) | AJ704811 |
| 48 | A/TY/18-2/2000 (H7N1) | AF497552 |
| 49 | A/swan/Potsdam/64/81 (H7N3) | AM922155 |
| 50 | A/duck/Alberta/48/76 (H7N3) | AF497554 |
| 51 | A/turkey/Italy/2043/03 (H7N3) | |
| 52 | A/Ck/BC/04 (H7N3) | AY611524 |
| 53 | A/Mall/AB/8734/2007 (H7N3) | |
| 54 | A/chicken/Germany/R28/03 (H7N7) | AJ620350 |
| 55 | A/Mallard/NVP/177 6-80/03 (H7N7) | |
| 56 | A/duck/Potsdam/15/80 (H7N7) | AJ704797 |
| 57 | A/turkey/Germany/R11/01 (H7N7) | AJ704812 |
| 58 | A/turkey/Ireland/PV8/98 (H7N7) | AJ704799 |
| 59 | A/turkey/Ontario/6118/68 (H8N4) | CY014659 |
| 60 | A/Anser spec./Germany/R44/2006 (H8) | AM922158 |
| 61 | A/turkey/Wisconsin/66 (H9N2) | CY014663 |
| 62 | A/turkey/Germany/176/95 (H9N2) | AF218101 |
| 63 | A/chicken/Emirates/R66/02 (H9N2) | AM922159 |
| 64 | A/Iran/541 (H9N2) | AJ781825 |
| 65 | A/chicken/Germany/90/95 (H9N2) | AF218099 |
| 66 | A/turkey/Germany/22/96 (H9N2) | AJ781820 |
| 67 | A/Moorhen/LKStendal/Wv1703-04 (H10N4) | |
| 68 | A/Mallard/NVP/1682-85 (H10N4) | |
| 69 | A/Mallard/NVP/Wv1677-81/03 (H10N4) | AM922160 |
| 70 | A/Mallard/Föhr/Wv1781-82/03 (H10N7) | |
| 71 | A/chicken/Germany/N/49 (H10N7) | CY014671 |
| 72 | A/guinea fowl/Hungary/1/6 (H10N8) | |
| 73 | A/quail/Italy/1117/65 (H10N8) | CY014644 |
| 74 | A/duck/England/56 (H11N6) | CY014679 |
| 75 | A/Mallard/Föhr/Wv1499-1503/03 (H11N9) | |
| 76 | R1128/06 (H11) | |
| 77 | A/Anas platyrhynchos/Germany/R2219/2006 (H11) | AM922161 |
| 78 | A/Anas platyrhynchos/Germany/R2843/2006 (H11) | |
| 79 | A/duck/Alberta/60/76 (H12N5) | AB288334 |
| 80 | A/pilot whale/Maine/328/84 (H13N2) | M26091 |
| 81 | A/gull/Maryland/704/77 (H13N6) | CY014694 |
| 82 | A/gull/Stralsund/Wv1136-40/03 (H13N6) | AM922163 |
| 83 | R2603/06 (H13N8) | |
| 84 | R2609/06 (H13N8) | |
| 85 | R2613/06 (H13) | |
| 86 | R2622/06 (H13) | |
| 87 | A/Larus ridibundus/Germany/R2064/2006 (H13) | AM922164 |
| 88 | A/Ma/Gur/263/82 (H14N3) | M35997 |
| 89 | A/She/WA/2576/79 (H15N6) | CY006010 |
| 90 | A/BHG/Sweden/5/99 (H16N3) | AY684891 |
| 91 | R2788/06 (H16) | |
| 92 | R2792/06 (H16) |
The NA subtypes were not determined for all isolates.
Acc. no., accession number from the EMBL/GenBank/DDBJ databases.
No sequence data were available for the HA cleavage site.
Isolation of RNA.
Viral RNA was purified from the allantoic fluids and cloacal swabs with a QIAamp viral RNA minikit (Qiagen), according to the manufacturer's instructions. The protocol was modified by addition of 5 μl in vitro-transcribed internal control RNA (2 × 105 copies/μl), after lysis of the sample to control the efficiency of RNA isolation and RT-PCR (15).
Target preparation.
One-step RT-PCR for amplification of the HA0 cleavage site of influenza A viruses of all 16 HA subtypes (pan-HA RT-PCR) was performed as described previously (13). In parallel, RT-PCR targeting the matrix (M) gene was conducted (34) by utilizing the same protocol with 20 pmol/μl each of primers IVA-M_1for (AGA TGA GTC TTC TAA CCG AGG TCG) and IVA-M1.1(647)-R (GCA GTA TAT CGC TTG ACA TGC AAA AAC ATC TTC AAG TYT CTG; a nonviral universal tail sequence is presented in italics; MWG Biotech AG, Germany). Pan-HA amplicons of 164 to 176 bp (+18 nucleotides of the universal tail) and M-gene amplicons of ∼101 bp (+18 nt nucleotides of the universal tail) were visualized by 3% agarose gel electrophoresis for 1 h at 120 V in TAE (Tris-acetate-EDTA) and submitted to microarray analysis without further purification.
Design of array probes.
A total of 1,997 influenza A virus HA sequences were derived from the EMBL/GenBank/DDBJ databases. ClustalW alignments (37) were prepared for each HA subtype with the software BioEdit (version 7.0.9.0) (14). Within the sequence of the pan-HA RT-PCR product, capture probes with a theoretical melting temperature of 60 to 65°C were designed by following the guidelines compiled by Barlaan et al. (4). For HA subtyping, probes targeting regions that are conserved within a given subtype but that are variable between subtypes were chosen. For pathotyping, probes targeting the HA0 cleavage site of HPAIV were designed. For some influenza A virus isolates, specific sequence data were generated as described previously (13) and specific probes for HA subtyping were deduced. Furthermore, the probe IVA-MA-Bio (34) was used for detection of the M gene. A nonspecific probe, the standard M13uni(−43)-bio primer sequence, was utilized for calculation of signal/noise ratios. All probes used in the array (see Table S1 in the supplemental material) were verified in silico with the software tool OligoCalc (19) as well as by a BLASTn search. Probes were biotinylated at their 5′ ends and synthesized by MWG Biotech AG.
Microarray processing.
The NanoChip 400 system (Nanogen Inc.), whose technical features and hybridization conditions were described by Borsting et al. (5) and Keen-Kim et al. (17), was used. Biotinylated capture probes (100 nM in 50 mM histidine buffer) were electronically addressed at 350 nA/pad for 15 s to streptavidin comprising individual sites of the NanoChip cartridge by use of the NanoChip 400 system (Nanogen Inc.). For each sample, 5 μl (each) of pan-HA and M-gene amplicon was mixed with 65 μl Capdown sample buffer A (114 mM histidine, 142.5 mM 1-thioglycerol; Nanogen Inc.) and electronically addressed at 995 nA/pad for 120 s to designated sites loaded with HA subtyping and pathotyping probes and the IVA-MA-Bio probe (34), respectively. In addition, each amplicon was also hybridized to the M13uni(−43)-bio probe for calculation of signal/noise ratios. A universal reporter probe mixture containing 0.8 μM Uni-Alexa555 (CTC AAT GTT CGG ACT CAG) labeled at the 5′ end with the fluorescent dye Alexa Fluor 555 and 0.5 μM Uni-Alexa647 (TGT CAA GCG ATA TAC TGC) labeled at the 5′ end with Alexa Fluor 647 (MWG Biotech AG) in high-salt buffer (Nanogen Inc.) was prepared. Hybridization was accomplished with the temperature profile 80 s at 60°C, 60 s at 56°C, 25 s at 53°C, 25 s at 47°C, 25 s at 44°C, 25 s at 41°C, and 25 s at 38°C, followed by two washes for 60 s at 37°C with high-salt buffer. Reporting was performed at 24°C by excitation with 525-nm and 626-nm lasers and scanning with a charge-coupled-device camera integrated in the NanoChip 400 system. The reporter probes and amplicons were removed by two washes for 60 s at 56°C with low-salt buffer (Nanogen Inc.) and subsequent electronic addressing of target prep buffer (Nanogen Inc.) at −995 nA/pad for 120 s. The microarray could be used for the simultaneous or sequential analysis of up to four samples.
Data analysis.
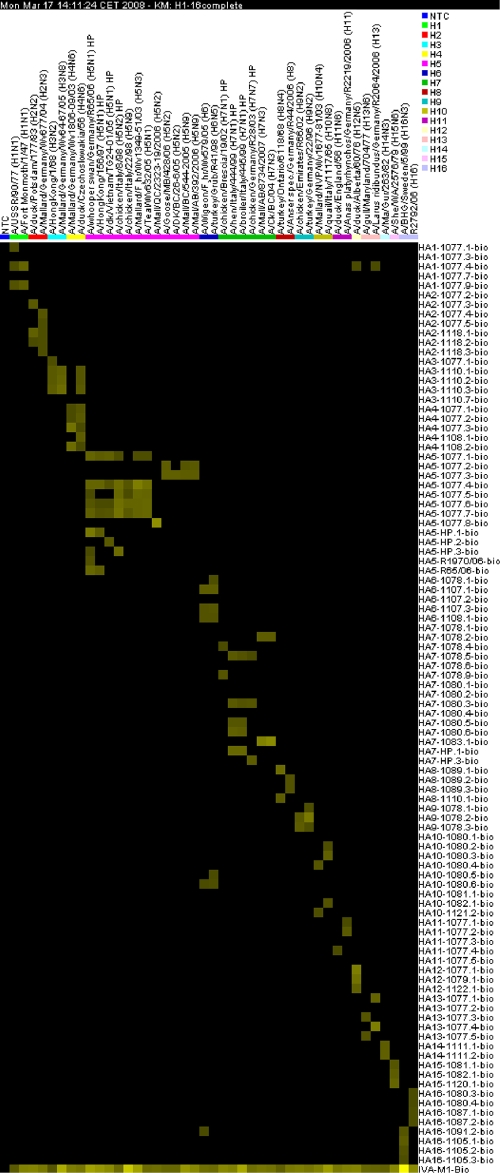
The acquired background reference images were subtracted from the raw images by the NanoChip 400 data analysis software (version 1.00.09). The processed data were analyzed with a script (see File S2 in the supplemental material) based on software R (version 2.5.1) and were summarized in an Excel program file. Signal/noise ratios were calculated for each spot by division of the signal derived from the influenza virus-specific probe by the signal from nonspecific probe M13uni(−43)-bio. The cutoff for a positive value was set at a signal/noise ratio of 4. The software GenePilot (version 1.29b) was used to visualize data sets as a heat map (Fig. 1). Thereby, signal/noise ratios were plotted on a continuous color spectrum. The upper limit (the brightest yellow color) was set for the spot with the highest signal/noise ratio, while the lower limit (black color) stands for spots with a signal/noise ratio of 0.
FIG. 1.
Characterization of 43 influenza A virus isolates. The heat map was created with the software GenePilot (version 1.29b). Signal/noise ratios of the fluorescence are plotted on a continuous color spectrum and are shown as pixels. The upper limit (the brightest yellow color) was set for the spot with the highest signal/noise ratio, while the lower limit (black) stands for spots with a signal/noise ratio of 0. Therefore, the brighter yellow that a spot is, the higher the signal/noise ratio is.
Duplex rRT-PCR.
Real-time RT-PCR (rRT-PCR) for detection of the M gene of influenza A virus was carried out by a previously published protocol (34), with some modifications, which included the integration of an internal control system (15). For absolute quantification, each run included four external standards of in vitro-transcribed RNA.
RESULTS
Selection of array probes.
After in silico analysis, each array probe was validated for its performance with the NanoChip 400 electronic microarray system. To this end, probes for HA subtyping and pathotyping were tested in checkerboards with pan-HA amplicons of the appropriate subtype and pathotype from a total of 92 influenza A virus isolates (Table 1). The panel of avian influenza A viruses used in this study comprised isolates of all known HA subtypes. One porcine isolate (H1N1) and six human isolates (four H1N1 isolates and one isolate each of H3N2 and H5N1) were also analyzed. The isolates were recovered over wide geographical and temporal ranges. All probes resulting in signal/noise ratios of greater than 3 were added to the microarray for AIV (see Table S3 in the supplemental material, in which the probes with signal/noise ratios of greater than 3 are shown in boldface type). Coverage was deemed sufficient when each influenza A virus positive by the pan-HA RT-PCR was detected with a minimum of two probes. The exceptions were A/Mall/QC/2323-19/2006 (H5N2) and R1128/06 (H11), which were each detected with one probe only. The probe IVA-MA-Bio detected the M-gene amplicon derived from each of the 92 influenza A virus isolates listed in Table 1 (data not shown). Moreover, the nonspecific probe M13uni(−43)-bio (standard primer sequence) used for calculation of signal/noise ratios scored negative for the pan-HA and M-gene amplicons of all 92 influenza A virus isolates (data not shown). Finally, a total of 99 probes were included in the microarray. Sequences, positions, and specific targets are listed in Table S1 in the supplemental material.
Characterization of influenza A virus isolates.
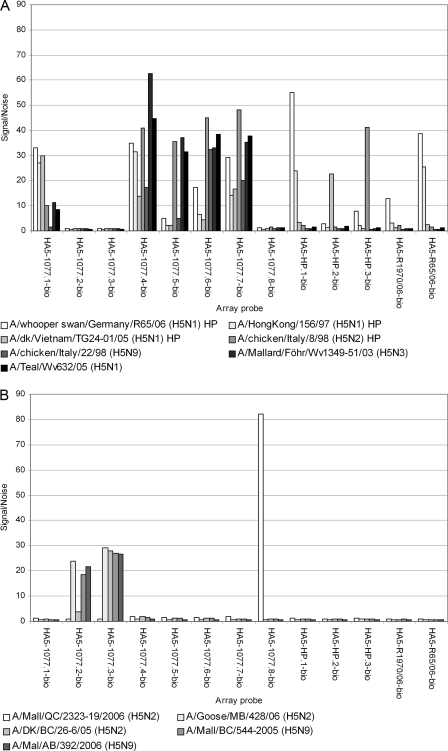
Among the panel of 92 influenza A virus isolates used in this study, a total of 43 reference strains were tested with the microarray for AIV. All were identified, and their HA subtypes as well as pathotypes were correctly determined (Fig. 1). No signals could be observed for the negative control. Detection of HPAIV H5N1/Asia, which has occurred in Germany since 2006 (35, 40), was also ensured by the use of specific probes. The geographic origin (Eurasia or North America) of influenza A viruses of the H5 and H7 subtypes could be differentiated by their specific reaction patterns. As an example, Fig. 2A displays the data as a bar graph for seven influenza A viruses of the H5 subtype from Eurasia. They reacted with probes HA5-1077.1-bio, HA5-1077.4-bio, HA5-1077.5-bio, HA5-1077.6-bio, and HA5-1077.7-bio. HPAIVs had positive reactions with probes HA5-HP.2-bio and HA5-HP.3-bio, whereas the HA0 cleavage site of HPAIV H5N1/Asia was specifically detected by HA5-HP.1-bio, HA5-R1970/06-bio, and HA5-R65/06-bio. Figure 2B shows the results for five H5 influenza A viruses from North America. They reacted with probes HA5-1077.2-bio, HA5-1077.3-bio, and HA5-1077.8-bio. Similar results were found for H7 influenza A viruses (data not shown; refer to Fig. 1 and to Table S4 in the supplemental material). Nevertheless, for isolate A/duck/Czechoslovakia/56 (H4N6), in addition to five probes specific for the H4 subtype, positive results were obtained with three H3-specific probes. The virus A/turkey/Grub/R41/98 (H6N5) had strong signals with four probes specific for the H6 subtype, besides (weaker) signals with two H10-specific probes. Figure 1 summarizes the results for all 43 influenza A viruses included as a heat map, whereas detailed data can be found in Table S4 in the supplemental material.
FIG. 2.
Differentiation of HPAIVs and LPAIVs among Eurasian and North American influenza A viruses of subtype H5. (A) Seven isolates (including LPAIV and HPAIV isolates) from Eurasia were tested. (B) Five LPAIV isolates from North America were analyzed. Only data for H5-specific probes are presented in the bar charts. The signal/noise ratios resulting from other probes can be found in Table S4 in the supplemental material.
Specificity.
Newcastle disease virus strain La Sota, reovirus strain 1133, infectious bursitis disease virus D78, infectious laryngotracheitis virus A489, Mycoplasma gallinarum, and Mycoplasma gallinaceum were not detected with the AIV microarray (see Table S5 in the supplemental material).
Sensitivity of the microarray compared to that of rRT-PCR.
The sensitivity of the microarray for the M genes of the influenza A viruses of the H5 subtype (A/whooper swan/Germany/R65/06 [H5N1]) and the H7 subtype (A/chicken/Italy/444/99 [H7N1]) selected was comparable to that of the M-gene-specific rRT-PCR (34). For HA subtyping and pathotyping, the sensitivity of the microarray depended on the pan-HA RT-PCR protocol used (13), and the microarray had detection limits of 1.6 × 103 copies (H5) and 1.5 × 102 copies (H7) (Table 2). For the H5N1 isolate selected, the sensitivity of the microarray was 100-fold lower than that of the rRT-PCR targeting the M gene, whereas it was 10-fold lower than that of the rRT-PCR targeting the M gene of the H7N1 virus tested.
TABLE 2.
Sensitivity for influenza A virus of subtypes H5 and H7
| Virus and dilution | M-gene-specific rRT-PCR result
|
RT-PCR resulta
|
Microarray assay result
|
|||
|---|---|---|---|---|---|---|
| CT (dR)b | No. of copies/reaction | M gene | Pan-HA | M gene | H5 and H7 | |
| A/whooper swan/Germany/R65/06 (H5N1) HPAIV | ||||||
| 10−4 | 28.09 | 1.51E + 04 | +++ | + | + | + |
| 10−5 | 31.4 | 1.63E + 03 | +++ | + | + | + |
| 10−6 | 34.83 | 1.63E + 02 | ++ | +− | + | − |
| 10−7 | 38.42 | 1.45E + 01 | ++ | − | + | − |
| A/hen/Italy/444/99 (H7N1) HPAIV | ||||||
| 10−4 | 28.19 | 1.41E + 04 | +++ | ++ | + | + |
| 10−5 | 31.59 | 1.44E + 03 | +++ | + | + | + |
| 10−6 | 34.91 | 1.54E + 02 | ++ | + | + | + |
| 10−7 | 38.12 | 1.77E + 01 | + | +− | + | − |
+++ to + −, different intensities of the bands by agarose gel electrophoresis; −, no band visible.
CT, threshold cycle; dR, baseline-corrected fluorescence intensity.
Characterization of influenza A viruses from diagnostic samples.
RNA from 19 cloacal swab specimens from wild and domestic birds was proven to contain the influenza A virus genome (threshold cycle value range, 20.43 to 33.10 by the M-gene-specific rRT-PCR [34]) (Table 3). By using the newly developed microarray, the presence of influenza A virus in all swabs was verified by detection of the M gene. HA subtyping succeeded for 14 field samples for which a pan-HA RT-PCR product (13) could be generated. The HA subtypes determined by the microarray assay corresponded to the subtypes obtained by sequencing of the HA2 fragment (31) and/or the HA0 cleavage site (13) in all cases.
TABLE 3.
Characterization of influenza A viruses from diagnostic samples
| Samplea | M-gene-specific rRT-PCR resultb | RT-PCR resultc
|
Microarray assay result | |
|---|---|---|---|---|
| M gene | Pan-HA | |||
| A/Anas platyrhynchos/Germany/R2322/07 (H3N8) | 23.49 | +++ | +++ | M, H3 |
| A/Dendrocygna viduata/Germany/R2484/07 (H3N8) | 28.86 | +++ | ++ | M, H3 |
| A/wild duck/Germany/R4/08 (H1N1) | 28.20 | +++ | ++ | M, H1 |
| A/Anas platyrhynchos/Germany/R91/08 (H3N8) | 31.32 | +++ | ++ | M, H3 |
| A/Branta canadanensis/Germany/R105/08 (H11N9) | 23.88 | +++ | ++ | M, H11 |
| A/Anas platyrhynchos/Germany/R1649/07 (H1N1) | 27.97 | +++ | + | M, H1 |
| A/Anas domesticus/Germany/R2583/07 (H9Nx) | 27.85 | +++ | + | M, H9 |
| A/Anas platyrhynchos/Germany/R2706/07 (H2N9) | 27.75 | +++ | + | M, H2 |
| A/Anas platyrhynchos/Germany/R2777/07 (H9Nx) | 26.54 | +++ | + | M, H9 |
| A/wild duck/Germany/R5/08 (H6Nx) | 27.69 | +++ | + | M, H6 |
| A/Branta canadanensis/Germany/R103/08 | 27.67 | +++ | + | M, H3 |
| A/Anas platyrhynchos/Germany/R3240/07 (H1N1) | 29.45 | +++ | + | M, H1 |
| A/Houbara/Dubai/R3218/07 (H9N2) | 20.43 | +++ | + | M |
| A/Anas platyrhynchos/Germany/R2709/07 (H2N9) | 27.13 | +++ | +− | M, H2 |
| A/Cygnus olor/Germany/R3310/07 (H6Nx) | 29.29 | +++ | +− | M, H6 |
| A/Numida meleagris/Germany/R2495/07 (H7N3LP) | 23.61 | +++ | +− | M |
| A/Numida meleagris/Germany/R2497/07 (H7N3LP) | 29.20 | +++ | +− | M |
| A/Anas platyrhynchos/Germany/R2619/07 | 33.10 | +++ | +− | M |
| A/Wild goose/Germany/R3250/07 (H6N1) | 28.29 | +++ | +− | M |
Subtyping was not successful for all samples. HA subtypes determined by sequencing of the HA2 gene (31) and/or the HA0 cleavage site (13).
The data represent the threshold cycle (dR [baseline-corrected fluorescence intensity]) values.
+++ to + −, different intensities of the bands by agarose gel electrophoresis; −, no band visible.
DISCUSSION
The rapid detection and the exact characterization of avian influenza viruses are fundamental for diagnostics during both outbreaks of highly pathogenic avian influenza and surveillance of infections with LPAIVs. The current epidemic of HPAIV H5N1 of Asian origin indicates the limitations of existing diagnostic methods. Microarrays that simultaneously detect multiple genes enable a novel approach to the common problem of AIV classification. Here we describe a low-density microarray assay for the simultaneous detection, HA subtyping, and pathotyping of AIV.
Every single probe used on the microarray for AIV was validated by testing of the microarray with a panel of 92 influenza A virus isolates, in addition to in silico analysis. Amplicons from a previously described pan-HA RT-PCR (13) that amplifies the HA0 cleavage site of influenza A viruses of all 16 HA subtypes as well as an M-gene-specific RT-PCR (34) were tested with appropriate probes on the NanoChip 400 electronic microarray. The microarray developed comprises 97 probes (from among the 115 probes verified in silico) for HA subtyping and pathotyping, 1 probe for the detection of the M gene, and 1 nonspecific probe for calculation of signal/noise ratios.
As our AIV microarray contains 99 probes, it is this redundancy that ensures that the diagnostic test has a high degree of reliability. Accordingly, all 43 influenza A virus isolates of all 16 HA subtypes tested were readily detected, and the HA subtypes were accurately determined. Microarrays for the detection and the characterization of influenza A viruses and other pathogens have been described previously (9, 10, 18, 20, 21, 22, 23, 24, 25, 27, 29, 32, 33, 38, 39). The assay reported here combines a one-step RT-PCR by use of a commercially available kit with a low-density microarray assay for characterization of the amplicons without further purification. This approach simplifies the assay, minimizes the turnaround time, and reduces the risk of cross contamination; therefore, it is suitable for use for routine diagnostics for AIV. Furthermore, the use of an electronic microarray allows the faster adaptation of the probe library to emerging virus strains because no spotting process is required. In addition to the detection and HA subtyping of AIV, the microarray reported here can verify the HPAIV H5N1 of the Qinghai lineage, representatives of which have occurred in Germany and other European countries since February 2006 (35, 40), by detection of its HA0 cleavage site. Proof-of-principle experiments indicated that it is also capable of determining pathotypes by targeting the HA0 cleavage sites of other HPAIV H5 and H7 isolates. Considering the variability of AIV and the occurrence of discrepancies between molecular and biological pathotypes (26), the results should be verified, however, by the use of conventional methods (1, 2, 3, 41). Furthermore, the specific reaction patterns of the selected probes can be used for the differentiation of Eurasian and North American viruses of the most important H5 and H7 subtypes. Only two cross-reactions between subtypes were observed. The viruses A/duck/Czechoslovakia/56 (H4N6) and A/turkey/Grub/R41/98 (H6N5) revealed positive signals from probes for the appropriate subtype and additional weaker signals from probes for a second subtype. This finding cannot be explained on the basis of the sequences and may be caused by the secondary structure or the formation of multimers. The results demonstrate that updates, extensions, and continuous adaption of the probe content to currently circulating strains are necessary. Accordingly, the reaction patterns of the probes used for the differentiation of Eurasian and North American viruses will have to be revalidated periodically.
The microarray for AIV presented here was shown to have a high specificity and a high sensitivity. Pathogens important for the differential diagnosis of avian influenza were included in the study (Newcastle disease virus, reovirus, infectious bursitis disease virus, infectious laryngotracheitis virus, Mycoplasma gallinarum, and Mycoplasma gallinaceum) and tested negative. For detection of the selected influenza A viruses, the microarray, combined with the conventional M-gene-specific RT-PCR, revealed the same sensitivity as the widely used and highly sensitive rRT-PCR targeting the M gene described by Spackman et al. (34). In contrast, the sensitivity of the microarray for HA subtyping and pathotyping was 10- to 100-fold lower due to the lower sensitivity of the pan-HA RT-PCR protocol (13). The detection limits ascertained in the study described here roughly correspond to those of the microarrays described by Lin et al. (22), who detected 101 to 102 copies, and Palacios et al. (29), who had a detection limit of 104 copies for influenza A viruses. Thus, the microarray technology has the potential to be a highly sensitive method but is dependent on the RT-PCR protocol used to generate the amplicons.
The microarray described here was further validated with clinical material represented by 19 cloacal swab specimens from wild and domestic birds. The presence of influenza A virus was verified in all samples, and the HA subtypes could be determined for 14 samples. Therefore, the feasibility of the assay for the detection and subtyping of AIV from diagnostic samples was demonstrated. However, successful HA subtyping again depended on the amount of the pan-HA RT-PCR product (13). Amplification from field samples may fail or may show only minor efficiency due to reduced viral loads, the insufficient removal of inhibitors of RT and/or PCR, or mutations in primer binding sites.
In conclusion, the newly developed microarray for the detection of influenza A virus and HA typing was comprehensively validated and can be implemented for routine diagnostics. It combines the sensitive and specific detection and the HA subtyping and pathotyping of AIV in a single approach. As such, the assay is cost-effective, allows the screening of PCR-positive samples, and accelerates the characterization of newly emerging isolates. Finally, the probe library described in the validation study presented here was successfully applied for spotting of a microarray for use with the ArrayTube system (Clondiag). The easy-to-use and inexpensive assay is currently being integrated into routine diagnostics.
Supplementary Material
Acknowledgments
We thank Anne Juengling and the laboratory team of the OIE and the German National Reference Laboratory for Avian Influenza for excellent technical assistance. We are grateful to colleagues within the Friedrich-Loeffler-Institut and at the OIE Reference Laboratories for Avian Influenza in Canada, Italy, and the United Kingdom for supplying reference material and to Nikolaus Osterrieder for critically reading the manuscript.
This work was supported by the Federal Ministry of Food, Agriculture and Consumer Protection, BMELV, Germany (FSI, project no. 1.1.), and the EU Network of Excellence, EPIZONE (contract no. FOOD-CT-2006-016236).
Footnotes
Published ahead of print on 3 December 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Anonymous. 2005. Avian influenza. In Manual of diagnostic tests and vaccines for terrestrial animals. OIE, Paris, France. http://www.oie.int/eng/normes/mmanual/A_00037.htm.
- 2.Anonymous. 2007. Avian influenza. In Terrestrial animal health code. OIE, Paris, France. http://www.oie.int/eng/normes/mcode/en_chapitre_2.7.12.htm.
- 3.Anonymous. 2006. Commission decision of 4 August 2006 approving a diagnostic manual for avian influenza as provided for in Council Directive 2005/94/EC (notified under document number C(2006) 3477) (2006/437/EC). Commission of the European Communities. http://eur-lex.europa.eu/LexUriServ/site/en/oj/2006/l_237/l_23720060831en00010027.pdf.
- 4.Barlaan, E. A., M. Sugimori, S. Furukawa, and K. Takeuchi. 2005. Electronic microarray analysis of 16S rDNA amplicons for bacterial detection. J. Biotechnol. 11511-21. [DOI] [PubMed] [Google Scholar]
- 5.Borsting, C., J. J. Sanchez, and N. Morling. 2004. Multiplex PCR, amplicon size and hybridization efficiency on the NanoChip electronic microarray. Int. J. Legal Med. 11875-82. [DOI] [PubMed] [Google Scholar]
- 6.Butt, K. M., G. J. D. Smith, H. Chen, L. J. Zhang, Y. H. C. Leung, K. M. Xu, W. Lim, R. G. Webster, K. Y. Yuen, J. S. Malik Peiris, and Y. Guan. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 435760-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capua, I., and D. J. Alexander. 2006. The challenge of avian influenza to the veterinary community. Avian Pathol. 35189-205. [DOI] [PubMed] [Google Scholar]
- 8.Cong, Y. L., J. Pu, Q. F. Liu, S. Wang, G. Z. Zhang, X. L. Zhang, W. X. Fan, E. G. Brown, and J. H. Liu. 2007. Antigenic and genetic characterization of H9N2 swine influenza viruses in China. J. Gen. Virol. 882035-2041. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, E. D., C. L. Moore, D. M. Dankbar, M. Mehlmann, M. B. Townsend, J. A. Smagala, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2007. Identification of A/H5N1 influenza viruses using a single gene diagnostic microarray. Anal. Chem. 79378-384. [DOI] [PubMed] [Google Scholar]
- 10.Dawson, E. D., C. L. Moore, J. A. Smagala, D. M. Dankbar, M. Mehlmann, M. B. Townsend, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2006. MChip: a tool for influenza surveillance. Anal. Chem. 787610-7615. [DOI] [PubMed] [Google Scholar]
- 11.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball. 2005. Virus taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier, Academic Press, Amsterdam, The Netherlands.
- 12.Fouchier, R. A., V. Munster, A. Wallensten, T. M. Bestebroer, S. Herfst, D. Smith, G. F. Rimmelzwaan, B. Olsen, and A. D. Osterhaus. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 792814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gall, A., B. Hoffmann, T. Harder, C. Grund, and M. Beer. 2008. Universal primer set for amplification and sequencing of HA0 cleavage sites of all influenza A viruses. J. Clin. Microbiol. 462561-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 15.Hoffmann, B., K. Depner, H. Schirrmeier, and M. Beer. 2006. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 136200-209. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 1462275-2289. [DOI] [PubMed] [Google Scholar]
- 17.Keen-Kim, D., W. W. Grody, and C. S. Richards. 2006. Microelectronic array system for molecular diagnostic genotyping: Nanogen NanoChip® 400 and molecular biology workstation. Expert Rev. Mol. Diagn. 6287-294. [DOI] [PubMed] [Google Scholar]
- 18.Kessler, N., O. Ferraris, K. Palmer, W. Marsh, and A. Steel. 2004. Use of the DNA Flow-Thru chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J. Clin. Microbiol. 422173-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kibbe, W. A. 25 May 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35(web server issue). doi: 10.1093/nar/gkm234. http://www.basic.northwestern.edu/biotools/oligocalc.html. [DOI] [PMC free article] [PubMed]
- 20.Li, H., M. A. McCormac, R. W. Estes, S. E. Sefers, R. K. Dare, J. D. Chappell, D. D. Erdman, P. F. Wright, and Y.-W. Tang. 2007. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J. Clin. Microbiol. 452105-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, B., K. M. Blaney, A. P. Malanoski, A. G. Ligler, J. M. Schnur, D. Metzgar, K. L. Russell, and D. A. Stenger. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J. Clin. Microbiol. 45443-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, R. H., M. J. Lodes, T. Nguyen, T. Siuda, M. Slota, H. S. Fuji, and A. McShea. 2006. Validation of a fully integrated microfluidic array device for influenza A subtype identification and sequencing. Anal. Chem. 784184-4193. [DOI] [PubMed] [Google Scholar]
- 24.Lodes, M. J., D. Suciu, M. Elliott, A. G. Stover, M. Ross, M. Caraballo, K. Dix, J. Crye, R. J. Webby, W. J. Lyon, D. L. Danley, and A. McShea. 2006. Use of semiconductor-based oligonucleotide microarrays for influenza A virus subtype identification and sequencing. J. Clin. Microbiol. 441209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodes, M. J., D. Suciu, J. L. Wilmoth, M. Ross, S. Munro, K. Dix, K. Bernards, A. G. Stover, M. Quintana, N. Iihoshi, W. J. Lyon, D. L. Danley, and A. McShea. 2007. Identification of upper respiratory tract pathogens using electrochemical detection on an oligonucleotide microarray. PLoS One 2e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Londt, B. Z., J. Banks, and D. Alexander. 2007. Highly pathogenic avian influenza viruses with low virulence for chickens in in vivo tests. Avian Pathol. 36347-350. [DOI] [PubMed] [Google Scholar]
- 27.Mehlmann, M., A. B. Bonner, J. V. Williams, D. M. Dankbar, C. L. Moore, R. D. Kuchta, A. B. Podsiad, J. D. Tamerius, E. D. Dawson, and K. L. Rowlen. 2007. Comparison of the MChip to viral culture, reverse transcription-PCR, and the QuickVue Influenza A+B test for rapid diagnosis of influenza. J. Clin. Microbiol. 451234-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, C. L., J. A. Smagala, C. B. Smith, E. D. Dawson, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2007. Evaluation of MChip with historic subtype H1N1 influenza A viruses, including the 1918 “Spanish flu” strain. J. Clin. Microbiol. 453807-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios, G., P.-L. Quan, O. J. Jabado, S. Conlan, D. L. Hirschberg, Y. Liu, J. Zhai, N. Renwick, J. Hui, H. Hegyi, A. Grolla, J. E. Strong, J. S. Towner, T. W. Geisbert, P. B. Jahrling, C. Büchen-Osmond, H. Ellerbrok, M. Paz Sanchez-Seco, Y. Lussier, P. Formenty, S. T. Nichol, H. Feldmann, T. Briese, and W. I. Lipkin. 2007. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 1373-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perdue, M. L., and D. L. Suarez. 2000. Structural features of the avian influenza virus hemagglutinin that influence virulence. Vet. Microbiol. 7477-86. [DOI] [PubMed] [Google Scholar]
- 31.Phipps, L. P., S. C. Essen, and I. H. Brown. 2004. Genetic subtyping of influenza A viruses using RT-PCR with a single set of primers based on conserved sequences within the HA2 coding region. J. Virol. Methods 122119-122. [DOI] [PubMed] [Google Scholar]
- 32.Quan, P.-L., G. Palacios, O. J. Jabado, S. Conlan, D. L. Hirschberg, F. Pozo, P. J. M. Jack, D. Cisterna, N. Renwick, J. Hui, A. Drysdale, R. Amos-Ritchie, E. Baumeister, V. Savy, K. M. Lager, J. A. Richt, D. B. Boyle, A. Garcia-Sastre, I. Casas, P. Perez-Brena, T. Briese, and W. I. Lipkin. 2007. Detection of respiratory viruses and subtype identification of influenza A viruses by GreeneChipResp oligonucleotide microarray. J. Clin. Microbiol. 452359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta, S., K. Onodera, A. Lai, and U. Melcher. 2003. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 414542-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spackman, E., D. A. Senne, T. J. Myers, L. L. Bulaga, L. P. Garber, M. L. Perdue, K. Lohman, L. T. Daum, and D. L. Suarez. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 403256-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starick, E., M. Beer, B. Hoffmann, C. Staubach, O. Werner, A. Globig, G. Strebelow, C. Grund, M. Durban, F. J. Conraths, T. Mettenleiter, and T. Harder. 2007. Phylogenetic analyses of highly pathogenic avian influenza virus isolates from Germany in 2006 and 2007 suggest at least three separate introductions of H5N1 virus. Vet. Microbiol. 128243-252. [DOI] [PubMed] [Google Scholar]
- 36.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 2581-20. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Townsend, M. B., E. D. Dawson, M. Mehlmann, J. A. Smagala, D. M. Dankbar, C. L. Moore, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2006. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J. Clin. Microbiol. 442863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, L.-C., C.-H. Pan, L. L. Severinghaus, L.-Y. Liu, C.-T. Chen, C.-E. Pu, D. Huang, J.-T. Lir, S.-C. Chin, M.-C. Cheng, S.-H. Lee, and C.-H. Wang. 2008. Simultaneous detection and differentiation of Newcastle disease and avian influenza viruses using oligonucleotide microarrays. Vet. Microbiol. 127217-226. [DOI] [PubMed] [Google Scholar]
- 40.Weber, S., T. Harder, E. Starick, M. Beer, O. Werner, B. Hoffmann, T. C. Mettenleiter, and E. Mundt. 2007. Molecular analysis of highly pathogenic avian influenza virus of subtype H5N1 isolated from wild birds and mammals in northern Germany. J. Gen. Virol. 88554-558. [DOI] [PubMed] [Google Scholar]
- 41.Wood, G. W., J. W. McCauley, J. B. Bashruddin, and D. J. Alexander. 1993. Deduced amino acid sequences at the haemagglutinin cleavage site of avian influenza A viruses of H5 and H7 subtypes. Arch. Virol. 130209-217. [DOI] [PubMed] [Google Scholar]
- 42.Wright, K. E., G. A. Wilson, D. Novosad, C. Dimock, D. Tan, and J. M. Weber. 1995. Typing and subtyping of influenza viruses in clinical samples by PCR. J. Clin. Microbiol. 331180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.