Abstract
We characterized 208 human Salmonella isolates from 2006 to 2007 and 27 human Salmonella enterica serovar Typhimurium isolates from 1987 to 1993 from Henan Province, China, by serotyping, by antimicrobial susceptibility testing, and, for the most common serovars, by pulsed-field gel electrophoresis (PFGE). The most common serovars among the 2006-2007 isolates were S. enterica serovar Typhimurium (27%), S. enterica serovar Enteritidis (17%), S. enterica serovar Derby (10%), S. enterica serovar Indiana (6%), and S. enterica serovar Litchfield (6%). A high percentage of the isolates were multiple-drug resistant, and 54% were resistant to both nalidixic acid and ciprofloxacin. Of these, 42% were resistant to a high level of ciprofloxacin (MIC > 4 μg/ml), whereas for the remaining isolates, the MICs ranged from 0.125 to 2 μg/ml. Five isolates (2%) were ceftiofur resistant and harbored blaCTX-M14 or blaCTX-M15. With the possible exception of the quinolones and cephalosporins, the 1987-1993 S. enterica serovar Typhimurium isolates were almost as resistant as the recent isolates. PFGE typing of S. enterica serovar Typhimurium showed that the most common cluster predominated over time. Two other clusters have emerged, and another cluster has disappeared.
Salmonella enterica is a common cause of human gastroenteritis and bacteremia worldwide, and a wide variety of animals, particularly food animals, have been identified as reservoirs for nontyphoidal Salmonella. Although >2,500 serovars of Salmonella enterica have been identified, most human infections are caused by a limited number of serovars. Salmonella enterica serovars Typhimurium and Enteritidis are the most common causes of human salmonellosis worldwide, although other serovars have been reported to be more prevalent in some regions (1, 13, 18, 20, 25). Shifts in the prevalence of specific strain types and serovars can reflect the influence of international travel and trade of animals and food products, and they can therefore serve as useful epidemiological markers.
In recent years, an increase in the occurrence of antimicrobial resistance, including resistance to quinolones, among salmonellae has been observed in many countries, particularly in Asia (2, 10, 19, 22, 23, 27, 29). The emergence of antimicrobial resistance is a matter of concern. Humans with infections caused by antimicrobial-resistant Salmonella, particularly quinolone-resistant Salmonella, have a higher fatality rate, are more likely to be hospitalized, and are hospitalized for longer periods than patients with infections caused by susceptible strains (16, 17, 28). Recently, a few reports on the occurrence of Salmonella serovars and the antimicrobial susceptibilities of Salmonella serovars from food products in China have been published (6, 30). A recent report also showed an alarmingly high frequency of fluoroquinolone resistance in S. enterica serovar Typhimurium (9). However, to our knowledge, no reports on the occurrence of different Salmonella serovars isolated from humans with infections in China, on their molecular types, or on the frequencies of antimicrobial resistance among them are available in the international scientific literature.
The World Health Organization Global Salm-Surv (WHO GSS) program launched in January 2000 is a global network of laboratories and individuals involved in the surveillance, isolation, identification, and antimicrobial susceptibility testing of Salmonella (http://www.who.int/salmsurv/en/). In 2005, the WHO GSS launched the Enhanced Salmonella Surveillance Project in China, including in Henan Province. This project is a cooperative program between the Chinese Center for Disease Control and Prevention (China CDC), the U.S. CDC, and the WHO GSS. Henan Province is located in mideastern China; it has an area of over 160,000 square kilometers and the highest population among the 30 provinces, with around 93.8 million people (7% of the Chinese population).
This study reports the first results from the surveillance of the occurrence of different Salmonella serovars and their antimicrobial susceptibilities in Henan Province in China. In addition, the molecular types of the most common serovars were determined.
MATERIALS AND METHODS
Bacterial isolates.
A total of 208 cross-sectional isolates of Salmonella enterica originated from infections in humans during 2006 to 2007 of Henan Province, China. The strains were isolated from 28 sentinel hospitals and six regional CDC diagnostic laboratories located in five different geographic regions in Henan Province. All these laboratories participated in the Enhanced Salmonella Surveillance Project of the WHO GSS for nontyphoid Salmonella. Six of the hospitals were located in rural areas and the remaining 22 in urban areas. Fresh fecal specimens collected from diarrhea patients of all ages were inoculated into Carry-Blair medium and forwarded to the regional CDC laboratories within 4 h. The stools samples were enriched in selenite brilliant-green broth (Becton Dickinson and Co., Sparks, MD) for 16 h at 37°C, followed by subcultivation onto CHROMagar Salmonella agar (CHROMagar Company, Paris, France) and salmonella-shigella agar (Oxoid, Hampshire, England). The plates were incubated at 37°C for 18 to 24 h. Purple colonies were screened by tests in triple-sugar-iron agar, motility indol-urea agar, l-lysine decarboxylase, and β-galactosidase (o-nitrophenyl-β-d-galactopyranoside [ONPG]). The presumptively positive Salmonella isolates were identified by using API 20E strips (bioMérieux, France). In addition, 27 serovar Typhimurium isolates from infections in humans of Henan Province during 1987 to 1993 were included for comparison.
Serotyping.
All isolates were shipped to the National Food Institute (NFI), Technical University of Denmark, as diagnostic samples cataloged under UN-3373-B. For all 235 isolates, O and H antigens were characterized by slide agglutination with hyperimmune sera (State Serum Institute, Denmark) and the serotype was assigned according to the Kauffmann-White scheme.
Antimicrobial susceptibility.
Susceptibility to antimicrobial agents was determined at the NFI for all 235 isolates as MIC determinations using a commercially prepared, dehydrated panel (Sensititre) according to CLSI standards (8). The following antimicrobial agents were tested: amoxicillin-clavulanate, ampicillin, apramycin, ceftiofur, chloramphenicol, ciprofloxacin, colistin, florfenicol, gentamicin, nalidixic acid, neomycin, spectinomycin, streptomycin, sulfamethoxazole, tetracycline, and tetracycline. Reduced susceptibility (low-level resistance) to ciprofloxacin was defined as a MIC of ≥0.125 μg/ml and high-level resistance as a MIC of ≥4 μg/ml. Extended-spectrum β-lactamase production was confirmed by the Kirby-Bauer disc diffusion method.
Detection of resistance genes.
Five isolates resistant to ceftiofur were examined for the presence of the blaTEM, blaCTX, and blaSHV genes, as previously described (15). Sixty ciprofloxacin-resistant isolates, including five isolates showing low-level resistance to ciprofloxacin but susceptibility to nalidixic acid, were examined for the presence of qnrA, qnrB, qnrS, qepA, and aac(6′)Ib-cr as previously described (3). All PCR amplifications were conducted with the buffer supplied by the manufacturers using 20 pmol of each primer, 0.5 U of Super Taq (HT Biotechnology, United Kingdom), and 50-μl reaction mixtures. Prior to being sequenced, the amplicons were purified using the GFX PCR DNA kit (Amersham Biosciences). The resulting nucleotide sequences were compared to sequences obtained from the GenBank database (http://www.lahey.org/studies/webt.html). The software Vector NTI suite 8 (InforMax, Inc.) was used for alignment.
PFGE.
All isolates of S. enterica serovar Typhimurium, S. enterica serovar Enteritidis, S. enterica serovar Derby, and S. enterica serovar Litchfield were analyzed for genetic relatedness by pulsed-field gel electrophoresis (PFGE) using XbaI according to the U.S. CDC PulseNet protocol (26). Electrophoresis was performed with a CHEF-DR III system (Bio-Rad Laboratories, Hercules, California) using 1% SeaKem agarose in 0.5× Tris-borate-EDTA at 180 V. Running conditions consisted of one phase from 2.2 to 63.8 s at a run time of 20 h. Comparison of the PFGE profiles was performed using Bionumerics software v3.5 (Applied Maths, Sint-Martens-Latem, Belgium).
RESULTS
Epidemiology.
From May to October in 2006, 2,661 stool specimens (1,449 from urban regions, 1,212 from rural regions) and from April to November in 2007, 3,061 stool specimens (1,666 from urban regions, 1,395 from rural regions) were collected from patients of all ages with diarrhea from 28 participating hospitals in Henan Province, China. Totals of 154 (5.8%) and 147 (4.8%) isolates of nontyphoid Salmonella were obtained in 2006 and 2007, respectively. The isolates from urban regions accounted for 5.7% (2006) and 3.3% (2007) and those from rural regions for 5.9% (2006) and 6.6% (2007) (P < 0.001, 2007). The isolation frequencies per month were from 1% in October to 14% in June (2006) and from 3% in October to 6% in July (2007).
Only 208 isolates were stored out of the 301 that were isolated. Of these 208 isolates (2006 to 2007), 98 (47%) were isolated from children less than 14 years of age and 110 (53%) from adolescents and adults. Twenty-five (12%) of the children were less than 10 months of age, and of these, 15 (60%) and 12 (48%) showed mucus and blood, respectively, in their feces and 6 (24%) had fever (38°C to 39.5°C). The male/female ratio was 1.7:1. The distributions relative to age and gender were nearly identical each year, with the highest incidence in young children aged less than 5 years (46 cases [37%] in 2006 and 32 cases [39%] in 2007) and the lowest in adults over 60 years (6 cases [5%] in 2006 and 4 cases [5%] in 2007). A difference in prevalence was also observed between the genders, particularly among children aged less than 5 years: 54 boys versus 24 girls (2.25:1). The highest number of infections with S. enterica serovar Typhimurium and S. enterica serovar Enteritidis were found in young children aged less than 2 years, among whom the incidence in boys of S. enterica serovar Typhimurium was almost thrice that in girls (Table 1).
TABLE 1.
Distribution relative to patient age and gender of 208 Salmonella enterica isolates from humans with infections in Henan Province, China, during 2006 and 2007
| Age (yr) | No. of isolates (% of total) | No. of:
|
No. of patients from:
|
No. of isolates (% of total) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Males | Females | Rural areas | Urban areas | Serovar Typhimurium | Serovar Enteritidis | Serovar Derby | Serovar Indiana | Serovar Litchfield | Remaining 28 serotypes | ||
| ≤2 | 59 (28.4) | 44 | 15 | 27 | 32 | 26 (46) | 13 (37) | 4 (20) | 1 (8) | 15 (21) | |
| 3-10 | 29 (13.9) | 17 | 12 | 7 | 22 | 9 (16) | 8 (23) | 3 (15) | 3 (25) | 1 (8) | 5 (7) |
| 11-18 | 22 (10.6) | 18 | 4 | 10 | 12 | 4 (7) | 4 (11) | 4 (20) | 1 (8) | 2 (17) | 7 (10) |
| 19-26 | 14 (6.7) | 10 | 4 | 5 | 9 | 3 (5) | 2 (6) | 2 (10) | 2 (17) | 1 (8) | 4 (6) |
| 27-34 | 17 (8.2) | 9 | 8 | 5 | 12 | 2 (4) | 1 (3) | 2 (10) | 1 (8) | 11 (15) | |
| 35-42 | 18 (8.7) | 11 | 7 | 14 | 4 | 2 (6) | 3 (15) | 3 (25) | 4 (33) | 6 (8) | |
| 43-50 | 22 (10.6) | 9 | 13 | 14 | 8 | 4 (7) | 3 (9) | 1 (5) | 1 (8) | 13 (18) | |
| 51-58 | 12 (5.8) | 7 | 5 | 10 | 2 | 4 (7) | 2 (6) | 1 (8) | 1 (8) | 4 (6) | |
| 59-66 | 7 (3.4) | 4 | 3 | 6 | 1 | 2 (4) | 1 (5) | 2 (17) | 2 (3) | ||
| 67-74 | 5 (2.4) | 2 | 3 | 1 | 4 | 1 (2) | 4 (6) | ||||
| 75-80 | 3 (1.4) | 3 | 1 | 2 | 1 (2) | 2 (3) | |||||
| Total | 208 (100.0) | 131 | 77 | 100 | 108 | 56 (100) | 35 (100) | 20 (100) | 12 (100) | 12 (100) | 73 (100) |
Serotype distribution.
Thirty-three different serotypes were identified among the 208 nontyphoid Salmonella isolates from 2006 to 2007. S. enterica serovar Typhimurium was the most common serotype (26.9%), followed by S. enterica serovar Enteritidis (16.8%), S. enterica serovar Derby (9.6%), S. enterica serovar Indiana (5.8%), S. enterica serovar Litchfield (5.8%), S. enterica serovar Thompson (2.9%), and S. enterica serovar Agona (2.9%) (Table 2).
TABLE 2.
Distribution of serovars of 208 Salmonella enterica isolates from humans with infections in Henan Province, China, in 2006 and 2007
| Salmonella enterica serovar or serotype | No. of isolates from indicated urban area
|
No. of isolates from indicated rural area
|
Total | % of total | |||
|---|---|---|---|---|---|---|---|
| Zhengzhou | Zhoukou | Shangqiu | Jiyuan | Suixian | |||
| Typhimurium | 10 | 12 | 9 | 11 | 14 | 56 | 26.9 |
| Enteritidis | 8 | 10 | 5 | 3 | 9 | 35 | 16.8 |
| Derby | 4 | 3 | 4 | 3 | 6 | 20 | 9.6 |
| Indiana | 3 | 6 | 3 | 12 | 5.8 | ||
| Litchfield | 12 | 12 | 5.8 | ||||
| Thompson | 1 | 1 | 4 | 6 | 2.9 | ||
| Agona | 4 | 1 | 1 | 6 | 2.9 | ||
| Mbandaka | 3 | 2 | 5 | 2.4 | |||
| Aberdeen | 1 | 2 | 1 | 4 | 1.9 | ||
| Kottbus | 1 | 1 | 2 | 4 | 1.9 | ||
| Senftenberg | 3 | 1 | 4 | 1.9 | |||
| Schwarzengrund | 1 | 1 | 1 | 1 | 4 | 1.9 | |
| 3,10:−:1,5 | 1 | 3 | 4 | 1.9 | |||
| Infantis | 3 | 3 | 1.4 | ||||
| Corvallis | 2 | 1 | 3 | 1.4 | |||
| Bovismorbificans | 1 | 2 | 3 | 1.4 | |||
| 6,7:e,h:− | 3 | 3 | 1.4 | ||||
| Amsterdam | 2 | 2 | 1.0 | ||||
| Meleagridis | 1 | 1 | 2 | 1.0 | |||
| Muenster | 2 | 2 | 1.0 | ||||
| Paratyphi B | 2 | 2 | 1.0 | ||||
| Reading | 1 | 1 | 2 | 1.0 | |||
| Saintpaul | 2 | 2 | 1.0 | ||||
| Kentucky | 1 | 1 | 2 | 1.0 | |||
| Stanley | 2 | 2 | 1.0 | ||||
| Choleraesuis | 1 | 1 | 0.5 | ||||
| II | 1 | 1 | 0.5 | ||||
| IV | 1 | 1 | 0.5 | ||||
| Newport | 1 | 1 | 0.5 | ||||
| Singapore | 1 | 1 | 0.5 | ||||
| Somone | 1 | 1 | 0.5 | ||||
| Wandsworth | 1 | 1 | 0.5 | ||||
| 4,5,12:b:− | 1 | 1 | 0.5 | ||||
| Total | 44 | 35 | 31 | 51 | 47 | 208 | 100.0 |
Antimicrobial susceptibility.
The overall levels of resistance to individual antimicrobial agents of the most common serotypes are presented in Table 3. High frequencies of resistance were observed in all 235 Salmonella isolates.
TABLE 3.
Antimicrobial susceptibility among 208 Salmonella enterica isolates from humans with infections in Henan Province, China, during 2006 and 2007 and 27 serovar Typhimurium isolates from 1987 to 1993
| S. enterica serovar or serotype (yr of isolation) | No. of isolates | No. of isolates resistant to indicated agent at the indicated breakpoint in mg/liter (% resistance)a
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP
|
NAL, ≥32 | STR, ≥32 | SMX, ≥512 | TET, ≥16 | TMP, ≥16 | SPE, ≥128 | AMP, ≥32 | CHL, ≥32 | GEN, ≥8 | NEO, ≥16 | FFN, ≥32 | APR, ≥32 | POD, ≥4 | XNL, ≥8 | |||
| ≥4 | 0.125-2 | ||||||||||||||||
| Typhimurium (2006-2007) | 56 | 41 (73) | 13 (23) | 53 (95) | 51 (91) | 53 (95) | 50 (89) | 54 (96) | 53 (95) | 53 (95) | 47 (84) | 53 (95) | 12 (21) | 8 (14) | 6 (11) | 5 (9) | 5 (9) |
| Typhimurium (1987-1993) | 27 | 2 (7) | 18 (67) | 20 (74) | 25 (93) | 25 (93) | 24 (89) | 25 (93) | 25 (93) | 25 (93) | 25 (93) | 25 (93) | 9 (33) | 3 (11) | 0 | 5 (19) | 0 |
| Enteritidis | 35 | 2 (6) | 32 (91) | 34 (97) | 4 (11) | 4 (11) | 5 (1) | 2 (6) | 2 (6) | 4 (11) | 3 (9) | 3 (9) | 1 (3) | 1 (3) | 1 (3) | 0 | 0 |
| Derby | 20 | 0 | 1 (5) | 1 (5) | 5 (25) | 6 (30) | 9 (45) | 2 (10) | 5 (25) | 0 | 7 (35) | 0 | 0 | 2 (10) | 0 | 0 | 0 |
| Indiana | 12 | 8 (67) | 4 (33) | 12 (100) | 9 (75) | 12 (100) | 12 (100) | 11 (92) | 11 (92) | 12 (100) | 12 (100) | 10 (83) | 8 (67) | 8 (67) | 8 (67) | 0 | 0 |
| Litchfield | 12 | 0 | 0 | 0 | 7 (59) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Corvallis | 3 | 0 | 0 | 0 | 3 (100) | 3 (100) | 3 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Infantis | 3 | 0 | 1 (33) | 1 (33) | 0 | 1 (33) | 0 | 1 (33) | 0 | 1 (33) | 1 (33) | 0 | 0 | 1 (33) | 0 | 0 | 0 |
| 6,7:e,h:− | 3 | 0 | 3 (100) | 3 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bovismorbificans | 3 | 0 | 3 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Kentucky | 2 | 0 | 2 (100) | 1 (50) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Choleraesuis | 1 | 0 | 1 (100) | 1 (100) | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 | 0 | 0 | 0 |
| Singapore | 1 | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Somone | 1 | 0 | 0 | 0 | 1 (100) | 1 (100) | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IV | 1 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Othersb | 55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (2006-2007) | 208 (27) | 51 (25) | 61 (29) | 107 (51) | 82 (39) | 82 (39) | 82 (39) | 72 (35) | 72 (35) | 71 (34) | 71 (34) | 67 (32) | 22 (11) | 20 (10) | 15 (7) | 5 (2) | 5 (2) |
All isolates were susceptible to colistin and amoxicillin plus clavulanate. CIP, ciprofloxacin; NAL, nalidixic acid; STR, streptomycin; SMX, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim; SPE, spectinomycin; AMP, ampicillin; CHL, chloramphenicol; GEN, gentamicin; NEO, neomycin; FFN, florfenicol; APR, apramycin; POD, cefpodoxime; XNL, ceftiofur.
Isolates belonging to the remaining 18 serovars were susceptible to all the antimicrobials tested.
Among the isolates from 2006 to 2007, we observed resistance to ciprofloxacin (54%), nalidixic acid (51%), streptomycin (39%), sulfamethoxazole (39%), tetracycline (39%), trimethoprim (35%), spectinomycin (35%), chloramphenicol (34%), ampicillin (34%), gentamicin (32%), neomycin (11%), florfenicol (10%), apramycin (7%), cefpodoxime (2%), and ceftiofur (2%), whereas all isolates were susceptible to colistin and amoxicillin-clavulanic acid. Especially among S. enterica serovar Typhimurium isolates, the frequencies of resistance to the following agents were very high among isolates from both 1987 to 1993 and 2006 to 2007: trimethoprim (93% in 1987 to 1993 and 96% in 2006 to 2007), sulfamethoxazole (93% and 95%), ampicillin (93% and 95%), spectinomycin (93% and 95%), gentamicin (93% and 95%), streptomycin (93% and 91%), ciprofloxacin (74% and 96%), nalidixic acid (74% and 95%), tetracycline (89% and 89%), chloramphenicol (93% and 84%), neomycin (33% and 21%), florfenicol (11% and 14%), apramycin (0% and 11%), and ceftiofur (0% and 9%). Forty-one S. enterica serovar Typhimurium isolates (49%) showed the most common multiple-drug resistance patterns against ampicillin, chloramphenicol, ciprofloxacin, gentamicin, nalidixic acid, spectinomycin, streptomycin, sulfamethoxazole, tetracycline, and tetracycline.
High rates of resistance to ciprofloxacin (100%), nalidixic acid (100%), ampicillin (100%), chloramphenicol (100%), sulfamethoxazole (100%), trimethoprim (92%), tetracycline (92%), spectinomycin (92%), streptomycin (84%), gentamicin (83%), florfenicol (67%), apramycin (67%), and neomycin (67%) were observed among the S. enterica serovar Indiana isolates.
PFGE.
A total 150 isolates of the Salmonella serovars Typhimurium, Enteritidis, Derby, and Litchfield were analyzed for genetic relatedness by PFGE using XbaI.
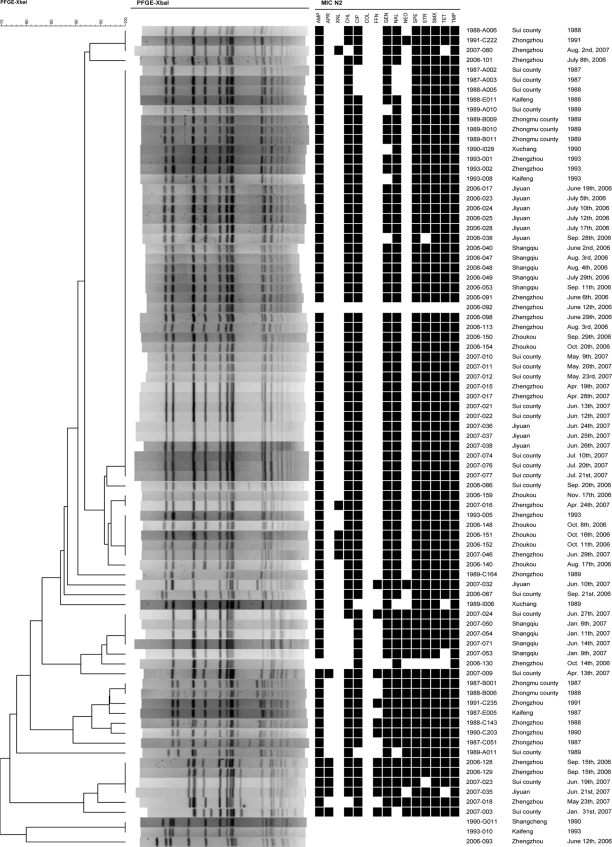
Twenty-seven different PFGE patterns were observed among the 83 isolates of S. enterica serovar Typhimurium from 1987 to 1993 (27 isolates) and 2006 to 2007 (56 isolates) (Fig. 1). A total of 42 isolates were assigned to the same type. Using a cutoff value of 85% similarity, four clusters and some individual types were observed. The most common cluster consisted of 59 isolates. This cluster was the most common for both historical and recent isolates comprising 17 of the 27 historical (63%) and 42 of the 56 recent (75%) isolates. The cluster was found in all districts and in both rural and urban areas. It was within the recent isolates of this cluster that four cephalosporin-resistant S. enterica serovar Typhimurium isolates were observed; three of these isolates were assigned to the same PFGE type, and all four originated from urban areas. Thirteen (76%) of the 17 historical isolates belonging to this cluster were resistant to ciprofloxacin, whereas this was the case for 41 (98%) of the recent isolates. Another cluster was comprised of seven recent, but no historical, isolates. Unlike with the most common cluster, almost all these isolates were resistant to neomycin and susceptible to chloramphenicol. A third cluster was comprised of seven historical, but no recent, isolates. All isolates were resistant to neomycin, and most were resistant to florfenicol. A fourth cluster was comprised of six recent, but no historical, isolates. These isolates were also mostly resistant to neomycin, florfenicol, and, in addition, apramycin.
FIG. 1.
Phylogeny of PFGE patterns of 83 Salmonella serovar Typhimurium isolates from humans with infections during the periods 1987 to 1993 and 2006 to 2007 in Henan Province, China. N2, panel used for susceptibility testing; AMP, ampicillin; APR, apramycin; XNL, ceftiofur; CHL, chloramphenicol; CIP, ciprofloxacin; FFN, florfenicol; GEN, gentamicin; NAL, nalidixic acid; NEO, neomycin; SPE, spectinomycin; STR, streptomycin; SMX, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim. Isolate numbers are noted to the right of the PFGE patterns.
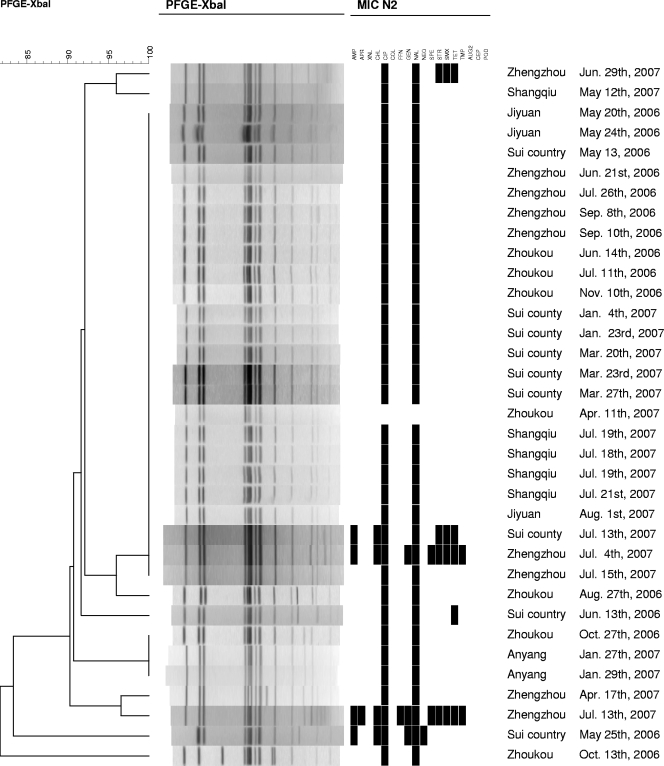
The 35 S. enterica serovar Enteritidis isolates were assigned to 10 different PFGE patterns. Twenty-four (69%) were assigned to the same type. The isolates were found in all regions, and an association between PFGE pattern and resistance could be observed (Fig. 2).
FIG. 2.
Phylogeny of PFGE patterns of 35 Salmonella serovar Enteritidis isolates from humans with infections during the period 2006 to 2007 in Henan Province, China. N2, panel used for susceptibility testing; AMP, ampicillin; APR, apramycin; XNL, ceftiofur; CHL, chloramphenicol; CIP, ciprofloxacin; COL, colistin, FFN, florfenicol; GEN, gentamicin; NAL, nalidixic acid; NEO, neomycin; SPE, spectinomycin; STR, streptomycin; SMX, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim; AUG2, amoxicillin plus clavulanic acid; CEP, cephalosporin; POD, cefpodoxime.
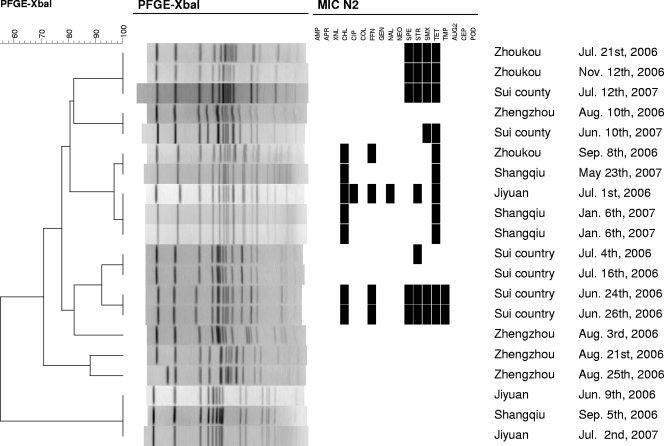
The 20 S. enterica serovar Derby isolates were assigned to 10 different PFGE patterns. Unlike with the other serovars, no predominant PFGE type was observed. Some clusterings of PFGE types and susceptibility patterns were obvious (Fig. 3).
FIG. 3.
Phylogeny of PFGE patterns of 20 Salmonella serovar Derby isolates from humans with infections during the period 2006 to 2007 in Henan Province, China. N2, panel used for susceptibility testing; AMP, ampicillin; APR, apramycin; XNL, ceftiofur; CHL, chloramphenicol; CIP, ciprofloxacin; COL, colistin, FFN, florfenicol; GEN, gentamicin; NAL, nalidixic acid; NEO, neomycin; SPE, spectinomycin; STR, streptomycin; SMX, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim; AUG2, amoxicillin plus clavulanic acid; CEP, cephalosporin; POD, cefpodoxime.
All 12 S. enterica serovar Litchfield isolates were assigned to the same PFGE pattern. All isolates were from the rural area of Jiyuan. Seven of the isolates were streptomycin resistant but were otherwise susceptible to all other antimicrobials tested.
Detection and identification of blaCTX and blaAAC.
Five ceftiofur-resistant isolates of S. enterica serovar Typhimurium (2.1%) gave positive reactions for blaCTX, which were identified as three blaCTX-M15-like and two blaCTX-M14 genes. Among the 60 isolates examined for the presence of transferable fluoroquinolone resistance, five isolates (8%) (four S. enterica serovar Typhimurium isolates and one S. enterica serovar Enteritidis isolate) gave positive reactions for aac(6′)-lb and were sequenced to blaAAC(6′)-lb-cr. Five isolates showed low-level ciprofloxacin resistance but were susceptible to nalidixic acid; none of them harbored a known qnr gene.
DISCUSSION
In this study, more isolates originated from urban than from rural regions, which might reflect access to or traditions of seeking medical care. The age distribution of nontyphoidal salmonellosis had peaks in two age groups: middle-aged adults (43 to 50 years of age [11%]) and children (less than 14 years of age [47%]), which is in agreement with the general knowledge of the epidemiology of salmonellosis (11). One anomaly in the latter age group was that infants less than 2 years old made up 28.4% of the patients. A major difference in isolation between the genders was observed. Thus, three times more salmonellae were isolated from boys than from girls. This might indicate that males in this age group are more likely than females to exhibit diarrhea when infected with nontyphoid Salmonella or that parents more often bring their boys to medical examinations. The male/female ratio in China might also have an influence.
In Henan Province, Salmonella serovar Typhimurium clearly dominated, followed by S. enterica serovar Enteritidis and S. enterica serovar Derby. Data from Asia, Europe, and Latin America have indicated that S. enterica serovar Enteritidis is the most common serovar, followed by S. enterica serovar Typhimurium, whereas in Africa and North America, S. enterica serovar Typhimurium seems to be more common than S. enterica serovar Enteritidis (1, 13, 18, 20, 25). S. enterica serovar Derby is also commonly found in other regions of the world (1, 13, 18, 20, 25) but not in as high a frequency as observed in Henan Province. The most common reservoir of S. enterica serovar Enteritidis infections are considered to be eggs and chickens (14). S. enterica serovar Typhimurium can be found in a wide range of animal species (20), but it is one of the most common types observed in pigs. Similarly, pigs are considered the main reservoir for S. enterica serovar Derby. Thus, the serovar distribution in Henan Province may indicate that pigs are the main reservoir for Salmonella infections in humans. Several other serovars that are common in many countries, such as S. enterica serovar Newport, S. enterica serovar Heidelberg, S. enterica serovar Virchow, and S. enterica serovar Weltevreden (common in Southeast Asia) (1, 13, 18, 20, 25), were rarely found in Henan Province.
We found a very high prevalence of antimicrobial resistance among the nontyphoid Salmonella isolates from outpatients in Henan Province compared to what has been reported in North America and Europe (2, 5, 10, 12, 19, 22, 23, 27, 29). In particular, we found that 54% of all isolates were resistant to ciprofloxacin and 23% were highly resistant. This was especially evident among S. enterica serovars Indiana, Enteritidis, and Typhimurium, among which 100%, 97%, and 89%, respectively, were ciprofloxacin resistant. Reports from other countries have found an increasing frequency of ciprofloxacin-resistant isolates (2, 10, 19, 22, 23, 27, 29) but not a frequency approaching that observed in Henan Province. Infections with ciprofloxacin-resistant isolates are associated with increased morbidity and mortality (16, 17, 28), and the alarmingly high frequency makes it difficult to use fluoroquinolones for treatment of Salmonella infections in humans. It is noteworthy that even among the isolates from 1987 to 1993, a very high frequency of resistance was found, indicating widespread and indiscriminate use. However, since no data on the consumption of antimicrobial agents for food animals are available for China, the reason for the high prevalence of ciprofloxacin resistance remains unknown. Five isolates had low-level resistance to ciprofloxacin but were susceptible to nalidixic acid. Cloning identified a novel qnrD gene in four of these isolates (4).
We found five isolates of S. enterica serovar Typhimurium that were resistant to cephalosporins, three of which had blaCTX-M-15-like and two had blaCTX-M-14. This is to our knowledge the first report of these two blaCTX-M variants among Salmonella isolates from China. The blaCTX-M-14 variants have previously been described to occur among isolates of Salmonella serovar Enteritidis and a single S. enterica serovar Typhimurium isolate (7, 21), whereas blaCTX-M-15-like has been reported from a single S. enterica serovar Typhimurium isolate in Hong Kong (21). Expanded-spectrum cephalosporins are commonly used to treat serious Salmonella infections. Therefore, expanded-spectrum-β-lactamase-producing strains are of concern (24). This is especially the case with the isolates observed in the present study, since a very high number of the isolates were already resistant to fluoroquinolones. The combination of fluoroquinolone and cephalosporin resistance in isolates already resistant to most other antimicrobial agents makes it almost impossible to find an effective treatment for some infections. Carbapenems and colistin seem to be the last available products.
PFGE typing of historical and recent S. enterica serovar Typhimurium isolates yielded some interesting results. Thus, the most common cluster seems to have been rather constant over time. In contrast, one smaller cluster seems to have disappeared and two other minor clusters to have emerged. Interestingly, these clusters differed also in their susceptibilities to antimicrobial agents. Thus, almost all isolates in these three changing clusters were resistant to neomycin and florfenicol, whereas apramycin resistance was observed only among the emerging clusters. PFGE typing of S. enterica serovar Enteritidis and S. enterica serovar Derby showed a relatively high diversity, indicating that the human cases were sporadic and not part of any major outbreaks.
In conclusion, this study showed that S. enterica serovar Typhimurium and S. enterica serovar Enteritidis are the most common serovars causing human salmonellosis in Henan Province, China, but also that other serovars are of importance. The study documents that a very high frequency of quinolone resistance was already present among isolates 20 years ago but that there was still a low frequency of resistance to expanded-spectrum cephalosporins. The resistance profiles of the most common S. enterica serovar Typhimurium clusters have been rather constant over time.
Acknowledgments
This work was supported by the WHO GSS Program and the Chinese CDC.
We thank Jacob Dyring Jensen and Birthe S. Rosenqvist Lund of the National Food Institute, Technical University of Denmark, for their excellent technical assistance with serotyping, PFGE, and MIC tests. We also thank all of the WHO GSS cooperative departments of the Zhengzhou CDC, the Jiyuan CDC, the Shangqiu CDC, the Zhoukou CDC, and the Suixian CDC. We are grateful to Chunrong Cheng (Zhengzhou CDC) and Qi Luo (Puyang CDC) for participating in this project.
Footnotes
Published ahead of print on 10 December 2008.
REFERENCES
- 1.Bangtrakulnonth, A., S. Pornreongwong, C. Pulsrikarn, P. Sawanpanyalert, R. S. Hendriksen, D. M. Lo Fo Wong, and F. M. Aarestrup. 2004. Salmonella serovars from humans and other sources in Thailand, 1993-2002. Emerg. Infect. Dis. 10131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cailhol, J., R. Lailler, P. Bouvet, S. La Vieille, F. Gauchard, P. Sanders, and A. Brisabois. 2006. Trends in antimicrobial resistance phenotypes in non-typhoid salmonellae from human and poultry origins in France. Epidemiol. Infect. 134171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavaco, L. M., N. Frimodt-Møller, H. Hasman, L. Guardabassi, L. Nielsen, and F. M. Aarestrup. 2008. Prevalence of quinolone resistance mechanisms and associations to minimum inhibitory concentrations in quinolone-resistant Escherichia coli isolated from humans and swine in Denmark. Microb. Drug Resist. 14163-169. [DOI] [PubMed] [Google Scholar]
- 4.Cavaco, L. M., H. Hasman, S. Xia, and F. M. Aarestrup. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovars Kentucky and Bovismorbificans of human origin. Antimicrob. Agents Chemother. 53603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention, U.S. Department of Health and Human Services. 2007. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2004. Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, Georgia. http://www.cdc.gov/narms/.
- 6.Chen, S., S. Zhao, D. G. White, C. M. Schroeder, R. Lu, H. Yang, P. F. McDermott, S. Ayers, and J. Meng. 2004. Characterization of multiple-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, T. K., Y. W. Chu, M. Y. Chu, C. H. Ma, R. W. Yung, and K. M. Kam. 2005. Plasmid-mediated resistance to ciprofloxacin and cefotaxime in clinical isolates of Salmonella enterica serotype Enteritidis in Hong Kong. J. Antimicrob. Chemother. 56586-589. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 2nd ed. CLSI document M7-A2. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Cui, S., J. Li, Z. Sun, C. Hu, S. Jin, Y. Guo, L. Ran, and Y. Ma. 2008. Ciprofloxacin-resistant Salmonella enterica serotype Typhimurium, China. Emerg. Infect. Dis. 14493-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, M. A., D. D. Hancock, T. E. Besser, D. H. Rice, J. M. Gay, C. Gay, L. Gearhart, and R. DiGiacomo. 1999. Changes in antimicrobial resistance among Salmonella enterica serovar Typhimurium isolates from humans and cattle in the Northwestern United States, 1982-1997. Emerg. Infect. Dis. 5802-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Wit, M. A., A. M. Hoogenboom-Verdegaal, E. S. Goosen, M. J. Sprenger, and M. W. Borgdorff. 2000. A population-based longitudinal study on the incidence and disease burden of gastroenteritis and Campylobacter and Salmonella infection in four regions of The Netherlands. Eur. J. Epidemiol. 16713-718. [DOI] [PubMed] [Google Scholar]
- 12.European Food Safety Authority. 2007. The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. EFSA J. 2007130. http://www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178671312912.htm. [Google Scholar]
- 13.Galanis, E., D. M. Lo Fo Wong, M. E. Patrick, N. Binsztein, A. Cieslik, T. Chalermchikit, A. Aidara-Kane, A. Ellis, F. J. Angulo, and H. C. Wegener for World Health Organization Global Salm-Surv. 2006. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg. Infect. Dis. 12381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guard-Petter, J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3421-430. [DOI] [PubMed] [Google Scholar]
- 15.Hasman, H., D. Mevius, K. Veldman, I. Olesen, and F. M. Aarestrup. 2005. β-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56115-121. [DOI] [PubMed] [Google Scholar]
- 16.Helms, M., P. Vastrup, P. Gerner-Smidt, and K. Mølbak. 2002. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg. Infect. Dis. 8490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helms, M., J. Simonsen, and K. Mølbak. 2004. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J. Infect. Dis. 1901652-1654. [DOI] [PubMed] [Google Scholar]
- 18.Herikstad, H., Y. Motarjemi, and R. V. Tauxe. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 1291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoge, C. W., J. M. Gambel, A. Srijan, C. Pitarangsi, and P. Echeverria. 1998. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin. Infect. Dis. 26341-345. [DOI] [PubMed] [Google Scholar]
- 20.Humphrey, T. J. 2000. Public-health aspects of Salmonella infections, p. 245-263. In C. Wray and A. Wray (ed.), Salmonella in domestic animals. CABI Publishing, CAB International, Wallingford, United Kingdom.
- 21.Jin, Y., and J. M. Ling. 2006. CTX-M-producing Salmonella spp. in Hong Kong: an emerging problem. J. Med. Microbiol. 551245-1250. [DOI] [PubMed] [Google Scholar]
- 22.Jones, Y. E., S. Chappell, I. M. McLaren, R. H. Davies, and C. Wray. 2002. Antimicrobial resistance in Salmonella isolated from animals and their environment in England and Wales from 1988 to 1999. Vet. Rec. 150649-654. [DOI] [PubMed] [Google Scholar]
- 23.Lauderdale, T. L., F. M. Aarestrup, P. C. Chen, J. F. Lai, H. Y. Wang, Y. R. Shiau, I. W. Huang, C. L. Hung, et al. 2006. Multidrug resistance among different serotypes of clinical Salmonella isolates in Taiwan. Diagn. Microbiol. Infect. Dis. 55149-155. [DOI] [PubMed] [Google Scholar]
- 24.Mulvey, M. R., G. Soule, D. Boyd, W. Demczuk, and R. Ahmed. 2003. Characterization of the first extended-spectrum beta-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 41460-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen, S. J., R. Bishop, F. W. Brenner, T. H. Roels, N. Bean, R. V. Tauxe, and L. Slutsker. 2001. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987-1997. J. Infect. Dis. 183753-761. [DOI] [PubMed] [Google Scholar]
- 26.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, B. S. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis (PFGE) protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 359-67. www.cdc.gov/pulsenet/protocols.htm. [DOI] [PubMed] [Google Scholar]
- 27.Van Duijkeren, E., W. J. Wannet, D. J. Houwers, and W. van Pelt. 2003. Antimicrobial susceptibilities of Salmonella strains isolated from humans, cattle, pigs, and chickens in the Netherlands from 1984 to 2001. J. Clin. Microbiol. 413574-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varma, J. K., K. Molbak, T. J. Barrett, J. L. Beebe, T. F. Jones, T. Rabatsky-Ehr, K. E. Smith, D. J. Vugia, H. G. Chang, and F. J. Angulo. 2005. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J. Infect. Dis. 191554-561. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J. Y., J. J. Hwang, C. N. Hsu, L. C. Lin, and P. R. Hsueh. 2006. Bacteraemia due to ciprofloxacin-resistant Salmonella enterica serotype Choleraesuis in adult patients at a university hospital in Taiwan, 1996-2004. Epidemiol. Infect. 134977-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, M., L. Ran, Z. Wang, and Z. Li. 2004. Study on national active monitoring for food borne pathogens and antimicrobial resistance in China 2001. Med. Hyg. 33(1)49-54. [In Chinese.] [PubMed] [Google Scholar]