Abstract
Isolates of the Mycobacterium tuberculosis Beijing lineage are associated with high rates of transmission, hypervirulence and drug resistance. The Beijing lineage has been shown to dominate the tuberculosis (TB) epidemic in East Asia; however, the diversity and frequency of M. tuberculosis genotypes from Myanmar are unknown. We present the first comprehensive study describing the M. tuberculosis isolates circulating in Yangon, Myanmar. Thus, 310 isolates from pulmonary TB patients from Yangon, Myanmar, were genotyped by spoligotyping and IS6110-based restriction fragment length polymorphism analysis (IS6110 RFLP). The most frequent lineages observed were the East African-Indian (EAI; 48.4%; n = 150) and Beijing (31.9%; n = 99) lineages. Isolates belonging to the most frequent shared types (STs), ST1 (n = 98; Beijing), ST292 (n = 28; EAI), and ST89 (n = 11; EAI), had ≥75% similarity in their IS6110 patterns. Five of 11 Beijing isolates comprising five clusters with identical IS6110 RFLP patterns could be discriminated by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) analysis. Of the 150 EAI isolates, 40 isolates (26.7%) had only one IS6110 copy, and 17 of these isolates could be discriminated by MIRU-VNTR analysis. The findings from this study suggest that although there is a predominance of the ancient EAI lineage in Yangon, the TB epidemic in Yangon is driven by clonal expansion of the ST1 genotype. The Beijing lineage isolates (21.4%) were more likely (P = 0.009) than EAI lineage isolates to be multidrug resistant (MDR) (1.3%; odds ratio, 3.2, adjusted for the patients' history of exposure to anti-TB drugs), suggesting that the spread of MDR Beijing isolates is a major problem in Yangon.
Despite global efforts to combat tuberculosis (TB), the causative agent of which is Mycobacterium tuberculosis, TB still remains the leading cause of death caused by a single pathogen. This disease is a major public health problem worldwide, but it is especially a problem in developing countries. Myanmar is 1 of 22 countries that together account for 80% of the world's new TB cases (38). Several studies have described the prevalence and distribution of M. tuberculosis genotypes in other parts of the Southeast Asia region (2-4, 13); however, very little is known about the genotypes prevalent in Myanmar. A high proportion of M. tuberculosis isolates in many Asian countries and all over Russia belong to the Beijing lineage (5, 12, 21, 33). These strains multiply faster than a laboratory M. tuberculosis strain in monocytes (20), and although some studies have shown an association with increased multidrug resistance (MDR) (1, 2, 17, 31, 33), others have not found this association (14). It has previously been shown that missense mutations in the DNA repair genes of the W-Beijing genotype have provided a true selective advantage for this lineage that allow it to adapt and persist, including the ability to acquire resistance to anti-TB drugs (25). However, a study by Werngren and Hoffner (37) suggests that the association of the Beijing genotype with MDR may not be due to an increased mutation rate of the Beijing strains. Genotyping of M. tuberculosis strains has been greatly facilitated by the discovery of highly repetitive DNA elements on the M. tuberculosis genome (10). Two molecular strain typing methods that take advantage of the polymorphisms caused by repetitive elements on the M. tuberculosis genome are IS6110-based restriction fragment length polymorphism (RFLP) analysis (IS6110 RFLP) (34) and spoligotyping (15). These two techniques are routinely used to discriminate between M. tuberculosis isolates and are useful tools in investigations that attempt to detect emerging epidemics caused by new and/or hypervirulent genotypes (4, 17, 33).
In previous studies (23, 24), we have shown that about one-third of isolates from new TB patients attending the National TB Program (NTP) of Myanmar were resistant to any of the first-line anti-TB drugs (streptomycin, isoniazid, rifampin [rifampicin], or ethambutol) and that about 4% and 18% of new and previously treated TB patients (in Yangon, Myanmar), respectively, had MDR TB. Yangon is the former capital of Myanmar, and with about 6 million inhabitants, it is the most densely populated area in Myanmar (22). The aim of this study was to genotypically characterize the M. tuberculosis isolates from patients with pulmonary TB in Yangon and determine if there is any association between the prevalent genotypes and MDR TB.
(The results of this study were presented in part at a poster session at the 6th International Meeting on Microbial Epidemiological Markers, 27 to 30 August 2003, Les Diablerets, Switzerland.)
MATERIALS AND METHODS
Setting and patients.
Sputum specimens from 567 patients with pulmonary TB attending the Yangon Division of NTP (2002) were collected at four district diagnostic and treatment centers run by the NTP. The centers are situated in four administrative districts of the Yangon Division, i.e., the east, west, south, and north districts, which cover 2 million, 1 million, 1.4 million, and 1.6 million inhabitants, respectively. Patients were diagnosed as having pulmonary TB on the basis of their medical histories, clinical signs, two acid-fast bacillus smear-positive sputum samples, and chest X rays, when necessary. From April to August 2002 and from October to December 2002, patients older than 14 years of age with pulmonary TB seen at one of the four district centers were included in the study. Following the provision of informed consent, all participants were interviewed by trained physicians who used structured questionnaires that provided options for open-ended answers. The information collected included socioeconomic and demographic characteristics, current status and history of M. tuberculosis infection, history of contact with a TB case, history of anti-TB treatment, and chest X-ray findings. The study was approved by the Ethical Committee of the Ministry of Health of Myanmar and the Regional Committee for Ethics in Medical Research in Bergen, Norway.
Sample collection.
Two smear-positive sputum samples from each patient were sent to the national TB reference laboratory in Yangon and were inoculated onto Ogawa medium by standard procedures (18). The isolates were transferred to Greave's medium (5% monosodium glutamate, 10% glycerol, 5% bovine serum albumin [pH 7]) and were kept at −20°C prior to further analysis. Three hundred ten of 447 isolates recovered from the cultures were available for this study.
Spoligotyping.
Genomic DNA was isolated from the M. tuberculosis isolates as previously described by van Embden et al. (35). Spoligotyping was performed with commercially available activated Biodyne C membranes that had 43 synthetic oligonucleotides covalently bound to the membranes (Isogen Bioscience BV, Maarssen, The Netherlands), as recommended by the manufacturer. The hybridization patterns were converted into binary and octal formats, as described by Dale et al. (8), and compared with the patterns for the strains previously reported in the SpolDB4 database (7).
IS6110 RFLP.
IS6110 RFLP was performed with PvuII-restricted DNA by the standardized procedure described by van Embden et al. (34, 35). An enhanced chemiluminescence direct nucleic acid labeling and detection system kit (Amersham Pharmacia) was used for labeling and detection of the probe. The membranes were autoradiographed by using Hyperfilm (Amersham Pharmacia) and were scanned with a ScanJet 5200C scanner (Hewlett-Packard). The banding patterns were analyzed by using the Bionumerics (version 3.0) program (Applied Maths, Kortrijk, Belgium). The positions of the bands in each lane were normalized by using an internal marker. The band position tolerance and optimization were set at 1.0% each so that the banding patterns of the reference strain (M. tuberculosis H37Rv) from the different gels were 100% similar. The Dice coefficient of similarity was calculated, and the unweighted pair group method with arithmetic averages was used for cluster analysis. A cluster was defined as two or more isolates sharing an identical IS6110 RFLP pattern. Isolates were assigned to groups, which were based on three or more isolates sharing >75% similarity in their IS6110 RFLP patterns.
MIRU-VNTR analysis.
Isolates with identical IS6110 RFLP patterns and isolates with a single copy of IS6110 were chosen for genotyping by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) analysis, as described by Supply et al. (32). Clustered isolates and isolates with a single copy of IS6110 were subjected to PCR of the MIRU4, MIRU10, MIRU16, MIRU26, MIRU31, and MIRU40 loci. These loci were selected on the basis of their ability to discriminate isolates of the Beijing and East African-Indian (EAI) lineages (unpublished data). Isolates which could be discriminated by two or more loci by six-locus MIRU-VNTR analysis were considered to be clonally different. Isolates with a difference in one locus or less were analyzed by analysis of an additional nine loci (Mtub04, ETR-A, ETR-C, Mtub30, Mtub39, QUB-4156c, QUB-11c, Mtub21, and QUB-26). Isolates that had identical MIRU-VNTR types after 15-locus MIRU-VNTR analysis or that differed by only 1 locus were considered clonally related. Amplification was performed in a total volume of 20 μl containing 1 μl DNA, 0.04 to 0.4 μM of all 15 primer sets, and HotStart Taq Plus polymerase master mix (Qiagen). All reactions were subjected to a cycle of 95°C for 5 min, followed by 30 cycles of 30 s at 94°C, 1 min at 55°C, and 1.5 min at 72°C and a final cycle of 7 min at 72°C. Analysis of the genotyping results was performed by multiplex PCR with an X-rhodamine-labeled MapMarker 1000 size standard (PE Applied Biosystems) for sizing of the PCR products. The PCR fragments were analyzed with a capillary-based electrophoresis sequencer (ABI 3700), and sizing of the various VNTR alleles was done with Peak Scanner (version 1.0) software (PE Applied Biosystems).
Data analysis.
Differences between proportions were analyzed by the chi-square exact test (with Yates' correction for continuity) and expressed as odds ratios (ORs) with 95% confidence intervals (CIs). Differences between continuous data were analyzed by the t test and are expressed as means ± standard deviations. A P value of <0.05 was taken to represent statistical significance. The patient's history of TB treatment was adjusted for establishment of a link between drug resistance and the Beijing lineage by using simple and multivariable logistic regression analyses with SPSS (version 14.0) software (Softonic).
RESULTS
Patient characteristics.
A total of 310 patients with culture-positive pulmonary TB from four district diagnostic and treatment centers in the Yangon Division were included in this study. Seventy percent of the patients included were male, and the mean age was 36.6 years, with 73.5% of the patients being <45 years of age. Forty-four percent of the patients had a history of contact with a TB case, and 33% had previously been treated for TB.
Prevalence of spoligotypes in Yangon.
The spoligotyping results obtained in this study were compared with the shared type (ST) number and the lineages and sublineages described in the SpolDB4 database (7), which provides the genotyping results for 39,295 M. tuberculosis complex strains. A total of 109 different spoligotypes were observed among the 310 isolates (Fig. 1), and 56 of these were previously described (7). Two hundred ninety-six (95.5%) isolates were classified into seven previously described lineages. The frequencies of the spoligotype lineages were as follows: 150 (48.4%) isolates belonged to the EAI lineage, 99 (31.9%) belonged to the Beijing lineage, 15 (4.8%) belonged to the Central Asian (CAS) lineage, 14 (4.5%) belonged to the Latin American-Mediterranean lineage, 11 (3.5%) belonged to the T lineage, 5 (1.6%) belonged to the MANU lineage, and 2 (0.6%) belonged to the X lineage. One isolate was classified as belonging to the Beijing-like sublineage, and 13 isolates could not be assigned to any of the previously described lineages.
FIG. 1.
Spoligotype patterns among the 310 M. tuberculosis isolates.
Table 1 shows the ST distributions of M. tuberculosis isolates belonging to the two most commonly circulating lineages (Beijing and EAI) in Yangon. ST1 (Beijing lineage) was the most prevalent ST among the isolates in this study (n = 98 isolates; 31.6%). One hundred forty-three of 150 EAI isolates were grouped into nine previously described EAI sublineages, whereas 7 isolates could not be assigned to any of the previously described sublineages (Table 1) (7). Of the 43 orphan isolates from the EAI lineage, 5 isolates sharing the same spoligotype pattern were assigned to a group designated YGN.
TABLE 1.
Distribution of STs of M. tuberculosis isolates belonging to the two most commonly circulating lineages (Beijing and EAI) in Yangon, Myanmar
| Lineage or sublineage | ST no. | No. of strains | Total no. (%) of strains in sublineage |
|---|---|---|---|
| Beijing | 1 | 98 | 99 (31.9) |
| 255 | 1 | ||
| EAI5 | 236 | 6 | 49 (15.8) |
| 126 | 4 | ||
| 340 | 4 | ||
| 951 | 4 | ||
| 256 | 2 | ||
| 138 | 1 | ||
| 234 | 1 | ||
| 413 | 1 | ||
| 763 | 1 | ||
| 342 | 1 | ||
| 1390 | 1 | ||
| Orphan | 23 | ||
| EAI6-BGD1 | 292 | 28 | 38 (12.3) |
| 591 | 5 | ||
| 1409 | 2 | ||
| 1425 | 1 | ||
| Orphan | 2 | ||
| EAI1-SOM | 48 | 4 | 16 (5.2) |
| 6 | 2 | ||
| 349 | 2 | ||
| 493 | 1 | ||
| 355 | 1 | ||
| 947 | 3 | ||
| Orphan | 3 | ||
| EAI3-IND | 11 | 7 | 15 (4.8) |
| 113 | 1 | ||
| 473 | 1 | ||
| 654 | 1 | ||
| 1097 | 1 | ||
| Orphan | 4 | ||
| EAI2-NTB | 89 | 11 | 11 (3.5) |
| EAI2-Nonthaburi-like | Orphan | 4 | 4 (1.3) |
| EAI, undefined | 72 | 4 | 4 (1.3) |
| EAI | 1391 | 3 | 3 (1.0) |
| EAI2-Manila | ST1512 | 1 | 3 (2.9) |
| 287 | 1 | ||
| 483 | 1 | ||
| Previously undescribed EAI sublineage | YGN | 5 | 7 (2.3) |
| Orphan | 2 |
IS6110 RFLP of Beijing and EAI lineage isolates.
Table 2 shows the most prevalent genotypes (ST1, ST292, and ST89) divided on the basis of the number of copies of IS6110. Isolates with less than seven copies were termed low-copy-number isolates, and isolates with seven or more copies were termed high-copy-number isolates. One isolate belonging to the Beijing lineage (ST255) also had more than seven IS6110 copies (data not shown in Table 2). The majority of the ST292 isolates (96.4%) and 100% of the ST89 isolates which belonged to the EAI lineage had seven or more IS6110 copies. Only one ST292 isolate had a low copy number.
TABLE 2.
Results of IS6110 RFLP analysis of the three most dominant M. tuberculosis STs from Yangon, Myanmar
| No. of IS6110 copiesa | No. of isolates
|
||
|---|---|---|---|
| ST1 (n = 98) | ST292 (n = 28) | ST89 (n = 11) | |
| 1 | 0 | 0 | 0 |
| 2-6 | 0 | 1 | 0 |
| 7-16 | 42 | 24 | 11 |
| ≥17 | 56 | 3 | 0 |
Isolates with less than seven IS6110 copies were considered low-copy-number isolates, and those with seven or more IS6110 copies were considered high-copy-number isolates.
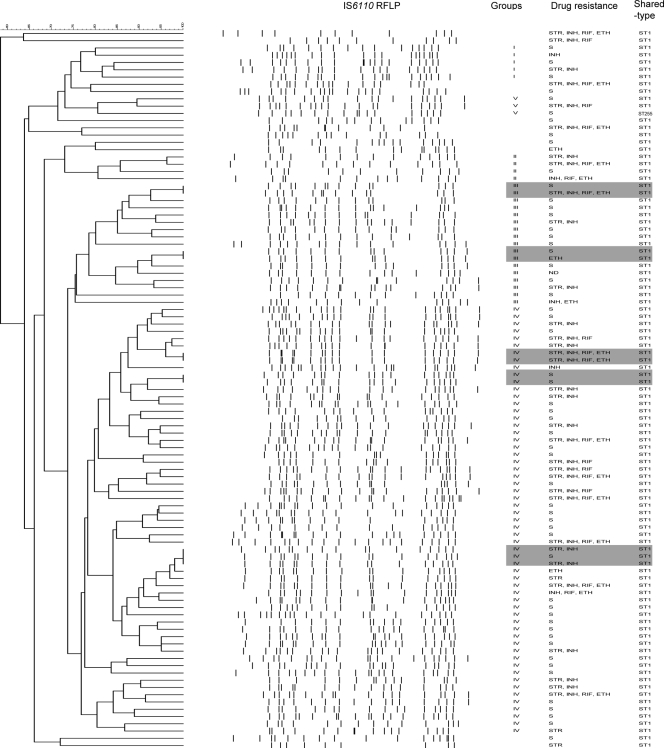
A hierarchical clustering of the IS6110 RFLP patterns for the Beijing lineage isolates showed that 11 isolates formed five clusters on the basis of the 100% similarity of their RFLP patterns. Four clusters had two isolates each, whereas one cluster comprised three isolates (Fig. 2). Of the 99 Beijing lineage isolates, 86 ST1 isolates (87.8%) were assigned to four groups with 75% similarity between the RFLP patterns. The last group (group V) comprised two ST1 isolates and one ST255 isolate, with all three isolates having ≥17 copies of IS6110 (Fig. 2). Groups II and V were the smallest groups and comprised 4 and 3 isolates, respectively, whereas the largest group (group IV) comprised 59 isolates.
FIG. 2.
Dendrogram of the 99 Beijing isolates produced by following the Dice and the unweighted pair group method with arithmetic average analysis of the IS6110 RFLP patterns. Gray highlights isolates with identical IS6110 RFLP patterns.
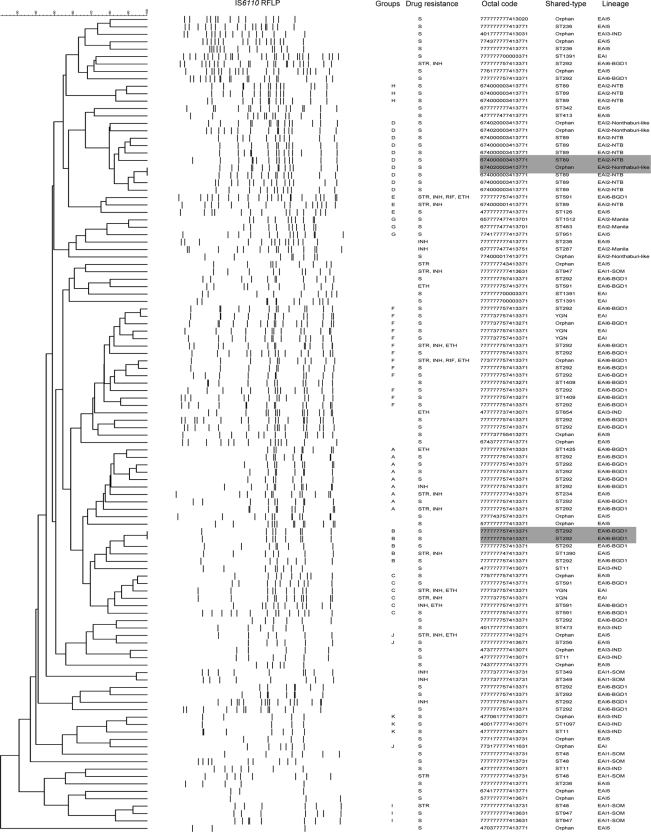
Since genotyping by IS6110 RFLP has a low discriminatory power for isolates with few IS6110 copies (36), RFLP computer analysis was not performed for isolates with one copy of IS6110 in this study. Forty of 150 EAI lineage isolates had only one IS6110 copy. Thus, Fig. 3 shows the RFLP patterns of 110 EAI lineage isolates with two or more IS6110 copies. Four isolates comprised two clusters of 100% similarity (Fig. 3). Two of these isolates shared an identical spoligotype pattern (ST292), whereas of the isolates in the other cluster, one belonged to the ST89 lineage and the other isolate had a previously unreported spoligotype (674020003413771). Sixty-one of the 110 EAI isolates with two or more IS6110 copies were assigned to 11 groups on the basis of 75% similarity between their RFLP patterns (Fig. 3). The largest group (group F) comprised 13 isolates, whereas groups E, G, H, I, J, and K comprised 3 isolates each. Of the 28 ST292 isolates, 17 (60.7%) were distributed among 3 of the 11 groups (group A, n = 7; group B, n = 4; group F, n = 6), whereas the remaining 11 isolates did not belong to any of the assigned groups. Of the 11 ST89 isolates, 10 (90.9%) were distributed among 2 of the 11 groups (group D, n = 7; group H, n = 3), whereas the remaining ST89 isolate belonged to group E (Fig. 3).
FIG. 3.
Dendrogram of 110 EAI isolates with two or more copies of IS6110 produced by following the Dice and the unweighted pair group method with arithmetic average analysis of the IS6110 RFLP patterns. Gray highlights the isolates with identical IS6110 RFLP patterns.
MIRU-VNTR analysis of isolates which clustered by IS6110 RFLP and isolates with one IS6110 copy.
Eleven Beijing isolates comprising five clusters with identical IS6110 RFLP patterns, 2 ST292 isolates with identical IS6110 RFLP patterns, and 40 EAI isolates with a single copy of IS6110 were analyzed by MIRU-VNTR analysis (Table 3). MIRU-VNTR analysis could discriminate between two of the five Beijing clusters, whereas seven Beijing isolates belonging to three different clusters could not be discriminated by 15-locus MIRU-VNTR analysis (Table 3). The two ST292 isolates and 17 of the 40 EAI isolates with one copy of IS6110 could also be discriminated by six-locus MIRU-VNTR analysis. Five isolates with one copy of IS6110 (three isolates belonging to the ST72 genotype and two isolates with orphan genotypes) could not be discriminated by 15-locus MIRU-VNTR analysis (Table 3).
TABLE 3.
MIRU-VNTR analysis of 53 isolates comprising seven clusters with identical IS6110 RFLP patterns
| Lineage and family | ST | Octal code for spoligotyping | No. of IS6110 copies | No. of alleles in the following MIRU-VNTR loci:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIRU4 | MIRU26 | MIRU40 | MIRU10 | MIRU16 | MIRU31 | Mtub04 | ETR-C | ETR-A | Mtub30 | Mtub39 | QUB-4156c | QUB-11b | Mtub21 | QUB-26 | ||||
| Beijing | ||||||||||||||||||
| TL68 | ST1 | 000000000003771 | 18 | 3 | 5 | 0.9 | 2 | 2 | 4 | |||||||||
| TL297 | ST1 | 000000000003771 | 18 | 3 | 6 | 2.6 | 1 | 2 | 4 | |||||||||
| TL117 | ST1 | 000000000003771 | 14 | 3 | 6 | 3.4 | 2 | 2 | 4 | |||||||||
| TL237 | ST1 | 000000000003771 | 14 | 3 | 7 | 2.6 | 2 | 2 | 3 | |||||||||
| TL120 | ST1 | 000000000003771 | 14 | 3 | 6 | 3.5 | 2 | 2 | 4 | 3 | 3 | 4 | 4 | 2 | 2 | 8 | 4 | 3 |
| TL194 | ST1 | 000000000003771 | 14 | 3 | 6 | 3.4 | 2 | 2 | 4 | 3 | 3 | 4 | 4 | 2 | 2 | 8 | 4 | 3 |
| TL174 | ST1 | 000000000003771 | 17 | 3 | 6 | 2.6 | 2 | 2 | 3 | 3 | 3 | 4 | 4 | 2 | 2 | 5 | 4 | 7 |
| TL181 | ST1 | 000000000003771 | 17 | 3 | 6 | 2.6 | 2 | 2 | 4 | 3 | 3 | 4 | 4 | 2 | 2 | 5 | 4 | 7 |
| TL261 | ST1 | 000000000003771 | 17 | 3 | 6 | 2.6 | 2 | 2 | 4 | 3 | 3 | 4 | 2 | 2 | 2 | 3 | 4 | 7 |
| TL205 | ST1 | 000000000003771 | 18 | 3 | 6 | 2.6 | 1 | 2 | 4 | 44 | 33 | 4 | 4 | 2 | 2 | 6 | 4 | 6 |
| TL264 | ST1 | 000000000003771 | 18 | 3 | 6 | 2.6 | 1 | 2 | 3 | 44 | 33 | 4 | 4 | 2 | 2 | 6 | 4 | 6 |
| EAI | ||||||||||||||||||
| TL166 | ST292 | 777777757413371 | 8 | 4 | 2 | 1.8 | 3 | 2 | 3 | |||||||||
| TL171 | ST292 | 777777757413371 | 8 | 5 | 2 | 2.6 | 3 | 2 | 4 | |||||||||
| TL45 | Orphan | 077737777413731 | 1 | 7 | 2 | 2.6 | 3 | 2 | 4 | |||||||||
| TL115 | Orphan | 077737777413731 | 1 | 4 | 5 | 1.8 | 1 | 2 | 2 | |||||||||
| TL64 | Orphan | 734177777413771 | 1 | 5 | 2 | 2.6 | 3 | 1 | 4 | 2 | 3 | 3 | 2 | 4 | 1 | 2 | 4 | 6 |
| TL100 | Orphan | 734177777413771 | 1 | 7 | 2 | 2.6 | 2 | 1 | 4 | 2 | 2 | 6 | 2 | 4 | 1 | 2 | 4 | 6 |
| TL267 | Orphan | 734177777413771 | 1 | 5 | 2 | 2.6 | 2 | 1 | 4 | 2 | 2 | 6 | 2 | 4 | 1 | 2 | 4 | 6 |
| TL244 | Orphan | 734177777413771 | 1 | 5 | 2 | 2.6 | 2 | 1 | 3 | 2 | 3 | 6 | 2 | 4 | 1 | 2 | 4 | 5 |
| TL126 | Orphan | 734177777413771 | 1 | 5 | 3 | 2.6 | 2 | 1 | 4 | |||||||||
| TL41 | ST11 | 477777777413071 | 1 | 2 | 2 | 3.4 | 2 | 2 | 4 | 2 | 3 | 6 | 1 | 5 | 1 | 3 | 5 | 6 |
| TL288 | ST11 | 477777777413071 | 1 | 7 | 2 | 3.4 | 3 | 2 | 3 | 2 | 3 | 6 | 1 | 3 | 1 | 3 | 5 | 4 |
| TL230 | ST11 | 477777777413071 | 1 | 5 | 2 | 3.4 | 2 | 2 | 3 | |||||||||
| TL19 | ST113 | 477777776413071 | 1 | 5 | 2 | 3.4 | 3 | 2 | 3 | |||||||||
| TL72 | ST126 | 477777774133771 | 1 | 5 | 2 | 1.8 | 3 | 2 | 4 | 2 | 3 | 6 | 1 | 4 | 1 | 3 | 5 | 6 |
| TL188 | ST126 | 477777774133771 | 1 | 5 | 2 | 1.8 | 3 | 2 | 4 | 2 | 3 | 5 | 1 | 3 | 1 | 3 | 4 | 6 |
| TL348 | ST138 | 477777774133771 | 1 | 6 | 2 | 1.8 | 3 | 2 | 4 | 2 | 3 | 6 | 1 | 4 | 1 | 3 | 5 | 3 |
| TL149 | ST138 | 777777777413700 | 1 | 5 | 2 | 3.4 | 4 | 2 | 4 | |||||||||
| TL219 | ST236 | 777777777413771 | 1 | 4 | 2 | 2.6 | 3 | 2 | 4 | 2 | 3 | 6 | 1 | 3 | 1 | 3 | 3 | 6 |
| TL409 | ST236 | 777777777413771 | 1 | 4 | 2 | 2.6 | 3 | 1 | 4 | 2 | 3 | 6 | 2 | 3 | 1 | 2 | 3 | 6 |
| TL360 | ST256 | 777777777413637 | 1 | 6 | 2 | 2.6 | 1 | 1 | 4 | |||||||||
| TL145 | ST340 | 474377777413771 | 1 | 5 | 2 | 3.4 | 3 | 2 | 5 | 2 | 3 | 6 | 1 | 2 | 1 | 1 | 10 | 6 |
| TL163 | ST340 | 474377777413771 | 1 | 5 | 2 | 2.6 | 2 | 2 | 4 | 2 | 2 | 6 | 1 | 3 | 1 | 3 | 12 | 5 |
| TL183 | ST340 | 474377777413771 | 1 | 4 | 2 | 2.6 | 3 | 2 | 4 | 2 | 3 | 6 | 1 | 3 | 1 | 2 | 9 | 6 |
| TL286 | ST340 | 474377777413771 | 1 | 5 | 2 | 2.6 | 3 | 2 | 4 | |||||||||
| TL217 | ST355 | 477777777413031 | 1 | 5 | 2 | 3.4 | 3 | 2 | 3 | |||||||||
| TL16 | ST493 | 774177777413731 | 1 | 5 | 2 | 2.6 | 1 | 1 | 3 | |||||||||
| TL139 | ST6 | 077777777413731 | 1 | 5 | 2 | 2.6 | 3 | 2 | 4 | 2 | 3 | 8 | 2 | 2 | 1 | 7 | 4 | 5 |
| TL161 | ST6 | 077777777413731 | 1 | 5 | 2 | 2.6 | 3 | 2 | 3 | 2 | 3 | 8 | 2 | 2 | 1 | 7 | 5 | 5 |
| TL247 | ST72 | 777577777003731 | 1 | 6 | 2 | 2.6 | 3 | 2 | 3 | 2 | 2 | 4 | 2 | 4 | 1 | 5 | 8 | 4 |
| TL260 | ST72 | 777577777003731 | 1 | 6 | 2 | 2.6 | 3 | 2 | 4 | 2 | 2 | 7 | 2 | 4 | 1 | 5 | 8 | 4 |
| TL293 | ST72 | 777577777003731 | 1 | 6 | 2 | 2.6 | 3 | 2 | 4 | 2 | 2 | 7 | 2 | 4 | 1 | 5 | 11 | 4 |
| TL294 | ST72 | 777577777003731 | 1 | 6 | 2 | 2.6 | 3 | 2 | 4 | 2 | 2 | 7 | 2 | 4 | 1 | 5 | 11 | 4 |
| TL7 | ST763 | 777700777413700 | 1 | 5 | 2 | 2.6 | 4 | 2 | 4 | |||||||||
| TL186 | ST951 | 774177777413771 | 1 | 5 | 2 | 2.6 | 3 | 1 | 4 | 3 | 3 | 6 | 2 | 5 | 1 | 2 | 4 | 6 |
| TL227 | ST951 | 774177777413771 | 1 | 5 | 2 | 2.6 | 2 | 1 | 4 | 2 | 3 | 6 | 2 | 5 | 1 | 2 | 4 | 3 |
| TL36 | ST951 | 774177777413771 | 1 | 5 | 2 | 3.4 | 3 | 1 | 3 | |||||||||
| TL213 | Orphan | 417777777413071 | 1 | 5 | 2 | 3.4 | 3 | 2 | 4 | |||||||||
| TL77 | Orphan | 474377707413771 | 1 | 5 | 2 | 2.6 | 3 | 2 | 4 | |||||||||
| TL23 | Orphan | 700001777413771 | 1 | 5 | 2 | 3.4 | 3 | 1 | 4 | |||||||||
| TL332 | Orphan | 730177777413631 | 1 | 5 | 2 | 3.4 | 2 | 1 | 4 | |||||||||
| TL197 | Orphan | 773700777413700 | 1 | 5 | 2 | 2.6 | 4 | 2 | 4 | |||||||||
| TL20 | Orphan | 777777777413611 | 1 | 6 | 1 | 2.6 | 4 | 2 | 2 | |||||||||
Drug resistance among isolates of the Beijing lineage and the EAI lineage.
A total of 41.4% and 25.3% of the patients infected with Beijing lineage isolates and patients infected with EAI lineage isolates had a history of exposure to anti-TB drugs, respectively (P = 0.006) (Table 4). The proportion of drug-resistant isolates was significantly higher (P = 0.01) among patients infected with Beijing lineage isolates (44.9%) than among patients infected with EAI lineage isolates (24.7%).
TABLE 4.
Characteristics of patients infected with Beijing versus EAI lineage isolates
| Characteristic | Patients infected with Beijing lineage isolates (n = 99) | Patients infected with EAI lineage isolates (n = 150) | P value |
|---|---|---|---|
| Mean age ± SD (yr) | 36.0 ± 12.6 | 37.6 ± 13.2 | 0.36 |
| No. (%) of patients | |||
| Age ≤45 yr | 73 (73.7) | 111 (74.0) | 0.54 |
| Male sex | 64 (64.6) | 107 (71.3) | 0.17 |
| History of TB contact | 47 (48.0)a | 60 (40.5)b | 0.15 |
| History of previous treatment for TB | 41 (41.4) | 38 (25.3) | 0.006 |
| Infected with drug-resistant isolatesc | 44 (44.9)a | 37 (24.7) | 0.01 |
n = 98.
n = 148.
The anti-TB drugs tested were streptomycin, isoniazid, rifampin, and ethambutol.
Twenty-one Beijing lineage isolates (21.4%) were resistant to both isoniazid and rifampin (Table 5) and, hence, were defined as MDR, whereas only 2 of 150 (1.3%) of the EAI lineage isolates were MDR. The Beijing lineage isolates were more likely (P = 0.009) than EAI lineage isolates to be MDR (OR, 3.2, adjusted for the patients previous history of exposure to anti-TB drugs).
TABLE 5.
Drug resistance among isolates of the Beijing and EAI lineages
| Resistant pattern | No. (%) of isolates
|
Crude OR (95% CI) | Adjusted ORa (95% CI) | P value | |
|---|---|---|---|---|---|
| Beijing lineage (n = 98) | EAI lineages (n = 150) | ||||
| Monoresistance to any one anti-TB drugb | 8 (8.2) | 19 (12.7) | 0.61 (0.26-1.46) | 1.88 (0.83-4.23) | 0.13 |
| Resistance to any two anti-TB drugsc | 14 (14.3) | 13 (8.7) | 1.76 (0.79-3.92) | 0.59 (0.23-1.53) | 0.28 |
| MDR TBd | 21 (21.4) | 2 (1.3) | 20.18 (4.61-88.33) | 3.20 (1.34-7.67) | 0.009 |
Adjusted for the patient's history of exposure to anti-TB drugs.
The anti-TB drugs tested were streptomycin, isoniazid, rifampin, and ethambutol.
Strains resistant to any two anti-TB drugs except a combination of isoniazid and rifampin.
Strains resistant to at least isoniazid and rifampin.
DISCUSSION
To our knowledge, this is the first comprehensive study describing the frequency and diversity of M. tuberculosis genotypes from pulmonary TB patients from Yangon, Myanmar. Representative sputum samples from TB patients from the entire city of Yangon were collected over a period of 8 months, and the M. tuberculosis isolates were characterized genotypically. Two hundred eighty-nine of 310 isolates (93.2%) could be classified into seven lineages previously described in the SpolDB4 database (7). The relative frequencies of M. tuberculosis spoligotype lineages in Yangon were quite similar to those described in countries in Far East Asia (7). The predominant lineages observed in this study were the EAI and Beijing lineages. Our study identified 56 different previously described STs that are spreading in Yangon and showed that the single most prevalent M. tuberculosis ST was ST1 (31.6%), which belonged to the Beijing lineage. The other highly prevalent genotypes in Yangon were ST292 (9.0%) and ST89 (3.5%), which are members of the EAI6-BGD1 and EAI2-NTB sublineages, respectively (Table 1). Five isolates of the ST26 genotype (CAS1-DELHI sublineage) were also found in Yangon. Additionally, this study identified 54 previously undescribed spoligotype patterns.
Some studies have indicated that there is a strong phylogeographical clustering of the M. tuberculosis bacillus population (7, 11, 16, 19, 28). The ST1 M. tuberculosis strains are prevalent in Far East Asia (China), Southeast Asia (Vietnam, Thailand), the Middle East-Central Asia, and Oceania (7). Isolates of the EAI lineage are reported to be prevalent in Southeast Asia, India, and East Africa (11, 16, 19, 28). Isolates belonging to EAI6-BGD and EAI2-NTB sublineages are phylogeographically specific for Bangladesh and Thailand, respectively (7). Isolates of the CAS1-DELHI sublineage, however, are mainly found in the Indian subcontinent (13, 28, 29). Myanmar has borders with China, India, Bangladesh, and Thailand. In addition, the top four countries of origin of foreigners living in Myanmar are China, India, Pakistan, and Bangladesh (22). Thus, it is not surprising to find phylogeographically specific isolates from China, Bangladesh, India, and Thailand in Myanmar.
Some studies (6, 13) have reported that the M. tuberculosis-specific deletion 1 (TbD1) can consistently be found in the Beijing lineage strains, which are considered to have evolved more recently than other, more ancient M. tuberculosis strains (6). Many M. tuberculosis strains from Bangladesh and India have a conserved TbD1 region and thus may be regarded as ancient (3, 13, 27). In a previous study, we have detected the TbD1 deletion in isolates of the ST1, ST26, ST42, ST53, and ST126 genotypes from Yangon, whereas all other non-Beijing lineage isolates, including ST89 and ST292, from Yangon had this region intact (30). In our study, IS6110 RFLP showed that 88 of 99 Beijing lineage isolates were assigned to five groups on the basis of 75% similarity, and the IS6110 RFLP patterns of 11 isolates were identical. Two sets of clustered isolates from the Beijing lineage had the same drug susceptibility pattern as well. Furthermore, three different Beijing clusters could not be differentiated by the MIRU-VNTR analysis method (Table 3). These findings suggest that the clonal expansion of the Beijing lineage strains may be associated with recent transmission in the community. Similarly, relatively high proportions of isolates belonging to ST292 (61%) and ST89 (100%) could be assigned to groups on the basis of a 75% similarity of their IS6110 RFLP patterns (Fig. 3). More than 70% of the patients infected with the Beijing and EAI lineage isolates were under the age of 45 years (Table 4), supporting the theory that the isolates from this study belong to an endemic strain pool (3, 9). Collectively, one can assume that TB in Yangon is shaped by the clonal expansion of both modern and ancient M. tuberculosis strains.
Our study showed that a significantly higher number (P = 0.009) of MDR isolates belonged to the Beijing genotype (n = 21) than to the EAI genotype (n = 2) (Table 5). Of 58 new TB patients infected with Beijing lineage isolates, 24 (42.4%) were infected with drug-resistant isolates, and 8 of these were MDR. Thus, there are a large number of drug-resistant Beijing lineage isolates circulating in Yangon.
A high proportion of M. tuberculosis isolates in southern India and Bangladesh have small numbers of or no IS6110 copies (26). Our study also showed a large number of EAI lineage isolates (n = 40, 12.9%) with one copy of IS6110. Thus, the use of a combination of the spoligotyping and MIRU-VNTR analysis methods for studying the molecular epidemiology of TB may provide a better means of fingerprinting, at least in Yangon, than a combination of the spoligotyping and IS6110 RFLP methods.
Our study is the first to show the frequency of M. tuberculosis genotypes in Yangon, Myanmar. The majority of the isolates belonged to the Beijing and EIA lineages and may reflect the M. tuberculosis populations introduced during past migrations from neighboring countries. In addition, in Yangon, Beijing lineage isolates were more likely than EAI lineage isolates to be MDR.
Acknowledgments
We thank the staff of the Myanmar reference laboratory (NTP) for the collection of isolates. We thank Robin Warren for valuable comments. We thank Kristi Øvreås, Sonal Patel, and Grete Hopland for technical assistance and Haukeland University Hospital for excellent laboratory facilities.
This study was supported by the Western Norway Regional Health Authority.
Footnotes
Published ahead of print on 26 November 2008.
REFERENCES
- 1.Agerton, T. B., S. E. Valway, R. J. Blinkhorn, K. L. Shilkret, R. Reves, W. W. Schluter, B. Gore, C. J. Pozsik, B. B. Plikaytis, C. Woodley, and I. M. Onorato. 1999. Spread of strain W, a highly drug-resistant strain of Mycobacterium tuberculosis, across the United States. Clin. Infect. Dis. 2985-92. [DOI] [PubMed] [Google Scholar]
- 2.Anh, D. D., M. W. Borgdorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banu, S., S. V. Gordon, S. Palmer, M. R. Islam, S. Ahmed, K. M. Alam, S. T. Cole, and R. Brosch. 2004. Genotypic analysis of Mycobacterium tuberculosis in Bangladesh and prevalence of the Beijing strain. J. Clin. Microbiol. 42674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhanu, N. V., D. van Soolingen, J. D. van Embden, L. Dar, R. M. Pandey, and P. Seth. 2002. Predominace of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis (Edinburgh) 82105-112. [DOI] [PubMed] [Google Scholar]
- 5.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 1045-52. [DOI] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 993684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. Van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 5216-219. [PubMed] [Google Scholar]
- 9.Dye, C. 2006. Global epidemiology of tuberculosis. Lancet 367938-940. [DOI] [PubMed] [Google Scholar]
- 10.Eisenach, K. D., J. T. Crawford, and J. H. Bates. 1988. Repetitive DNA sequences as probes for Mycobacterium tuberculosis. J. Clin. Microbiol. 262240-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdinand, S., C. Sola, S. Chanteau, H. Ramarokoto, T. Rasolonavalona, V. Rasolofo-Razanamparany, and N. Rastogi. 2005. A study of spoligotyping-defined Mycobacterium tuberculosis clades in relation to the origin of peopling and the demographic history in Madagascar. Infect. Genet. Evol. 5340-348. [DOI] [PubMed] [Google Scholar]
- 12.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez, M. C., N. Ahmed, E. Willery, S. Narayanan, S. E. Hasnain, D. S. Chauhan, V. M. Katoch, V. Vincent, C. Locht, and P. Supply. 2006. Predominance of ancestral lineages of Mycobacterium tuberculosis in India. Emerg. Infect. Dis. 121367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanekom, M., G. D. van der Spuy, E. Streicher, S. L. Ndabambi, C. R. McEvoy, M. Kidd, N. Beyers, T. C. Victor, P. D. van Helden, and R. M. Warren. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 451483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 372607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruuner, A., S. E. Hoffner, H. Sillastu, M. Danilovits, K. Levina, S. B. Svenson, S. Ghebremichael, T. Koivula, and G. Kallenius. 2001. Spread of drug-resistant pulmonary tuberculosis in Estonia. J. Clin. Microbiol. 393339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudoh, S., and T. Kudoh. 1974. A simple technique for culturing tubercle bacilli. Bull. W. H. O. 5171-82. [PMC free article] [PubMed] [Google Scholar]
- 19.Kulkarni, S., C. Sola, I. Filliol, N. Rastogi, and G. Kadival. 2005. Spoligotyping of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in Mumbai, India. Res. Microbiol. 156588-596. [DOI] [PubMed] [Google Scholar]
- 20.Li, Q., C. C. Whalen, J. M. Albert, R. Larkin, L. Zukowski, M. D. Cave, and R. F. Silver. 2002. Differences in rate and variability of intracellular growth of a panel of Mycobacterium tuberculosis clinical isolates within a human monocyte model. Infect. Immun. 706489-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mokrousov, I. 22 July 2008. Genetic geography of Mycobacterium tuberculosis Beijing genotype: A multifacet mirror of human history? Infect. Genet. Evol. doi: 10.1016/j.meegid. 2008.07.003. (Subsequently published, Infect. Genet. Evol. 8:777-785, 2008.) [DOI] [PubMed]
- 22.Myanmar Central Statistical Organization. 2001. Ministry of national planning and economic development: statistical yearbook 2001. Ministry of Immigration and Population, Yangon, Myanmar.
- 23.Phyu, S., T. Lwin, T. Ti, W. Maung, W. W. Mar, S. S. Shein, and H. M. S. Grewal. 2005. Drug-resistant tuberculosis in Yangon, Myanmar. Scand. J. Infect. Dis. 37846-851. [DOI] [PubMed] [Google Scholar]
- 24.Phyu, S., T. Ti, R. Jureen, T. Hmun, H. Myint, A. Htun, H. M. S. Grewal, and B. Bjorvatn. 2003. Drug-resistant Mycobacterium tuberculosis among new tuberculosis patients, Yangon, Myanmar. Emerg. Infect. Dis. 9274-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rad, M. E., P. Bifani, C. Martin, K. Kremer, S. Samper, J. Rauzier, B. Kreiswirth, J. Blazquez, M. Jouan, D. van Soolingen, and B. Gicquel. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radhakrishnan, I., K. My, R. A. Kumar, and S. Mundayoor. 2001. Implications of low frequency of IS6110 in fingerprinting field isolates of Mycobacterium tuberculosis from Kerala, India. J. Clin. Microbiol. 391683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao, K. R., F. Kauser, S. Srinivas, S. Zanetti, L. A. Sechi, N. Ahmed, and S. E. Hasnain. 2005. Analysis of genomic downsizing on the basis of region-of-difference polymorphism profiling of Mycobacterium tuberculosis patient isolates reveals geographic partitioning. J. Clin. Microbiol. 435978-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh, U. B., J. Arora, N. Suresh, H. Pant, T. Rana, C. Sola, N. Rastogi, and J. N. Pande. 2007. Genetic biodiversity of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in India. Infect. Genet. Evol. 7441-448. [DOI] [PubMed] [Google Scholar]
- 29.Singh, U. B., N. Suresh, N. V. Bhanu, J. Arora, H. Pant, S. Sinha, R. C. Aggarwal, S. Singh, J. N. Pande, C. Sola, N. Rastogi, and P. Seth. 2004. Predominant tuberculosis spoligotypes, Delhi, India. Emerg. Infect. Dis. 101138-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stavrum, R., H. Valvatne, T. H. Bo, I. Jonassen, J. Hinds, P. D. Butcher, and H. M. S. Grewal. 2008. Genomic diversity among Beijing and non-Beijing Mycobacterium tuberculosis isolates from Myanmar. PLoS One 3e1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, Y. J., A. S. Lee, S. Y. Wong, H. Heersma, K. Kremer, D. van Soolingen, and N. I. Paton. 2007. Genotype and phenotype relationships and transmission analysis of drug-resistant tuberculosis in Singapore. Int. J. Tuberc. Lung Dis. 11436-442. [PubMed] [Google Scholar]
- 32.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toungoussova, O. S., P. Sandven, A. O. Mariandyshev, N. I. Nizovtseva, G. Bjune, and D. A. Caugant. 2002. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J. Clin. Microbiol. 401930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Embden, J. D., L. M. Schouls, and D. van Soolingen. 1995. Molecular techniques: applications in epidemiologic studies, p. 15-27. In C. O. Thoen and J. H. Steele (ed.), Mycobacterium bovis infection in animals and humans. Iowa State University Press, Ames.
- 36.van Soolingen, D., P. E. de Haas, P. W. Hermans, P. M. Groenen, and J. D. van Embden. 1993. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol. 311987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werngren, J., and S. E. Hoffner. 2003. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J. Clin. Microbiol. 411520-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. 2007. Global tuberculosis control: surveillance, planning, financing. World Health Organization Report. World Health Organization, Geneva, Switzerland.