Abstract
Identification of fowl adenovirus (FAdV) serotypes is of importance in epidemiological studies of disease outbreaks and the adoption of vaccination strategies. In this study, real-time PCR and subsequent high-resolution melting (HRM)-curve analysis of three regions of the hexon gene were developed and assessed for their potential in differentiating 12 FAdV reference serotypes. The results were compared to previously described PCR and restriction enzyme analyses of the hexon gene. Both HRM-curve analysis of a 191-bp region of the hexon gene and restriction enzyme analysis failed to distinguish a number of serotypes used in this study. In addition, PCR of the region spanning nucleotides (nt) 144 to 1040 failed to amplify FAdV-5 in sufficient quantities for further analysis. However, HRM-curve analysis of the region spanning nt 301 to 890 proved a sensitive and specific method of differentiating all 12 serotypes. All melt curves were highly reproducible, and replicates of each serotype were correctly genotyped with a mean confidence value of more than 99% using normalized HRM curves. Sequencing analysis revealed that each profile was related to a unique sequence, with some sequences sharing greater than 94% identity. Melting-curve profiles were found to be related mainly to GC composition and distribution throughout the amplicons, regardless of sequence identity. The results presented in this study show that the closed-tube method of PCR and HRM-curve analysis provides an accurate, rapid, and robust genotyping technique for the identification of FAdV serotypes and can be used as a model for developing genotyping techniques for other pathogens.
Fowl adenoviruses (FAdVs), belonging to the Aviadenovirus genus of the family Adenoviridae (19), have been grouped into five species based on their molecular structure and further divided into 12 serotypes, based largely on cross-neutralization assays (18). There are several strains in each serotype. FAdVs are endemic worldwide and known to cause inclusion body hepatitis, quail bronchitis, hydropericardium syndrome (18, 28), gizzard erosion, and pancreatic necrosis (33, 34, 43, 44, 56).
Diagnosis of FAdV infections can be made from the observation of gross and histopathological changes in the liver and the use of electron microscopy (3, 11) and various serological tests, such as enzyme-linked immunosorbent assay (3, 18, 45), agar gel immunodiffusion, counterimmunoelectrophoresis, indirect hemagglutination, immunofluorescence (3), and Southern hybridization (10, 12). Identification of the serotype(s) involved is very useful for epidemiological tracing and is of critical importance where vaccination is to be used for the control of the disease (3, 20). Typing of the virus conventionally requires isolation in cell culture, followed by a virus neutralization assay (16); however, the implementation of this method is lengthy and labor intensive, and cross-reactivity between serotypes can sometimes render results inconclusive. Tests using PCR together with DNA sequencing (22, 59) and/or restriction enzyme analysis (29, 48, 50) have recently been used for comparatively faster FAdV typing. However, these methods are still time consuming, often require extensive interpretation, and can be relatively expensive to use as a routine typing tool.
Recent research has found that the technique of combining PCR with high-resolution melting (HRM)-curve analysis provides a useful and cost-effective alternative for the direct analysis of genetic variation, particularly when large numbers of samples are to be analyzed. HRM-curve analysis of PCR amplicons has been shown to aid in clinical (52) and/or epidemiological (5, 36) investigations, by providing means for rapid and effective genotyping, variation scanning, microbial detection, and species determination (13, 21, 26, 53). When melted in the presence of a saturating intercalating fluorescent dye, such as LCGreen I or Styo 9 (17, 31, 46), and acquiring fluorescence data over small temperature increments, amplicons containing different sequences can be discriminated based on the melting transition of the PCR product and the resulting melt curve shape.
Commonly, amplicons of fewer than 200 bp have been used for detection of single nucleotide polymorphisms, mutation scanning, and genotyping (26), as slight shifts in the melting domain are easily detected. The use of larger amplicons has been reported to reduce sensitivity and specificity of HRM-curve analysis (6, 24, 25, 60). However, some studies have reported that the melting of larger amplicons, up to 1,000 bp, may produce multiple melting domains and can be used for genotyping (8, 21, 46, 47). The presence of more than one melting domain, as depicted by multiple peaks in the conventional melt curve profile, adds further variation to the normalized HRM-curve profile, thus increasing the power of the HRM-curve analysis technique for sequence differentiation and genotyping (21, 26, 58).
The adenovirus hexon is the major capsid protein of FAdV and contains type-, group-, and subgroup-specific antigenic determinants (18, 28, 35). Several reports have used PCR of different regions of the FAdV viral gene for detection, differentiation, and phylogenetic analysis of FAdVs (29, 48, 50, 59). The hexon loop 1 (L1) region, flanked by pedestal 1 (P1), represents the most variable region and has, when used in PCR coupled with DNA sequencing or restriction enzyme analysis, proved most successful in identifying and differentiating some or all of the 12 FAdV serotypes (2, 29, 30, 37, 48).
The aim of the present study was to investigate the application of HRM-curve analysis of PCR amplicons of different sizes from the hexon gene L1 and P1 region of representative strains of the 12 FAdV serotypes, using the DNA intercalating dye Syto 9, to establish a single closed-tube test method for FAdV serotype differentiation. Both conventional and normalized dissociation plots were generated, and genotyping was applied to differentiate the serotypes, and the results were compared with those of existing FAdV typing methods of PCR and restriction enzyme analysis of the hexon gene and DNA sequencing of PCR amplicons.
MATERIALS AND METHODS
Viral strains.
Representative strains of the 12 known serotypes of FAdV-1 to -7, -8a, -8b, and -9 to -11) were provided by Intervet International (Boxmeer, The Netherlands) (Table 1). Each live unattenuated FAdV strain was propagated in chicken embryo liver cells from 14-day-old specific-pathogen-free embryos (Charles River Laboratories, Australia). Cells in culture flasks (25 cm2) were inoculated with 1 × 106 cells per ml in BioWhittaker medium 199 (Cambrex Bioscience Walkersville, Inc.) containing 5% calf serum (Invitrogen), to a total volume of 10 ml, and incubated at 37°C with 5% CO2. Medium was changed every 1 to 2 days, until the cell monolayer became confluent (approximately 3 to 4 days). Chicken embryo liver cell culture monolayer was inoculated with 495 μl medium 199 with 1% calf serum and 5 μl of live virus, incubated for 1 h at 37°C with 5% CO2 and agitated every 10 to 15 min. Medium was made up to 10 ml and cell culture incubated until more than 80% cytopathic effect was observed. Propagated virus was harvested by freezing (−70°C)/thawing the culture twice, removing the infected medium, and centrifuging at 1,000 × g for 5 min to separate cell debris. The supernatant was stored in aliquots at −70°C.
TABLE 1.
FAdV species, serotypes (according to the current ICTV nomenclature system and the previous European Union and United States systems), strains used in this study, and other known strains within each serotype (19, 28, 48)
| Species | Serotype
|
Strain used in this study | Other strain within each serotype | ||
|---|---|---|---|---|---|
| ICTV | European Union | United States | |||
| A | 1 | 1 | 1 | CELO | 112, QBV, Ote, H1 |
| B | 5 | 5 | 8 | 340 | TR22, M2, Tiptron, IBH-2A |
| C | 3 | 3 | 3 | SR49 | 75, H5, 75-1A-1 |
| 4 | 4 | 4 | KR5 | 506, H2, K31, 61, J2-A | |
| 10 | 11 | 10 | C2B | M11, CFA20, SA2, C-2B | |
| D | 2 | 2 | 2 | SR48 | 685, H3, P7-A, GA1-1, Z7 |
| 9 | 10 | 9 | A02 | 90, CFA19, A2-A | |
| 11 | 12 | 12 | UF71 | 380 | |
| E | 6 | 6 | 5 | CR119 | 168 |
| 7 | 7 | 11 | YR36 | X11, X11-A, 122 | |
| 8a | 8 | 6 | TR59 | 58, CFA40, T8-A | |
| 8b | 9 | 7 | 764 | VRI-33, B-3A | |
Additionally, an Australian field strain (FI-W888-05) isolated from the liver of a 30-day-old broiler, was propagated as described above and the serotype established as ICTV serotype 8b by Intervet International (Boxmeer, The Netherlands). The commercially available Australian FAdV-8b vaccine strain (Intervet Pty Ltd.) was reconstituted according to the manufacturer's instructions and used for DNA extraction.
Extraction of viral DNA.
Viral DNA was extracted from virus propagated in cultures and from Australian FAdV vaccine. A 10-μl volume of each virus sample was solubilized in 300 μl of RLT lysis buffer (Qiagen, Valencia, CA) containing 3 μl of β-mercaptoethanol (1.12 g/ml) and incubated at 4°C overnight. A volume of 15 μl of Qiaex II suspension (Qiagen) and 300 μl of 70% ethanol was combined and then added to the sample mixture. This mixture was loaded into a multispin MSK-100 column (Axygen, Inc., Hayward, CA) and placed in a wash tube. Columns were centrifuged for 30 s at 13,500 × g in a microcentrifuge, and the flowthrough was discarded. The matrix was washed once with 600 μl buffer RW1 (Qiagen) and twice with 500 μl buffer RPE (Qiagen). For each wash, columns were centrifuged for 30 s at 13,500 × g and the flowthrough was discarded, and then the columns were centrifuged for 90 s at 19,000 × g to dry the matrix. The matrix was overlaid with 30 μl of diethyl pyrocarbonate-treated water and incubated at room temperature for 1 min, and DNA was eluted by centrifugation of the column for 60 s at 13,500 × g. The DNA elution step was repeated to obtain a final elution volume of 60 μl. The elutions containing extracted viral DNA were used immediately or stored at −20°C prior to use in PCR.
Real-time PCR for amplification of three regions of FAdV hexon gene.
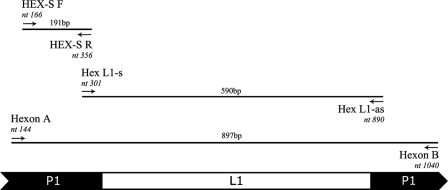
Three sets of forward and reverse degenerate oligonucleotide primers (Table 2) binding to the conserved P1 region of the FAdV hexon gene were used to amplify different regions of P1 and L1 (Fig. 1). The HEX-S forward and reverse primers were designed by comparison of the published hexon gene nucleotide sequences of all 12 FAdV serotypes (National Center for Biotechnology Information, GenBank [4], sequence accession numbers AF339914, AF508946, AF508948, AF508951, AF508952, AF508954, AF508955, AF508956, AF508958, AF508959, AF339920, and AF339925) to amplify a region of P1 containing as small as 2-bp changes between serotypes. All primers were manufactured by Invitrogen Pty Ltd.
TABLE 2.
Primers used in this study
| Primers | Sequence (5′ to 3′) | Positiona | PCR product size (bp) | Reference or source |
|---|---|---|---|---|
| Hexon A | CAARTTCAGRCAGACGGT | 144-161 | 897 | 19 |
| Hexon B | TAGTGATGMCGSGACATCAT | 1040-1021 | ||
| Hex L1-s | ATGGGAGCSACCTAYTTCGACAT | 301-323 | 590 | 4 |
| Hex L1-as | AAATTGTCCCKRAANCCGATGTA | 890-868 | ||
| HEX-S F | GCGCCBACYCGVAAYGTCA | 166-184 | 191 | This study |
| HEX-S R | TTGAARGAVGGHCCBCKGTC | 356-337 |
Nucleotide positions of primers relevant to the FAdV-1 CELO genes for hexon proteins, GenBank accession number Z67970, nt 1380 to 4208 (1).
FIG. 1.
Schematic presentation of Pl and L1 regions of the FAdV-1 CELO hexon gene and the primers used in this study. The base-pair numbering of the primer positions is based on the FAdV-1 CELO genes for hexon proteins, GenBank accession number Z67970, nt 1380 to 4208 (1).
Amplification of target sequences was carried out using a Rotor-Gene 6000 thermal cycler (Corbett Life Science Pty Ltd). A 25-μl reaction mixture consisted of 4 μl of deoxynucleoside triphosphates at 1.25 mM, 2 μl of each primer at 25 μM, 5 μl of 5× GoTaq Green Flexi reaction buffer (Promega), 2 μl of 5 μM Syto 9 green fluorescent nucleic acid stain (Invitrogen), 1.25 U of GoTaq Flexi DNA polymerase, 2 μl of extracted viral DNA, and 1, 2, and 3 μl of 25 mM MgCl2 for Hexon A/B, Hex L1, and HEX-S PCR, respectively.
The reaction mixtures were subjected to 94°C for 2 min, and then, for the Hexon A/B and Hex L1 primer sets, 40 cycles of 94°C for 20 s, 62°C (56°C for Hex L1 primer set) for 20 s, and 72°C for 30 s, and for the HEX-S primer set, 50 cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 20 s. Optical measurements in green channel (excitation at 470 nm and detection at 510 nm) were recorded during the extension step. After completion of PCR cycles, a final extension of 72°C for 2 min was performed.
The PCR products were analyzed by electrophoresis in 1.5% agarose gels stained with GelRed (Biotium, Hayward, CA), and visualized by UV transillumination.
Restriction enzyme analysis.
Restriction enzyme analysis of Hexon A/B PCR products was performed as described previously (29), with some modifications. Briefly, 4 μl of PCR product from each reference serotype (except for FAdV-5, which yielded very low-level or no amplification with Hexon A/B primers) were digested for 3 h with 10 U of each restriction enzyme (New England BioLabs) in a total volume of 20 μl. Restriction fragments were separated in 2% agarose gel stained with GelRed and visualized by UV transillumination.
HRM-curve acquisition and analysis.
HRM-curve analysis was performed on the Rotor-Gene 6000 using identical volumes of PCR products subjected to a temperature increasing from 80 to 94°C at intervals (ramps) of 0.1°C/s or 0.15°C/s. HRM-curve analysis was performed using Rotor-Gene 6000 1.7.87 software and the HRM algorithm provided. All amplicons were tested in triplicate to detect variations induced by technical errors.
The conventional melt curves were generated automatically and contained one or more peaks, depending on the amplicon tested. To generate the normalized HRM curves, normalization regions were applied to the curves as follows: HEX-S, 84.97 to 85.47 and 90.48 to 91.00; Hex L1, 84.50 to 85.00 and 90.50 to 91.00; and Hexon A/B, 81.00 to 81.40 and 90.50 to 90.90. Genotypes were defined by selecting a representative sample from each serotype. The software then auto-called the genotype of each sample and provided confidence percentages (C%) as an integrity check. A confidence threshold was not applied to the analysis in order to view the genotype and C% assigned to each sample replicate. Profiles giving a C% of less than 95% to any of the existing profiles were considered distinctly different profiles. The means of the C% of the sample replicates correctly assigned to a representative genotype, together with the standard error (SE), were calculated using Microsoft Office Excel (2003).
DNA sequence analysis.
Amplicons were gel purified using the QIAquick gel purification kit (Qiagen) according to the manufacturer's instructions, eluted in 30 μl of buffer E, and then subjected to automated sequencing (BigDye Terminator version 3.1, Applied Biosystems) in both directions using the same primers used for PCR. The sequences in the approximately 590-bp region (amplified by primers Hex L1-s and Hex L1-as) were analyzed using ClustalW and DNAdist in BioManager (Australian National Genomic Information Service, Sydney Bioinformatics) and BioEdit Sequence Alignment Editor (version 6.0.9.0).
Nucleotide sequence accession number.
GenBank accession numbers were assigned to the nucleotide sequences of the FAdV reference strains as follows: FAdV-1 CELO, EU979367; FAdV-2 SR48, EU979368; FAdV-3 SR49, EU979369; FAdV-4 KR5, EU979370; FAdV-5 340, EU979371; FAdV-6 CR119, EU979372; FAdV-7 YR36, EU979373; FAdV-8a TR59, EU979374; FAdV-8b 764, EU979375; FAdV-9 A02, EU979376; FAdV-10 C2B, EU979377; and FAdV-11 UF71, EU979378.
RESULTS
PCR amplification of different regions of the hexon gene from FAdV reference serotypes.
Three sets of forward and reverse oligonucleotide primers, HEX-S, Hex L1, and Hexon A/B, were used to amplify approximately 191-bp, 590-bp, and 897-bp fragments of the hexon gene from the FAdV reference serotypes, respectively (Fig. 1).
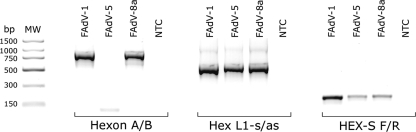
The three primer sets all generated detectable amplicons (by agarose gel electrophoresis) from each of the 12 reference serotypes, with the exception of FAdV-5, which showed low-level or no amplification with the Hexon A/B primer set (Fig. 2).
FIG. 2.
Agarose gel electrophoresis of Hexon A/B, Hex L1, and HEX-S PCR products of the hexon gene from FAdV serotypes 1, 5, and 8a, plus a nontemplate control (NTC). MW, molecular weight marker (PCR marker, Sigma).
The amplification of FAdV-5 with Hexon A/B primers did not vary with repeated PCRs performed on different days using different quantities of DNA as templates, DNA extracted from different samples of propagated virus, or primers manufactured on different dates (data not shown). Examination of the nucleotide sequence of the FAdV-5 strain 340 hexon gene, GenBank accession number AF508952 (30), revealed no mismatch with the primers used.
Restriction enzyme analysis of Hexon A/B PCR products failed to classify some FAdV reference serotypes.
The region between nucleotides (nt) 144 and 1040 (amplified by primers Hexon A and B) of the FAdV reference serotypes used in this study were subjected to restriction enzyme analysis using the method described previously (29). Serotypes 1, 3, 6, 7, 8a, and 9 were differentiated by successive use of the restriction endonucleases BsiWI, StyI, and MluI. An additional enzyme (BglI) was used to differentiate serotypes 4 and 10. Table 3 shows the different restriction enzyme digestion patterns exhibited, denoted by profiles A to J.
TABLE 3.
Results of restriction enzyme analysis, denoted by profiles A to J, and normalized HRM-curve analysis, indicating genotype and mean C% of sample replicates for 12 FAdV reference serotypes
| Serotype | Restriction enzyme profile | HEX-S
|
Hexon A/B
|
Hex L1
|
|||
|---|---|---|---|---|---|---|---|
| Genotypea | Mean C% ± SEb | Genotype | Mean C% ± SE | Genotype | Mean C% ± SE | ||
| FAdV-1 | A | 1 | 99.6 ± 0.16 | 1 | 99.5 ± 0.21 | 1 | 99.6 ± 0.18 |
| FAdV-2 | B | 2 | 99.5 ± 0.21 | 2 | 99.1 ± 0.21 | 2 | 99.2 ± 0.35 |
| FAdV-3 | C | 3 | 98.9 ± 0.47 | 3 | 99.1 ± 0.34 | 3 | 99.8 ± 0.07 |
| FAdV-4 | D | 4 | 99.5 ± 0.20c | 4 | 99.1 ± 0.30 | 4 | 99.8 ± 0.10 |
| 10 | 99.3d | ||||||
| FAdV-5 | NAe | 5 | 99.2 ± 0.36 | NAe | NAe | 5 | 99.8 ± 0.06 |
| FAdV-6 | E | 6 | 99.2 ± 0.02c | 6 | 99.7 ± 0.09 | 6 | 99.9 ± 0.03 |
| 11 | 98.9d | ||||||
| FAdV-7 | F | 7 | 99.0 ± 0.44 | 7 | 99.5 ± 0.18 | 7 | 99.1 ± 0.42 |
| FAdV-8a | G | 8a | 98.5 ± 0.72 | 8a | 99.5 ± 0.18 | 8a | 99.5 ± 0.23 |
| FAdV-8b | H | 8b | 99.8 ± 0.08 | 8b | 98.8 ± 0.56 | 8b | 99.1 ± 0.19 |
| FAdV-9 | I | 9 | 99.5 ± 0.36 | 9 | 99.4 ± 0.24 | 9 | 99.3 ± 0.33 |
| FAdV-10 | J | 10 | 99.4 ± 0.70c | 10 | 99.6 ± 0.20 | 10 | 99.8 ± 0.05 |
| 4 | 99.4d | ||||||
| FAdV-11 | B | 11 | 99.6 ± 0.16 | 11 | 99.8 ± 0.07 | 11 | 99.5 ± 0.24 |
Genotypes were defined by selecting a representative sample from each serotype.
The mean C% of three replicate samples per serotype is shown (unless otherwise indicated), plus or minus the SE.
The mean C% of two replicate samples per serotype is shown, plus or minus the SE.
One replicate sample of the serotype was assigned to a representative genotype of a different serotype; therefore, the mean C% and SE were not calculated.
Primers Hexon A and Hexon B did not produce a PCR amplicon for FAdV-5; therefore, restriction enzyme analysis and HRM-curve analysis were not performed on these samples. NA, not available.
FAdV serotypes 2 and 11 showed identical restriction patterns (as indicated by profile B in Table 3), even when digested with a fourth enzyme, Tth111I (also known as AspI), as recommended (29). Digestion with the enzyme ScaI was not required to differentiate any strains.
Due to low-level or no amplification by the Hexon A/B primers, FAdV-5 was not included in the restriction enzyme analysis.
HRM-curve analysis of the Hex L1 PCR product differentiated all FAdV serotypes from each other.
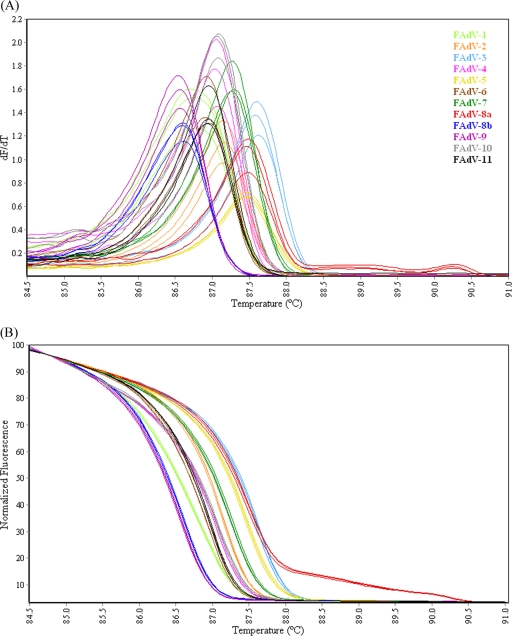
PCR products of the region of the hexon P1 gene amplified by the HEX-S primers (as defined in Table 2) from FAdV reference serotypes were subjected to HRM-curve analysis. In the conventional melt curve analysis, all serotypes generated one peak between 86°C and 88°C, with FAdV-8a generating two additional shoulder peaks between 88.5°C and 90.5°C (Fig. 3A). Examination of the FAdV-8a PCR product by agarose gel electrophoresis resulted in a single stained band (Fig. 2), indicating that the additional peaks were not due to other amplifications in the PCR. The melt curve profiles were consistently produced from PCR products generated on different days and from templates belonging to different DNA extractions. Some serotypes exhibited very similar melt curve profiles and peak temperatures, making it difficult to distinguish them from each other (Fig. 3A). These included FAdV-4 and -10, -8b and -9, and -6 and -11. Analysis of the normalized HRM curves (Fig. 3B) revealed that while some serotypes could easily be distinguished, for example FAdV-1 and -8a, the curve profiles of other serotypes were very similar and could therefore not be visually differentiated. When genotyping was applied to the normalized HRM curves (Table 3), a single replicate of some serotypes was auto-called as a different genotype at a C% between 98.9 and 99.4% (e.g., one replicate of FAdV-4 genotyped as FAdV-10 with a C% of 99.3). Therefore the normalized HRM-curve profiles of these serotypes could not be considered to represent distinctly different genotypes.
FIG. 3.
(A) Conventional melt curves of HEX-S PCR products of the P1 region of the hexon gene from 12 FAdV reference serotypes. (B) Normalized HRM curves of HEX-S PCR products of the P1 region of the hexon gene from 12 FAdV reference serotypes.
HRM-curve analysis of the approximately 897-bp Hexon A/B PCR product generated conventional melt curves containing one to four peaks between 81°C and 91°C for each FAdV serotype (Fig. 4A), except for serotype 5, which did not produce a PCR amplicon (Fig. 2). The curve profiles were visually distinct, making each serotype easy to distinguish from others. In the normalized HRM graph (Fig. 4B), each serotype (excluding FAdV-5) displayed a distinct curve profile; when genotyping was applied, replicates of each serotype were correctly genotyped with a C% equal to or greater than 98.8% (Table 3).
FIG. 4.
(A) Conventional melt curves of Hexon PCR products of the P1 and L1 regions of the hexon gene from 12 FAdV reference serotypes. (B) Normalized HRM curves of Hexon PCR products of the P1 and L1 regions of the hexon gene from 12 FAdV reference serotypes.
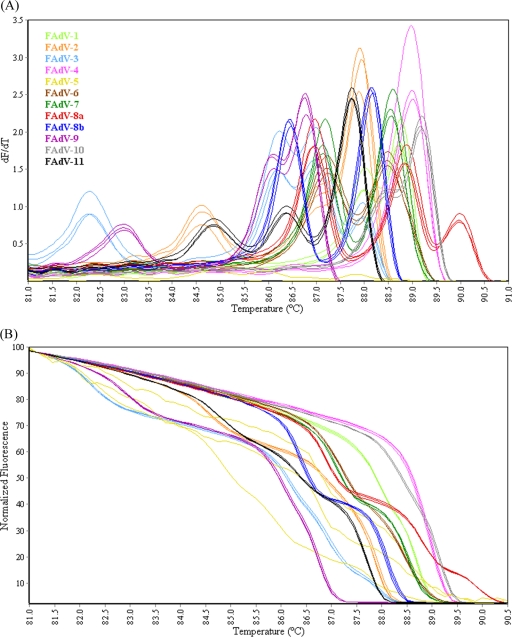
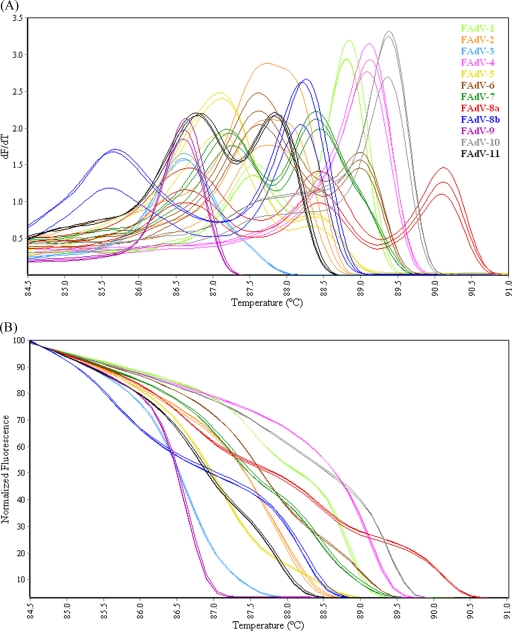
The conventional melt curve profiles generated by HRM-curve analysis of the Hex L1 amplicons from FAdV serotypes (Fig. 5A) contained one, two, or three peaks between 85°C and 90.5°C. Based on visual analysis, each strain produced a distinctly different melt curve profile. FAdV-3 and FAdV-9 contained a single peak at 86.6°C; however, visual interpretation differentiated the two serotypes, with the FAdV-3 curve shape distinctly wider than that of FAdV-9. The normalized HRM graph (Fig. 5B) showed that each serotype produced a distinct curve profile, with the genotyping C% for replicates of each serotype ranging from 99.1% to 99.9% (Table 3).
FIG. 5.
(A) Conventional melt curves of Hex L1 PCR products of the P1 and L1 regions of the hexon gene from 12 FAdV reference serotypes. (B) Normalized HRM curves of Hex L1 PCR products of the P1 and L1 regions of the hexon gene from 12 FAdV reference serotypes.
HRM-curve analysis for all three regions of the hexon gene showed slight shifts in melting temperatures between PCRs run on different days using templates from different DNA extractions; however, the conventional melt curve shapes were unchanged. For amplicons from the Hexon A/B and Hex L1 PCRs, defining representative genotypes in the normalized HRM graph consistently resulted in replicates of each serotype being correctly genotyped, with C% well above the 95% threshold.
Differences in melting curve profiles were not necessarily related to nucleotide sequence identities but mainly to distribution of GC contents throughout the amplicons.
In order to determine if differences in melting profiles correlated with sequence identities, the homologies of the nucleotide sequence of the Hex L1 PCR products from each FAdV serotype were compared (Table 4). Sequence identities ranged from 28% to 99%. More than 94% identity was found between FAdV-3 and -9 and FAdV-4 and -10, with an identity of 99% found between serotypes 2 and 11. The lowest identity (28%) was between FAdV-10 and -11.
TABLE 4.
Sequence identity matrix and percentages of GC content corresponding to the region of nt 301 to 890 of the FAdV-1 CELO genes for hexon proteinsa
| FAdV serotype (strain) | % Identity with FAdV serotype (strain):
|
GC content (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (CELO) | 2 (SR48) | 3 (SR49) | 4 (KR5) | 5 (340) | 6 (CR119) | 7 (YR36) | 8a (TR59) | 8b (764) | 9 (A02) | 10 (C2B) | 11 (UF71) | ||
| 1 (CELO) | ID | 58.0 | |||||||||||
| 2 (SR48) | 40.7 | ID | 54.0 | ||||||||||
| 3 (SR49) | 44.9 | 69.6 | ID | 51.9 | |||||||||
| 4 (KR5) | 56.2 | 29.2 | 29.8 | ID | 58.8 | ||||||||
| 5 (340) | 42.5 | 45.7 | 48.6 | 40.6 | ID | 55.2 | |||||||
| 6 (CR119) | 43.7 | 63.1 | 59.9 | 37.3 | 55.1 | ID | 56.4 | ||||||
| 7 (YR36) | 44.0 | 58.6 | 56.9 | 38.6 | 54.6 | 84.5 | ID | 56.2 | |||||
| 8a (TR59) | 40.5 | 58.9 | 53.4 | 38.1 | 53.6 | 79.7 | 80.4 | ID | 57.2 | ||||
| 8b (764) | 38.3 | 56.3 | 56.2 | 31.7 | 48.0 | 77.6 | 87.3 | 77.5 | ID | 54.0 | |||
| 9 (A02) | 43.2 | 70.6 | 94.9 | 30.3 | 47.3 | 58.8 | 56.9 | 51.3 | 55.6 | ID | 51.4 | ||
| 10 (C2B) | 56.0 | 29.5 | 30.5 | 95.3 | 40.5 | 37.3 | 38.2 | 38.8 | 32.0 | 31.9 | ID | 58.7 | |
| 11 (UF71) | 41.2 | 99.0 | 70.3 | 28.6 | 46.0 | 62.4 | 57.3 | 58.6 | 55.9 | 70.8 | 28.0 | ID | 53.6 |
The FAdV-1 CELO genes for hexon proteins are assigned GenBank accession number Z67970, nt 1380 to 4208 (1) (Hex L1 PRC product). The matrix was calculated using the DNAdist program in BioManager, with the Kimura distance method, and GC contents were calculated using BioEdit Sequence Alignment Editor. ID, identical.
Serotypes that shared more than 90% sequence identity but were greatly different in HRM-curve analyses were compared further using pair-wise alignment, and their GC contents were assessed. The pair-wise alignment of FAdV-3 and -9 (data not shown) revealed 29 nt differences between the sequences and a 3-nt insertion at nt position 121 of the FAdV-9 amplicon. Of the differences, nine were inverted (i.e., A::T or C::G) and did not influence the GC/AT content. The overall GC contents of the FAdV-3 and -9 amplicons were 51.9% and 51.4%, respectively (Table 4). However, the distribution levels of the GC content varied along the length of the amplicon. By examining the sequences in three equal sections, the middle section of FAdV-3 (from nt 196 to 393 of the 590-bp Hex L1 amplicon) was found to have a GC content of 54.0% compared with the equivalent region of FAdV-9, which had a GC content of 51.3%.
Similarly, the pair-wise alignment of FAdV-4 and -10 showed 23 nt differences, of which 7 were inverted. The overall GC compositions of the amplicons were almost identical, with 58.8% for FAdV-4 and 58.7% for FAdV-10 (Table 4). FAdV-4 and -10 shared similar GC contents of 58.6% and 58.0%, respectively, for the region spanning nt 1 to 193 of the 578-bp amplicon. However FAdV-10 had a higher GC content than FAdV-4 in the middle region (nt 194 to 386) with 62.2% and 60.6%, respectively. In the last third of the amplicon (nt 387 to 578), FAdV-10 had a lower GC content than FAdV-4, with 55.7% and 57.3%, respectively.
The serotypes sharing the highest identity, FAdV-2 and -11, were shown to differ by only six nucleotides, with no inverted differences. The overall GC contents of the 593-bp amplicons differed slightly, with 54.0% for FAdV-2 and 53.6% for FAdV-11 (Table 4). The GC contents of the region spanning nt 1 to 300 of the amplicons were similar with 55.0% and 55.3% for FAdV-2 and -11, respectively. The following 150 bp (nt 301 to 450), however, contained four of the six nucleotide differences, all of which contained a G or C nucleotide for FAdV-2 and an A or T nt for FAdV-11; therefore, FAdV-2 had a higher GC content (54.7%) than FAdV-11 (52.0%) in this region. The remaining 143-bp region of the amplicon (nt 451 to 593) contained similar low GC contents for FAdV-2 (51.1%) and FAdV-11 (51.8%).
Hex-L1 PCR HRM-curve analysis genotyped a vaccine and a field strain according to their serotype.
In order to assess the capacity of the newly developed Hex-L1 PCR HRM-curve analysis for genotyping additional FAdV isolates, two viruses of known serotype (8b) were subjected to HRM-curve analyses. The viruses used were the live attenuated Australian FAdV vaccine and FAdV field isolate FI-W888-05. Analysis of the conventional melt curve revealed that these viruses generated identical patterns, two peaks at 86.2 and 88.5°C, which were most similar to reference strain FAdV-8b. Examination of the normalized HRM curves also revealed that the curves generated from the Australian vaccine and field strains had resemblance to FAdV-8b. When genotyping was applied in the presence of all reference strains, replicates of both the vaccine and field isolate were genotyped as FAdV-8b, with mean confidence percentages of 68.9% ± 1.89 SE and 57.6% ± 0.81 SE, respectively.
DISCUSSION
This study provides a direct comparison between restriction enzyme analysis, nucleotide sequencing, and HRM-curve analysis for the detection of differences in the P1 and L1 regions of the FAdV hexon gene for the identification and differentiation of each of the 12 FAdV serotypes.
In previous studies the use of techniques such as microtiter virus neutralization tests (11, 16, 33, 38-40), PCR alone (49, 50, 59) or in combination with either sequencing (2, 14, 15, 30, 32, 48) or restriction fragment polymorphism (RFLP) (29, 44, 54), and a combination of these techniques (7, 9, 10, 20, 37, 41, 42, 57) have been used to identify FAdV serotypes. Although each of these techniques is readily available to most laboratories, they have limitations, including the lengthy processes involved, need for extensive interpretation, and potential for indeterminate results, for example, the cross-neutralization of serum samples (7, 10, 16, 27).
The results of this study indicate that restriction enzyme analysis, or RFLP, failed to classify FAdV serotypes 2 and 11 (Table 3) using enzymes previously shown to distinguish 12 FAdV serotypes (29, 30). Therefore, an additional restriction enzyme would be required to differentiate these two serotypes by this method. It should be noted that the representative strain of each serotype used in this study did not conform with those used previously (29). The ambiguity of the nomenclature of FAdV serotypes under two different systems, European Union and U.S. (Table 1), has made the correlation of data relating to serotypes sourced from different geographical locations difficult. The International Committee on Taxonomy of Viruses recently implemented a single international nomenclature system for FAdVs (19). The results of the current study suggest that RFLP may not be able to distinguish serotypes from the same species and/or strains from the same serotype, which may be of importance in epidemiological studies of disease outbreaks. In addition, RFLP requires a high quantity of PCR product, which was shown not to be feasible in this study for FAdV-5 amplification by primers Hexon A and B (Fig. 2).
In this study HRM-curve analysis was applied to amplicons of three different regions of the FAdV hexon gene, using conventional PCR reagents, oligonucleotide primers, and Syto 9. Amplicons of the 191-bp HEX-S region from FAdV serotypes could not easily be discriminated from each other by analysis of their HRM curves (Table 3). This may have resulted from small amplicons containing limited sequence variations.
The amplicons of the approximately 897-bp region generated by primers Hexon A and B produced conventional melt curves with two or more major and minor peaks, with or without shoulders (Fig. 4A), with most serotypes distinguishable from others using the normalized HRM curves (Table 3). However, as these primers yielded low-level or no amplification of FAdV-5, further analysis was not feasible for this serotype. Therefore, HRM-curve analysis of the Hexon A/B PCR product was deemed not suitable for genotyping of FAdV reference serotypes.
In contrast, HRM-curve analysis of Hex L1 PCR products proved a highly sensitive and specific means of genotyping all 12 FAdV serotypes. The complexity of the conventional melt curves of the approximately 590-bp amplicons increased the power of the normalized HRM-curve analysis to differentiate serotypes. All serotypes generated one or more major peaks and were visually distinct from each other in conventional melt curve profiles (Fig. 5A), and replicates of each serotype were correctly genotyped using the normalized HRM curves with C% of more than 99% (Table 3). Even where two serotypes exhibited a single major peak with an identical melting temperature (FAdV-3 and -9 in Fig. 5A), in the normalized HRM graph these serotypes could be clearly differentiated (Fig. 5B) and replicates were correctly genotyped with more than 99% confidence (Table 3). The capacity of the newly developed Hex L1 PCR HRM-curve analysis to genotype nonreference strains was also demonstrated by correctly genotyping two further strains (an Australian FAdV vaccine and a field strain) according to their known serotypes in the presence of all 12 reference strains. Even though the design of the Hex L1 primers was based on the GenBank sequences of 27 different FAdV strains (48), further proof of the universality of these primers would become evident if they were utilized in further research involving a number of other FAdV strains.
In this study, FAdV-2 and -11 sequences shared 99.0% identity in the region amplified by the Hex L1 primers (Table 4) but nevertheless produced different conventional melt curve profiles (Fig. 5A), and replicates in the normalized HRM graph were correctly genotyped with close to 100% confidence (Table 3). This demonstrates the sensitivity of the technique developed here and its capacity for detecting very small sequence variations.
The melting profile of a PCR product is dependent upon length, sequence, GC content, and heterozygosity (17, 51, 53). The GC composition of a DNA fragment is not necessarily a good predictor of the melting temperature; however, the actual DNA melting analysis can detect finer-scale differences in sequences, such as localized domains of different GC composition (31). It has been suggested that melting domains are usually 50- to 300-bp in length; therefore, amplicons up to 800-bp may have multiple melting domains (58). Thus, it can be assumed that multiple and/or shoulder peaks in the conventional melt curve profile of amplicons of the L1 region of the hexon gene of some FAdV serotypes, as seen in Fig. 5A, may be due to the presence of multiple melting domains differing in GC content. For example, the melting temperatures of the three peaks for FAdV-8a, 86.7°C, 88.5°C, and 90.2°C (Fig. 5A), may be attributable to the different GC compositions of each third of the amplified sequence (51.5%, 57.3%, and 62.8%, respectively). Interestingly, the peak with the lowest melting temperature closely matches that of FAdV-3 and -9, which have overall GC contents (51.8% and 51.4%, respectively) close to that of the FAdV-8a region (51.5%) from nt 399 to 596. Even where sequence differences were small and localized, as seen between FAdV-2 and -11, the conventional melt curve profiles (Fig. 5A) were distinctly different as a result of contrasting GC compositions within a confined region of each amplicon. This supports the notion that the composition and distribution of GC nucleotides in an amplicon directly influences the shape and complexity of the conventional melt curve, and in turn, the power of the normalized HRM curve as a tool for genotyping.
The melt curve profiles generated in this study were highly reproducible and in agreement with those predicted by the web-based DNA melting simulation program POLAND (Heinrich-Heine University in Dusseldorf, Germany, Institute of Biophysics [http://www.biophys.uni-duesseldorf.de/local/POLAND/poland.html]) (55) (results not shown), a program that has been used to aid in the design of primers to amplify a region of DNA that will result in certain melting-curve characteristics (8, 23). This also confirmed that the complex melting profile of PCR amplicons was solely the result of DNA dissociation (47). The variability between different strains within each FAdV serotype could potentially be assessed using sequences from the GenBank database in the POLAND program. The Hex L1 nucleotide sequences from some FAdVs with known serotype identities were obtained from GenBank (AF508957, FAdV-8 strain 58; DQ323986, FAdV-9 isolate Stanford; U26221, FAdV serotype 10; and AF339925, FAdV-12 strain 380) and analyzed using POLAND as part of this study. The resultant melt curve shapes were highly similar, with regard to the number, height, and temperature of the peaks, to the respective reference strains used in this study (results not shown). Combining this technique with HRM-curve analysis could therefore provide serotype identification and potentially strain identification in laboratories that can perform HRM-curve analysis but do not have access to the reference strains of FAdV serotypes. However, as genotyping is performed using the normalized HRM data and further evaluated by the resultant confidence percentages in the RotorGene software, the use of the POLAND program to assess and compare sequences from GenBank is limited to visual comparisons of conventional melt curve shapes and is therefore not definitive.
This study presents for the first time a robust genotyping technique based on the closed-tube method of real-time PCR and HRM-curve analysis for FAdVs. The method is an accurate, rapid, and cost-effective alternative to existing serotype identification methods, and it can be used as a model for developing genotyping techniques for other pathogens. In addition, during this study, the basis for differences in melt curve profiles resulting from reference FAdVs were elucidated.
Acknowledgments
This work was supported by the Chicken Meat Program from the Rural Industries Research and Development Corporation (RIRDC) of the Australian government.
We thank Intervet International (Boxmeer, Netherlands) for providing the FAdV reference serotypes and Tom Grimes for his assistance and input.
Footnotes
Published ahead of print on 26 November 2008.
REFERENCES
- 1.Akopian, T. A., K. K. Doronin, V. A. Karpov, and B. S. Naroditsky. 1996. Sequence of the avian adenovirus FAV 1 (CELO) DNA encoding the hexon-associated protein pVI and hexon. Arch. Virol. 1411759-1765. [DOI] [PubMed] [Google Scholar]
- 2.Alvarado, I. R., P. Villegas, J. El-Attrache, E. Jensen, G. Rosales, F. Perozo, and L. B. Purvis. 2007. Genetic characterization, pathogenicity, and protection studies with an avian adenovirus isolate associated with inclusion body hepatitis. Avian Dis. 5127-32. [DOI] [PubMed] [Google Scholar]
- 3.Balamurugan, V., and J. M. Kataria. 2004. The hydropericardium syndrome in poultry-a current scenario. Vet. Res. Commun. 28127-148. [DOI] [PubMed] [Google Scholar]
- 4.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2008. GenBank. Nucleic Acids Res. 36D25-D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, J. C., C. L. Huang, C. C. Lin, C. C. Chen, Y. C. Chang, S. S. Chang, and C. P. Tseng. 2006. Rapid detection and identification of clinically important bacteria by high-resolution melting analysis after broad-range ribosomal RNA real-time PCR. Clin. Chem. 521997-2004. [DOI] [PubMed] [Google Scholar]
- 6.Chou, L. S., E. Lyon, and C. T. Wittwer. 2005. A comparison of high-resolution melting analysis with denaturing high-performance liquid chromatography for mutation scanning: cystic fibrosis transmembrane conductance regulator gene as a model. Am. J. Clin. Pathol. 124330-338. [DOI] [PubMed] [Google Scholar]
- 7.Dahiya, S., R. N. Srivastava, M. Hess, and B. R. Gulati. 2002. Fowl adenovirus serotype 4 associated with outbreaks of infectious hydropericardium in Haryana, India. Avian Dis. 46230-233. [DOI] [PubMed] [Google Scholar]
- 8.Do, H., M. Krypuy, P. L. Mitchell, S. B. Fox, and A. Dobrovic. 2008. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer 8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Attrache, J., and P. Villegas. 2001. Genomic identification and characterization of avian adenoviruses associated with inclusion body hepatitis. Avian Dis. 45780-787. [PubMed] [Google Scholar]
- 10.Erny, K., J. Pallister, and M. Sheppard. 1995. Immunological and molecular comparison of fowl adenovirus serotypes 4 and 10. Arch. Virol. 140491-501. [DOI] [PubMed] [Google Scholar]
- 11.Ganesh, K., R. Raghavan, R. N. Gowda, M. L. Satyanarayana, and V. V. Suryanarayana. 2002. Purification and characterization of the aetiological agent of hydropericardium hepatitis syndrome from infected liver tissues of broiler chickens. Trop. Anim. Health Prod. 347-17. [DOI] [PubMed] [Google Scholar]
- 12.Ganesh, K., V. V. Suryanarayana, and R. Raghavan. 2002. Detection of fowl adenovirus associated with hydropericardium hepatitis syndrome by a polymerase chain reaction. Vet. Res. Commun. 2673-80. [DOI] [PubMed] [Google Scholar]
- 13.Giglio, S., P. T. Monis, and C. P. Saint. 2005. Legionella confirmation using real-time PCR and SYTO9 is an alternative to current methodology. Appl. Environ. Microbiol. 718944-8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomis, S., A. R. Goodhope, A. D. Ojkic, and P. Willson. 2006. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. 50550-555. [DOI] [PubMed] [Google Scholar]
- 15.Grgic, H., C. Philippe, D. Ojkic, and E. Nagy. 2006. Study of vertical transmission of fowl adenoviruses. Can. J. Vet. Res. 70230-233. [PMC free article] [PubMed] [Google Scholar]
- 16.Grimes, T. M., and D. J. King. 1977. Serotyping avian adenoviruses by a microneutralization procedure. Am. J. Vet. Res. 38317-321. [PubMed] [Google Scholar]
- 17.Herrmann, M. G., J. D. Durtschi, L. K. Bromley, C. T. Wittwer, and K. V. Voelkerding. 2006. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin. Chem. 52494-503. [DOI] [PubMed] [Google Scholar]
- 18.Hess, M. 2000. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. 29195-206. [DOI] [PubMed] [Google Scholar]
- 19.ICTVdB Management. 2006. 00.001. Adenoviridae. In C. Büchen-Osmond (ed.), ICTVdB-the universal virus database, version 3. Columbia University, New York, NY.
- 20.Jadhao, S. J., J. N. Deepak, J. M. Kataria, R. S. Kataria, A. K. Tiwari, R. Somvanshi, P. Sangamithra, and K. C. Verma. 2003. Characterisation of fowl adenoviruses from chickens affected with infectious hydropericardium during 1994-1998 in India. Indian J. Exp. Biol. 41321-327. [PubMed] [Google Scholar]
- 21.Jeffery, N., R. B. Gasser, P. A. Steer, and A. H. Noormohammadi. 2007. Classification of Mycoplasma synoviae strains using single-strand conformation polymorphism and high-resolution melting-curve analysis of the vlhA gene single-copy region. Microbiology 1532679-2688. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, P., D. Ojkic, T. Tuboly, P. Huber, and E. Nagy. 1999. Application of the polymerase chain reaction to detect fowl adenoviruses. Can. J. Vet. Res. 63124-128. [PMC free article] [PubMed] [Google Scholar]
- 23.Krypuy, M., A. A. Ahmed, D. Etemadmoghadam, S. J. Hyland, A. DeFazio, S. B. Fox, J. D. Brenton, D. D. Bowtell, and A. Dobrovic. 2007. High resolution melting for mutation scanning of TP53 exons 5-8. BMC Cancer 7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krypuy, M., G. M. Newnham, D. M. Thomas, M. Conron, and A. Dobrovic. 2006. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer 6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liew, M., R. Pryor, R. Palais, C. Meadows, M. Erali, E. Lyon, and C. Wittwer. 2004. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 501156-1164. [DOI] [PubMed] [Google Scholar]
- 26.Lin, J. H., C. P. Tseng, Y. J. Chen, C. Y. Lin, S. S. Chang, H. S. Wu, and J. C. Cheng. 2008. Rapid differentiation of influenza A virus subtypes and genetic screening for virus variants by high-resolution melting analysis. J. Clin. Microbiol. 461090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazaheri, A., C. Prusas, M. Vob, and M. Hess. 1998. Some strains of serotype 4 fowl adenoviruses cause inclusion body hepatitis and hydropericardium. Avian Pathol. 27269. [DOI] [PubMed] [Google Scholar]
- 28.McFerran, J. B., and B. M. Adair. 2003. Adenovirus infections: group I adenovirus infections, p. 213-227. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), 11th ed. Diseases of Poultry, Iowa State University Press, Ames.
- 29.Meulemans, G., M. Boschmans, T. P. van den Berg, and M. Decaesstecker. 2001. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenovirus. Avian Pathol. 30655-660. [DOI] [PubMed] [Google Scholar]
- 30.Meulemans, G., B. Couvreur, M. Decaesstecker, M. Boschmans, and T. P. van den Berg. 2004. Phylogenetic analysis of fowl adenoviruses. Avian Pathol. 33164-170. [DOI] [PubMed] [Google Scholar]
- 31.Monis, P. T., S. Giglio, and C. P. Saint. 2005. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal. Biochem. 34024-34. [DOI] [PubMed] [Google Scholar]
- 32.Moscoso, H., J. J. Bruzual, H. Sellers, and C. L. Hofacre. 2007. FTA liver impressions as DNA template for detecting and genotyping fowl adenovirus. Avian Dis. 51118-121. [DOI] [PubMed] [Google Scholar]
- 33.Muroga, N., S. Taharaguchi, H. Ohta, K. Yamazaki, and K. Takase. 2006. Pathogenicity of fowl adenovirus isolated from gizzard erosions to immuno-suppressed chickens. J. Vet. Med. Sci. 68289-291. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, K., H. Tanaka, M. Mase, T. Imada, and M. Yamada. 2002. Pancreatic necrosis and ventricular erosion in adenovirus-associated hydropericardium syndrome of broilers. Vet. Pathol. 39403-406. [DOI] [PubMed] [Google Scholar]
- 35.Norrby, E. 1969. The relationship between the soluble antigens and the virion of adenovirus type 3. IV. Immunological complexity of soluble components. Virology. 37565-576. [DOI] [PubMed] [Google Scholar]
- 36.Odell, I. D., J. L. Cloud, M. Seipp, and C. T. Wittwer. 2005. Rapid species identification within the Mycobacterium chelonae-abscessus group by high-resolution melting analysis of hsp65 PCR products. Am. J. Clin. Pathol. 12396-101. [DOI] [PubMed] [Google Scholar]
- 37.Ojkic, D., E. Martin, J. Swinton, J. P. Vaillancourt, M. Boulianne, and S. Gomis. 2008. Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol. 3795-100. [DOI] [PubMed] [Google Scholar]
- 38.Okuda, Y., M. Ono, I. Shibata, and S. Sato. 2004. Pathogenicity of serotype 8 fowl adenovirus isolated from gizzard erosions of slaughtered broiler chickens. J. Vet. Med. Sci. 661561-1566. [DOI] [PubMed] [Google Scholar]
- 39.Okuda, Y., M. Ono, S. Yazawa, Y. Imai, I. Shibata, and S. Sato. 2001. Pathogenicity of serotype 1 fowl adenovirus in commercial broiler chickens. Avian Dis. 45819-827. [PubMed] [Google Scholar]
- 40.Okuda, Y., M. Ono, S. Yazawa, I. Shibata, and S. Sato. 2001. Experimental infection of specific-pathogen-free chickens with serotype-1 fowl adenovirus isolated from a broiler chicken with gizzard erosions. Avian Dis. 4519-25. [PubMed] [Google Scholar]
- 41.Ono, M., Y. Okuda, I. Shibata, S. Sato, and K. Okada. 2004. Pathogenicity by parenteral injection of fowl adenovirus isolated from gizzard erosion and resistance to reinfection in adenoviral gizzard erosion in chickens. Vet. Pathol. 41483-489. [DOI] [PubMed] [Google Scholar]
- 42.Ono, M., Y. Okuda, I. Shibata, S. Sato, and K. Okada. 2007. Reproduction of adenoviral gizzard erosion by the horizontal transmission of fowl adenovirus serotype 1. J. Vet. Med. Sci. 691005-1008. [DOI] [PubMed] [Google Scholar]
- 43.Ono, M., Y. Okuda, S. Yazawa, Y. Imai, I. Shibata, S. Sato, and K. Okada. 2003. Adenoviral gizzard erosion in commercial broiler chickens. Vet. Pathol. 40294-303. [DOI] [PubMed] [Google Scholar]
- 44.Ono, M., Y. Okuda, S. Yazawa, I. Shibata, N. Tanimura, K. Kimura, M. Haritani, M. Mase, and S. Sato. 2001. Epizootic outbreaks of gizzard erosion associated with adenovirus infection in chickens. Avian Dis. 45268-275. [PubMed] [Google Scholar]
- 45.Philippe, C., H. Grgic, D. Ojkic, and E. Nagy. 2007. Serologic monitoring of a broiler breeder flock previously affected by inclusion body hepatitis and testing of the progeny for vertical transmission of fowl adenoviruses. Can. J. Vet. Res. 7198-102. [PMC free article] [PubMed] [Google Scholar]
- 46.Price, E. P., H. Smith, F. Huygens, and P. M. Giffard. 2007. High-resolution DNA melt curve analysis of the clustered, regularly interspaced short-palindromic-repeat locus of Campylobacter jejuni. Appl. Environ. Microbiol. 733431-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasmussen, J. P., C. P. Saint, and P. T. Monis. 2007. Use of DNA melting simulation software for in silico diagnostic assay design: targeting regions with complex melting curves and confirmation by real-time PCR using intercalating dyes. BMC Bioinformatics 8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raue, R., H. Gerlach, and H. Müller. 2005. Phylogenetic analysis of the hexon loop 1 region of an adenovirus from psittacine birds supports the existence of a new psittacine adenovirus (PsAdV). Arch. Virol. 1501933-1943. [DOI] [PubMed] [Google Scholar]
- 49.Raue, R., H. M. Hafez, and M. Hess. 2002. A fiber gene-based polymerase chain reaction for specific detection of pigeon adenovirus. Avian Pathol. 3195-99. [DOI] [PubMed] [Google Scholar]
- 50.Raue, R., and M. Hess. 1998. Hexon based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus. J. Virol. Methods 73211-217. [DOI] [PubMed] [Google Scholar]
- 51.Reed, G. H., J. O. Kent, and C. T. Wittwer. 2007. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 8597-608. [DOI] [PubMed] [Google Scholar]
- 52.Reed, G. H., and C. T. Wittwer. 2004. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 501748-1754. [DOI] [PubMed] [Google Scholar]
- 53.Robinson, B. S., P. T. Monis, and P. J. Dobson. 2006. Rapid, sensitive, and discriminating identification of Naegleria spp. by real-time PCR and melting-curve analysis. Appl. Environ. Microbiol. 725857-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, A., M. S. Oberoi, G. S. Grewal, H. M. Hafez, and M. Hess. 2002. The use of PCR combined with restriction enzyme analysis to characterize fowl adenovirus field isolates from northern India. Vet. Res. Commun. 26577-585. [DOI] [PubMed] [Google Scholar]
- 55.Steger, G. 1994. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 222760-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taharaguchi, S., H. Ito, H. Ohta, and K. Takase. 2006. Characterization of monoclonal antibodies against fowl adenovirus serotype 1 (FAV1) isolated from gizzard erosion. Avian Dis. 50331-335. [DOI] [PubMed] [Google Scholar]
- 57.Toro, H., C. Prusas, R. Raue, L. Cerda, C. Geisse, C. Gonzalez, and M. Hess. 1999. Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile. Avian Dis. 43262-270. [PubMed] [Google Scholar]
- 58.Wittwer, C. T., G. H. Reed, C. N. Gundry, J. G. Vandersteen, and R. J. Pryor. 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 49853-860. [DOI] [PubMed] [Google Scholar]
- 59.Xie, Z., A. A. Fadl, T. Girshick, and M. I. Khan. 1999. Detection of avian adenovirus by polymerase chain reaction. Avian Dis. 4398-105. [PubMed] [Google Scholar]
- 60.Zhou, L., L. Wang, R. Palais, R. Pryor, and C. T. Wittwer. 2005. High-resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin. Chem. 511770-1777. [DOI] [PubMed] [Google Scholar]