Abstract
We evaluated the capacity of the Mycoplasma agalactiae p40 gene as a diagnostic marker for contagious agalactia in sheep by quantitative real-time PCR. The p40 gene encodes an immunodominant adhesin that plays a key role in cytoadhesion of M. agalactiae. The assay was 100% specific, with an analytical sensitivity of 1 genome equivalent (GE), a quantification that is highly linear (R2 > 0.992) and efficient (PCR efficiency, >0.992) over a 6-log dynamic range, down to 10 GE. We evaluated the capacity of the assay to detect Mycoplasma agalactiae in 797 milk samples (373 raw sheep milk samples from refrigerated tanks of different farms and 424 milk samples from individual sheep of a flock positive for M. agalactiae). In parallel, we also tested the samples by using microbiological isolation coupled with microscopy identification and by a PCR method recommended by the World Organization for Animal Health. While our assay was able to detect 57 (15.28%) positive samples of the 373 milk samples from different farms, identification by microbiological isolation coupled with microscopy detected only 36 (9.65%) samples, and the conventional PCR detected 31 (8.31%) samples. These findings showed that our assay based on the p40 gene is more specific and sensitive for the detection of M. agalactiae in actual natural samples and, thus, can be a promising alternative tool for diagnosis and epidemiological studies of M. agalactiae infection.
Mycoplasma agalactiae is the principal etiological agent of contagious agalactia in small ruminants. In male and female sheep, this syndrome is produced mostly by Mycoplasma agalactiae subsp. agalactiae, but other mycoplasmas are also implicated (M. capricolum subsp. capricolum, M. mycoides subsp. capri, M. mycoides subsp. mycoides LC, and M. putrefaciens). Contagious agalactia is a serious infectious process characterized by three major signs: mastitis, arthritis, and keratoconjunctivitis (17). It causes the reduction and even the suppression of milk production and occasionally causes abortion and death in up to 70% of young animals, resulting in serious economic loss (16). Nowadays, this syndrome affects animals in most countries that have a high production of sheep and goats. It is found across several continents, including mainly North America, western Asia, North Africa, and Europe, and is endemic in most Mediterranean countries (3). Consequently, this disease represents a very important animal health problem and is included in the list of notifiable diseases listed by the OIE (World Organization for Animal Health) because of its economic impact and widespread distribution. In fact, governments are implementing a compulsory diagnosis scheme for assessing the presence M. agalactiae in milk tanks or farms by PCR (7).
The principal sources of M. agalactiae infection are the ingestion of contaminated feed, water, or milk and the urine, feces, and nasal or ocular liquids of infected animals. Ewes can be infected through the udder, and lambs can be infected by the consumption of colostrum. Thus, it is recommended that newborn animals be removed from the dam immediately after birth and fed only by using pasteurized colostrum. The period of incubation of this syndrome varies between 7 and 56 days. Most cases of infection occur in the summer, during birth and peak lactation periods. Several mycoplasmas can be isolated from milk and blood for a short period of time during the infectious process (5).
The routine analysis method used by diagnostic laboratories for identifying M. agalactiae in clinical samples or milk tanks is based on microbiological culture in selective enrichment media including antibiotics in moist anaerobic chambers (5% CO2) for 5 to 7 days. However, this method represents a time-consuming and complicated process, as mycoplasmas grow very slowly. Mycoplasmas form typical “fried-egg”-shaped small colonies that, although visible under the microscope, are very difficult to enumerate. Moreover, these analyses need to be complemented with species identification by biochemical or immunofluorescence tests, making this an expensive procedure. Alternative DNA amplification-based methods for directly detecting this animal pathogen in milk or in other fluids have been devised to overcome this problem. There are several methods that identify M. agalactiae by PCR. Some of these methods are based on amplification of the 16S rRNA gene (2, 4, 12, 13). However, 16S rRNA gene sequences in M. agalactiae and M. bovis share 99.8% similarity, affecting the specificity of methods based on the amplification of this gene. Other diagnostic strategies are based on the amplification of unknown sequences (6, 27, 28) or specific genes, like uvrC (26) or the mb-mp81 gene encoding the membrane protein P81 (9). However, the last two methods are based on the PCR-restriction fragment length polymorphism technique, which is more laborious and time-consuming and is not quantitative.
The absolute quantification of contaminant microbiota by real-time quantitative PCR (Q-PCR) is becoming increasingly common for diagnostic purposes in clinical and food microbiology (24). This technique provides a higher specificity and analytical sensitivity and reduces the risk of cross-contamination; at the same time, this technique is faster than conventional PCR and is totally adapted to automation. Lorusso and coworkers (15) have developed a real-time PCR detection method using molecular beacon chemistry. This method targets a 117-bp region of the M. agalactiae mb-mp81 gene encoding the membrane lipoprotein P81, which is also present in the M. bovis genome. There is a general consensus that the addition of an internal amplification control (IAC) in each reaction mixture, to assess the potential effects of PCR inhibitors or the malfunction of thermocyclers, must be mandatory. An IAC is a chimeric nontarget DNA fragment that is present in every reaction mixture and can be coamplified with the target sequence (11). However, the method described by Lorusso and coworkers incorporates an alternate template (canine parvovirus type 2 [CPV-2] DNA), which is run in separate PCR wells (15).
It is clearly convenient for diagnostic laboratories to have a large battery of alternative analytical methods for the quantitative detection of M. agalactiae, due to the economic impact and public health relevance of this pathogen. An alternative method can be adopted by diagnostics laboratories only if the method has been evaluated and validated with samples collected from animals. Thus, in this study, we report the design and assessment of a real-time PCR assay for the quantitative detection of M. agalactiae and its evaluation and in-house validation in the analysis of milk samples. This method includes an IAC that is coamplified in the same reaction mixture to assess the PCR performance of each reaction, thus ensuring the diagnostic efficiency of this method. Finally, we evaluated the performance of the assay with natural samples, i.e., sheep milk samples. We tested 373 natural, raw sheep milk samples from refrigerated tanks from different sheep farms and 424 milk samples from individual sheep from a flock found positive for this pathogen.
Design and optimization of the M. agalactiae-specific duplex p40-IAC Q-PCR assay.
The assay targets an M. agalactiae species-specific region of the p40 gene (GenBank accession no. AJ344229) encoding an immunodominant adhesin that plays a key role in cytoadhesion of M. agalactiae (8). Bacterial adhesion is a key mechanism of mycoplasma virulence, and the protein P40 displays a strong and persistent signal in response to antibodies; thus, the M. agalactiae p40 gene has been proposed as a good candidate for the development of future diagnostic assays (8). The p40 gene sequence displayed regions that were 100% specific to M. agalactiae by nucleotide sequence comparison using BLAST-N version 2.2.14 (National Centre for Biotechnology Information; www.ncbi.nlm.nih.gov). We selected M. agalactiae-specific regions that were identical among various isolates of M. agalactiae by aligning all p40 sequences available in public databases, using a CLUSTALW multiple-alignment tool (EMBL, European Bioinformatics Institute; www.ebi.ac.uk); this was done to ensure no variation in sequences occurred as a result of different strains. Subsequently, we designed the PCR primers MAP40127F, MAP40235R, and MAP40160P by using Primer Express version 3.0 software (Applied Biosystems, Foster City, CA) (Table 1). These PCR oligonucleotides amplify a 109-bp fragment from the coding sequence of the M. agalactiae p40 gene corresponding to positions 127 to 235. The specificity of the oligonucleotides was confirmed in silico using BLAST-N version 2.2.14 software, as none of the selected oligonucleotides or amplicon sequences showed any similarity outside of the M. agalactiae-specific region.
TABLE 1.
Oligonucleotides used in this study
| Target | Primer | Type | Sequence |
|---|---|---|---|
| M. agalactiae p40 genea | MAP40127F | Forward | 5′-TCATTTACAGCAGTGCCTTTATTAG-3′ |
| MAP40235R | Reverse | 5′-CACCTAATGCTTGTTTTTCAACC-3′ | |
| MAP40160P | TaqMan probe | 5′-FAM-TGTGATGATAAGAACGAAAATTCACAAA-BHQ1-3′b | |
| IAC chimeric DNA | MAIACP | TaqMan MGB probe | 5′-VIC-CCATACACATAGGTCAGG-MGB-NFQ-3′ |
| L. monocytogenes prfA gene | MAIACF | Forward | 5′-TCATTTACAGCAGTGCCTTTATTAGGGCTCTATTTGCGGTC-3′ |
| MAIACR | Reverse | 5′-CACCTAATGCTTGTTTTTCAACCTCTTGATGCCATCAGGA-3′ |
The amplicon size is 109 bp, and the melting temperature of the amplicon is 73.0°C ± 0.6°C.
BHQ1, Black Hole Quencher 1.
One of the major barriers to the systematic introduction of Q-PCR-based methods for routine microbiology diagnostics is the common occurrence of false-negative results. These results occur due to the presence of PCR inhibitors in clinical and environmental samples. This is particularly relevant in analyses performed for detecting M. agalactiae cells in raw sheep milk. Raw sheep milk contains various substances that may cause the bacterial counts to be underestimated or the amplification reaction to be totally abolished (1). Our assay was designed to monitor this problem, using an IAC. This IAC consisted of a nontarget nucleic acid fragment that is coamplified with the target sequence. The same primers are used for the test reaction, including a probe labeled differently, for the simultaneous assessment of PCR performance. The IAC was constructed as previously described (22) and consists of a 118-bp chimeric DNA molecule containing a portion (nucleotide positions 421 to 491) of the Listeria monocytogenes-positive regulatory factor A (prfA) gene; the gene is flanked by the M. agalactiae-specific p40 gene sequences targeted by the MAP40127F and MAP40235R primers. If a negative signal is obtained for the target p40 signal, the absence of a positive IAC signal indicates that amplification has failed.
The M. agalactiae-specific PCR oligonucleotides were optimized for Q-PCR assays in which 1 ng of DNA from M. agalactiae strain ATCC 35890 was used as a template. The assays were carried out in a reaction mixture volume of 20 μl containing 1× Universal PCR Master Mix (Applied Biosystems). We used a model 7500 real-time PCR system platform (Applied Biosystems) with the following program: 2 min at 50°C, 10 min at 95°C, and 50 cycles of 15 s at 95°C and 1 min at 60°C. Optimal conditions (300 nM for primers MAP40127F and MAP40235R and 200 nM for the 6-carboxyfluorescein [FAM]-labeled MAP40160P probe) were the minimum primer and probe concentrations giving the lowest cycle threshold (CT) value and the highest fluorescence intensity for a normalized reporter value. The optimal IAC probe concentration was determined experimentally by carrying out Q-PCRs in the presence of 10,000 IAC molecules, no M. agalactiae DNA, 200 nM FAM-labeled p40 probe and with various amounts (from 25 to 250 nM) of VIC-labeled IAC probe (Table 1). The minimum probe concentration that did not result in an increase in CT values was 100 nM. As excess IAC may inhibit the target-specific reaction, Q-PCRs were also carried out in the presence of various amounts of IAC (10,000, 1,000, 100, and 10 molecules per reaction) and various amounts of M. agalactiae DNA (equivalent to approximately 1 × 104, 1 × 103, 100, 10, and 1 genome equivalents, in which there are 0.88 Mb per genome, according to Sirand-Pugnet and coworkers [25], and one genome equivalent corresponds to 0.972 fg of M. agalactiae DNA). The maximum amount of IAC with no inhibitory effect on the p40-specific FAM signal was 100 copies of chimeric DNA.
Selectivity of the p40-IAC assay.
The capacity of the p40-IAC Q-PCR assay to discriminate between target and nontarget bacteria was assessed using 1 ng of genomic DNA from various sources, as follows: 75 Mycoplasma species, including 26 M. agalactiae isolates (24 of them were sheep milk isolates) and 49 other Mycoplasma species; and 110 strains from 39 non-Mycoplasma species, including various genera frequently found in milk and dairy products (see Table S1 and S2 in the supplemental material). Bacterial DNA was isolated using a QIAamp DNA minikit (Qiagen, Hilden, Germany). The p40-IAC Q-PCR assay was inclusive for M. agalactiae and 100% exclusive for nontarget bacteria: all M. agalactiae strains tested gave a positive p40 signal, whereas none of the 138 nontarget bacteria produced a positive signal. All reactions generated a positive IAC (VIC) signal, excluding the possibility that the absence of a p40 (FAM) signal observed for non-M. agalactiae isolates was due to PCR failure.
Analytical sensitivity and quantification range of the p40-IAC Q-PCR assay.
The achievement of low detection and quantification limits is a critical aspect in the design of molecular diagnostic methods for microbial pathogens in clinical and environmental samples. The detection and quantification limits of the PCR assays were determined by using genomic DNA isolated from M. agalactiae strain ATCC 35890. Three independent PCRs were performed, with a range of DNA concentrations equivalent to approximately 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 10, and 1 target molecules. Table 2 shows the mean CT values for a total of 9 PCR replicates (30 replicates for 10 and 1 genome equivalents) from three independent experiments. Positive amplification of all PCR replicates of each DNA dilution was achieved if 10 or more target molecules were present, and 1 target molecule could be detected in at least 16 of the 30 replicates (Table 2). PCR efficiency is related to the slope of the linear regression curve calculated over a 6-log range (down to 10 target molecules) and was similar to that of the theoretical optimum of −3.32 (p40, −3.3216, efficiency = 100%; p40-IAC, −3.3416, efficiency = 99.2%). The linearity of calibration curves represented by the regression coefficient (R2) showed values close to 1 (p40, 0.9921; p40-IAC, 0.9931), indicating that the assay was highly linear. These results are similar to those for other Q-PCR methods used for other bacterial and eukaryotic organisms (10, 19-21, 23).
TABLE 2.
Detection and quantification limits of Q-PCR assay with genomic DNA from M. agalactiae strain ATCC 35890a
| Approximate genome equivalents/reaction |
p40 Q-PCR
|
p40-IAC Q-PCR
|
||
|---|---|---|---|---|
| Signal ratio | CT value ± SEM | Signal ratio | CT value ± SEM | |
| 1 × 107 | 9/9 | 17.13 ± 0.09 | 9/9 | 17.16 ± 0.06 |
| 1 × 106 | 9/9 | 20.77 ± 0.06 | 9/9 | 20.81 ± 0.07 |
| 1 × 105 | 9/9 | 24.49 ± 0.11 | 9/9 | 24.31 ± 0.04 |
| 1 × 104 | 9/9 | 27.98 ± 0.13 | 9/9 | 28.08 ± 0.04 |
| 1 × 103 | 9/9 | 31.40 ± 0.12 | 9/9 | 31.42 ± 0.12 |
| 1 × 102 | 9/9 | 33.31 ± 0.16 | 9/9 | 33.42 ± 0.17 |
| 1 × 101 | 30/30 | 36.15 ± 0.20 | 30/30 | 36.88 ± 0.40 |
| 1 | 16/30 | 38.45 ± 0.40 | 15/30 | 38.56 ± 0.20 |
Nontemplate controls for both Q-PCR systems were negative (CT values of 50 for all the replicates). The overall slopes of the regression curve were −3.3216 and −3.3406 for the TaqMan p40 and p40-IAC Q-PCR TaqMan systems, respectively, indicating PCR efficiency values of 1.000 and 0.992, respectively, and the regression coefficients (R2) were 0.9921and 0.9931, respectively. Signal ratio indicates the number of positive results out of 9 reactions, 30 reactions for those containing 10 and 1 genome equivalents per reaction. CT value ± standard error of the mean (SEM) indicates the mean CT value at which fluorescence intensity was equal to a fixed threshold. Experimental results were statistically significant (P < 0.05), taking into account the unavoidable error associated with serial dilutions.
Quantitative detection of M. agalactiae in raw sheep milk.
Samples that possess a strong capacity to inhibit PCR amplification pose the risk of false-negative results (1). A complex matrix like raw sheep milk directly transferred from the farm milk tank was selected to illustrate the implementation of the p40-IAC PCR method and to evaluate the capacity for quantitative detection of M. agalactiae. In three independently replicated experiments, 1-ml portions of phosphate-buffered saline containing approximately 2.5 × 107, 2.5 × 106, 2.5 × 105, 2.5 × 104, 2.5 × 103, 250, 25, or 3 M. agalactiae cells were added to 50-ml centrifuge tubes, each containing 25 ml of raw sheep milk. DNA was purified from milk by the procedure described by López-Enríquez and coworkers (14), with the only modification being homogenization for 10 min in a homogenizer (Pulsifier) prior to DNA extraction. We consistently detected as few as 250 M. agalactiae cells per 25 ml of milk (Table 3). This detection limit corresponds to approximately 1 genomic unit per reaction, similar to the limits obtained if genomic DNA isolated from M. agalactiae was used as a PCR template. We then evaluated the use of this method for quantifying M. agalactiae cells in sheep milk. We constructed regression curves with the CT values obtained with artificially contaminated milk samples and the corresponding numbers of M. agalactiae cells used for the inoculation. The overall correlation coefficient (R2 = 0.9989) demonstrated that the Q-PCR assay was linear over a range of 5 logs, down to 2,500 cells/25 ml of milk (Table 3). The PCR efficiencies obtained (E = 0.996) indicated that the performance of the Q-PCR assay was excellent. Moreover, these values were similar to those obtained with the analysis of purified M. agalactiae genomic DNA.
TABLE 3.
Quantitative detection of M. agalactiae in raw sheep milka
| Approximate M. agalactiae cells/25 ml | Approximate M. agalactiae genome equivalents/reactionb | Signal ratioc | CT value ± SEMd | Relative accuracye |
|---|---|---|---|---|
| 2.5 × 107 | 1 × 105 | 9/9 | 24.20 ± 0.02 | 87.17 |
| 2.5 × 106 | 1 × 104 | 9/9 | 27.12 ± 0.06 | 115.14 |
| 2.5 × 105 | 1 × 103 | 9/9 | 30.50 ± 0.05 | 110.98 |
| 2.5 × 104 | 1 × 102 | 9/9 | 34.09 ± 0.06 | 91.81 |
| 2.5 × 103 | 10 | 9/9 | 37.33 ± 0.23 | 97.43 |
| 2.5 × 102 | 1 | 9/9 | 38.63 ± 0.32 | NA |
| 25 | 0.1 | 0/9 | NAf | NA |
| 3 | 0.01 | 0/9 | NA | NA |
| 0f | 0 | 0/9 | NA | NA |
Raw milk was collected directly from a sheep farm. Data show results from three independent experiments with three PCR replicates used in each. The overall efficiency was 0.996, and the linearity (R2) was 0.9989. NA, not applicable.
Estimated numbers of M. agalactiae genome equivalents in each PCR run, assuming 100% DNA extraction efficiency, are shown. Each reaction mixture contained 2 μl of a DNA preparation taken from the 1:10 dilution of the initial 50 μl extracted from 25 ml of raw sheep milk.
Number of positive results out of nine reactions.
Mean CT value ± standard error of the mean (SEM) at which fluorescence intensity was equal to a fixed threshold. The experimental results were statistically significant (P < 0.05) taking into account unavoidable errors associated with serial dilutions.
Degree of correspondence between results obtained with the standard plating technique (M. agalactiae CFU/25 ml) and those obtained with the p40 Q-PCR method (M. agalactiae genome equivalents/25 ml).
Noncontaminated milk.
Validation of the p40-IAC Q-PCR assay for the detection of M. agalactiae in naturally contaminated sheep milk samples.
The p40-IAC Q-PCR assay was performed with 373 natural raw sheep milk samples from refrigerated tanks from different farms and with 424 milk samples from individual sheep from a flock that tested positive for this pathogen. In every experiment, we included negative controls to verify the accuracy of the results. In parallel, we also tested the samples by using two procedures recommended by the World Organization for Animal Health: microbiological isolation coupled with microscopy observation and a PCR for milk samples developed by Tola and coworkers (27). If it was not possible to process samples within 24 h, they were frozen at −20°C, after it was determined whether the mycoplasma cells could survive for at least 3 weeks at −20°C without the numbers of cells decreasing. All the samples were cultivated in liquid and on solid selective media before testing, as follows: mycoplasma broth base with mycoplasma supplement G (Oxoid), a medium specific for this genus; and MA1A, a medium specific for M. agalactiae (Mycoplasma Experience, Surrey, United Kingdom). Aliquots of samples were cultivated in Mycoplasma broth with Mycoplasma supplement G at 37°C in moist anaerobic chambers in a 5% CO2 atmosphere for 3 days. The samples were then vortexed, and 1 ml was transferred into a 1.5-ml Eppendorf tube and centrifuged at 13,000 × g for 15 min. The supernatant was discarded, and the cell pellet was resuspended in 100 μl of 10 mM Tris-HCl for further incubation at 95°C for 20 min. Finally, the tube was centrifuged at maximum speed for 10 min, and the supernatant was transferred into a new tube. We used 2 μl of the lysate for PCR. The DNA was then tested by using the conventional PCR (27) and the p40-IAC Q-PCR methods. In parallel, enriched samples from mycoplasma broth were also streaked on agar mycoplasma medium containing supplement G and on MA1A agar medium and cultivated at 37°C in moist anaerobic chambers in a 5% CO2 atmosphere for 6 days. The resulting, typical “fried-egg” Mycoplasma colonies were examined by light microscopy at 10× magnification. In order to confirm that the isolated colonies were M. agalactiae, we tested the enrichment broth cultures for the presence of strains of Mycoplasma species involved in contagious agalactia in small ruminants (i.e., M. mycoides cluster and M. putrefaciens), using PCR methods previously published (18, 29).
If the sample tested PCR positive, we then assayed DNA extracted directly from milk without culture (Fig. 1). In 373 tank milk samples, a total of 57 samples tested positive by the p40-IAC Q-PCR method after culture (Table 4). Of 57 samples, 31 were identified as Mycoplasma positive by microscopy observation (none was PCR positive for the M. mycoides cluster or M. putrefaciens, thus confirming that they were only M. agalactiae); these samples also tested positive with both of the PCR methods after culture. In addition, five samples, which could not be confirmed by microscopy observation, tested positive with both of the PCR methods. An additional 21 samples tested positive only by our real-time PCR detection system. Thus, the positivity of M. agalactiae in 373 milk samples found by the p40-IAC Q-PCR method was 15.28% (29 samples had a CT value of <25, 19 samples had CT values of 25 to 34, and 9 had CT values of ≥35) (Table 4). However, the positivity of M. agalactiae using microscopy observation or conventional PCR (27) only was 9.65% or 8.31%, respectively. These results indicated that these two procedures (i.e., microscopy observation and the conventional PCR) (27) underestimate the actual presence of M. agalactiae in natural samples by almost twofold (1.58 and 1.85 times, respectively) in comparison with our assay. Among the samples extracted directly from milk, 44 samples tested positive by the p40-IAC Q-PCR assay developed for this study (all of the samples were also positive after culture) (Table 4). However, the 13 samples not detected prior to incubation exhibited low positive signals when analyzed after culture (CT values above 32), suggesting very low M. agalactiae numbers in milk after the enrichment and, thus, probably below the limit of detection in the moment of direct bacterial DNA extraction prior to the enrichment. By contrast, conventional PCR (27) detected only 20 positive samples prior to the 6-day cultivation (Table 4). Thus, our p40-IAC Q-PCR assay was more sensitive, particularly if DNA was extracted directly from milk. Consequently, p40-IAC Q-PCR was capable of detecting realistic levels of M. agalactiae contamination in milk samples but was faster and the levels were quantifiable.
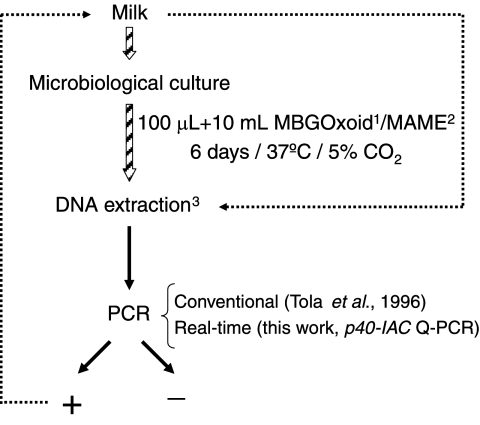
FIG. 1.
Raw sheep milk sample analysis scheme. Milk samples (797) were received, and 100 μl was cultured in 10 ml of specific microbiological media (1, MBG Mycoplasma broth base with supplement G from OXOID; and 2, MA1A medium from Mycoplasma Experience) at 37°C in 5% CO2 chambers for 6 days. DNA was then extracted using thermolysis (3) and analyzed by PCR using the p40-IAC Q-PCR assay and the method described by Tola and coworkers (27). If the PCR tested positive, DNA was directly purified from 25 ml of milk to demonstrate direct detection by avoiding cultivation and to improve the analytical performance.
TABLE 4.
Detection of M. agalactiae in naturally contaminated milk samplesa
| Time | No. of positives samples (%) usingb:
|
||
|---|---|---|---|
| p40-IAC Q-PCR | Conventional PCRc | Microscopyd | |
| Prior to 6-day culture | 44 (11.80) | 20 (5.36) | No observation |
| After 6-day culture | 57 (15.28) | 36 (9.65) | 31 (8.31) |
Natural raw sheep milk samples (373) from refrigerated tanks from different farms were investigated using three different methods (p40-IAC Q-PCR, conventional PCR [26], and microscopy identification) prior to and after a 6-day culture at 37°C in moist anaerobic chambers in a 5% CO2 atmosphere.
Data show numbers of positive results obtained per each method for 373 samples tested and percentages of prevalence of M. agalactiae, using the three different methods.
Conventional PCR method as published previously by Tola and coworkers (26).
Examination by light microscopy at 10× magnification. Samples were considered positive when typical “fried-egg” colonies were observed.
We also collected 424 milk samples from individual sheep from a flock for which the corresponding tank tested positive. Only four samples were positive: two exhibited a CT value below 25, one sample had CT values in the range of 26 to 34, and one sample's CT value was above 34. Moreover, all IAC results showed positive amplification in all samples that were negative by the p40-IAC Q-PCR method, indicating that the PCRs did not fail. The absence of inhibition in any of the reactions also suggests that the preamplification procedure used is effective in these types of samples. This result provides new insights into the contagious agalactia process in sheep flocks, since, whereas the refrigerated tank of the sheep farm tested positive, resulting in the belief that the flock was affected by contagious agalactia, only 4 out of 424 animals (0.94%) tested positive.
In conclusion, we describe a reliable and sensitive real-time PCR-based method for quantifying M. agalactiae cells in raw sheep milk taken directly from the refrigeration tanks from farms or from animals. Our method incorporates an IAC to assist in the interpretation of the results obtained. This method provides a significant quantification capacity, as defined by its wide dynamic quantification range (at least 6 orders of magnitude), linearity (R2 > 0.9921), PCR efficiency (E > 0.992), and quantification limit (down to 10 M. agalactiae genome equivalents). If the assay is combined with simple detergent and enzymatic treatments of samples before centrifugation and nucleic acid extraction, it provides a highly effective quantitative analysis for large volumes of milk. It is able to detect 250 cells in 25 ml of raw sheep milk, with an excellent accuracy relative to that of the reference microbiological method. We evaluated and validated the Q-PCR assay's capacity to detect M. agalactiae in 797 raw sheep milk samples and showed an average positivity for M. agalactiae of 15.28%. This method therefore provides a promising alternative to procedures currently recommended by the World Organization for Animal Health for the rapid, sensitive, and quantitative detection of M. agalactiae in milk, which may be easily adopted by animal health laboratories.
Supplementary Material
Acknowledgments
This study was supported by research grant RTA2008-00073 and the European Union's Marie-Curie mobility program (contract MERG-CT-2007-209050).
K.O. received a National Scholarship of the Slovak Republic Government and a FEMS research fellowship. L.L.-E. received a Ph.D. studentship from the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), and M.H. holds a contract from the INIA, cofunded by the European Social Fund (ESF).
Footnotes
Published ahead of print on 19 November 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Al-Soud, W. A., and P. Radstrom. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashiruddin, J. B., J. Frey, M. H. Königsson, K. E. Johansson, H. Hotzel, R. Diller, P. de Santis, A. Botelho, R. D. Ayling, R. A. Nicholas, F. Thiaucourt, and K. Sachse. 2005. Evaluation of PCR systems for the identification and differentiation of Mycoplasma agalactiae and Mycoplasma bovis: a collaborative trial. Vet. J. 169268-275. [DOI] [PubMed] [Google Scholar]
- 3.Bergonier, D., X. Bertholet, and F. Poumarat. 1997. Contagious agalactia of small ruminants: current knowledge concerning epidemiology, diagnosis and control. Rev. Off. Int. Epizoot. 16848-873. [DOI] [PubMed] [Google Scholar]
- 4.Chávez González, Y. R., C. Ros Bascuñana, G. Bölske, J. G. Mattsson, C. Fernández Molina, and K. E. Johansson. 1995. In vitro amplification of the 16S rRNA genes from Mycoplasma bovis and Mycoplasma agalactiae by PCR. Vet. Microbiol. 47183-190. [DOI] [PubMed] [Google Scholar]
- 5.DaMassa, A. J., P. S. Wakenell, and D. L. Brooks. 1992. Mycoplasmas of goats and sheep. J. Vet. Diagn. Invest. 4101-113. [DOI] [PubMed] [Google Scholar]
- 6.Dedieu, L., V. Mady, and P. C. Lefevre. 1995. Development of two PCR assays for the identification of mycoplasmas causing contagious agalactia. FEMS Microbiol. Lett. 129243-249. [DOI] [PubMed] [Google Scholar]
- 7.Director General de Producción Agropecuaria. 2006. Resolución de 24 de mayo de 2007, por la que desarrollan los programas sanitarios a realizar por las Agrupaciones de Defensa Sanitaria Ganaderas, definidos en la Orden AYG/1131/2006, de 30 de junio. Comunidad Autónoma de Castilla y León, Spain.
- 8.Fleury, B., D. Bergonier, X. Berthelot, E. Peterhans, J. Frey, and E. M. Vilei. 2002. Characterization of P40, a cytadhesin of Mycoplasma agalactiae. Infect. Immun. 705612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foddai, A., G. Idini, M. Fusco, N. Rosa, C. de la Fe, S. Zinellu, L. Corona, and S. Tola. 2005. Rapid differential diagnosis of Mycoplasma agalactiae and Mycoplasma bovis based on a multiplex-PCR and a PCR-RFLP. Mol. Cell. Probes 19207-212. [DOI] [PubMed] [Google Scholar]
- 10.Hernández, M., M. Pla, T. Esteve, S. Prat, P. Puigdomènech, and A. Ferrando. 2003. A specific real-time quantitative PCR detection system for event MON810 in maize YieldGard based on the 3′-transgene integration sequence. Transgenic Res. 12179-189. [DOI] [PubMed] [Google Scholar]
- 11.Hoorfar, J., N. Cook, B. Malorny, M. Wagner, D. De Medici, A. Abdulmawjood, and P. Fach. 2004. Diagnostic PCR: making internal amplification control mandatory. J. Appl. Microbiol. 96221-222. [DOI] [PubMed] [Google Scholar]
- 12.Johansson, K. E., M. U. Heldtander, and B. Pettersson. 1998. Characterization of mycoplasmas by PCR and sequence analysis with universal 16S rDNA primers. Methods Mol. Biol. 104145-165. [DOI] [PubMed] [Google Scholar]
- 13.Königsson, M. H., G. Bölske, and K. E. Johansson. 2002. Intraspecific variation in the 16S rRNA gene sequences of Mycoplasma agalactiae and Mycoplasma bovis strains. Vet. Microbiol. 85209-220. [DOI] [PubMed] [Google Scholar]
- 14.López-Enríquez, L., D. Rodríguez-Lázaro, and M. Hernández. 2007. Quantitative detection of Clostridium tyrobutyricum in milk by real-time PCR. Appl. Environ. Microbiol. 733747-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorusso, A., N. Decaro, G. Greco, M. Corrente, A. Fasanella, and D. A. Buonavoglia. 2007. A real-time PCR assay for detection and quantification of Mycoplasma agalactiae DNA. J. Appl. Microbiol. 103918-923. [DOI] [PubMed] [Google Scholar]
- 16.Madanat, D., Z. Zendulková, and I. L. Pospí. 2001. Contagious agalactia of sheep and goats: a review. Acta Vet. Brno 70403-412. [Google Scholar]
- 17.Nicolas, R. A. J. 2004. Contagious agalactia, p. 607-614. In OIE manual of diagnostic tests and vaccines for terrestrial animals (mammals, birds and bees), vol. 2. Paris, France. [Google Scholar]
- 18.Peyraud, A., S. Woubit, J. B. Poveda, C. De la Fe, P. Mercier, and F. Thiaucourt. 2003. A specific PCR for the detection of Mycoplasma putrefaciens, one of the agents of the contagious agalactia syndrome of goats. Mol. Cell. Probes 17289-294. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez-Lázaro, D., M. Hernández, T. Esteve, J. Hoorfar, and M. Pla. 2003. A rapid and direct real time PCR-based method for identification of Salmonella spp. J. Microbiol. Methods 54381-390. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Lázaro, D., M. Hernández, M. Scortti, T. Esteve, J. A. Vázquez-Boland, and M. Pla. 2004. Quantitative detection of Listeria monocytogenes and Listeria innocua by real-time PCR: assessment of hly, iap, and lin02483 targets and AmpliFluor technology. Appl. Environ. Microbiol. 701366-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Lázaro, D., M. D'Agostino, A. Herrewegh, M. Pla, N. Cook, and J. Ikonomopoulos. 2005. Real-time PCR-based methods for quantitative detection of Mycobacterium avium subsp. paratuberculosis in water and milk. Int. J. Food Microbiol. 10193-104. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Lázaro, D., M. Pla, M. Scortti, H. J. Monzó, and J. A. Vázquez-Boland. 2005. A novel real-time PCR for Listeria monocytogenes that monitors analytical performance via an internal amplification control. Appl. Environ. Microbiol. 719008-9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Lázaro, D., D. A. Lewis, A. A. Ocampo-Sosa, U. Fogarty, L. Makrai, J. Navas, M. Scortti, M. Hernández, and J. A. Vázquez-Boland. 2006. An internally controlled real-time PCR method for quantitative species-specific detection and vapA genotyping of Rhodococcus equi. Appl. Environ. Microbiol. 724256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Lázaro, D., B. Lombard, H. V. Smith, A. Rzezutka, M. D'Agostino, R. Helmuth, A. Schroeter, B. Malorny, A. Miko, B. Guerra, J. Davison, A. Kobilinsky, M. Hernández, Y. Bertheau, and N. Cook. 2007. Trends in analytical methodology in food safety and quality: monitoring microorganisms and genetically modified organisms. Trends Food Sci. Technol. 18306-319. [Google Scholar]
- 25.Sirand-Pugnet, P., C. Lartigue, M. Marenda, D. Jacob, A. Barre, V. Barbe, C. Schenowitz, S. Mangenot, A. Couloux, B. Segurens, A. de Daruvar, A. Blanchard., and C. Citti. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 3E75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramaniam, S., D. Bergonier, F. Poumarat, S. Capaul, Y. Schlatter, J. Nicolet, and J. Frey. 1998. Species identification of Mycoplasma bovis and Mycoplasma agalactiae based on the uvrC genes by PCR. Mol. Cell. Probes 12161-169. [DOI] [PubMed] [Google Scholar]
- 27.Tola, S., G. Idini, D. Manunta, G. Galleri, A. Angioi, A. M. Rocchigiani, and G. Leori. 1996. Rapid and specific detection of Mycoplasma agalactiae by polymerase chain reaction. Vet. Microbiol. 5177-84. [DOI] [PubMed] [Google Scholar]
- 28.Tola, S., A. Angioi, A. M. Rocchigiani, G. Idini, D. Manunta, G. Galleri, and G. Leori. 1997. Detection of Mycoplasma agalactiae in sheep milk samples by polymerase chain reaction. Vet. Microbiol. 5417-22. [DOI] [PubMed] [Google Scholar]
- 29.Woubit, S., L. Manso-Silván, S. Lorenzon, P. Gaurivau, F. Poumarat, M. P. Pellet, V. P. Singh, and F. Thiaucourt. 2007. A PCR for the detection of mycoplasmas belonging to the Mycoplasma mycoides cluster: application to the diagnosis of contagious agalactia. Mol. Cell. Probes 21391-399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.