Abstract
We report the first diagnostic test for the identification of Staphylococcus pseudintermedius involving a simple PCR-restriction fragment length polymorphism approach. The method allows discrimination of S. pseudintermedius from the closely related members of the Staphylococcus intermedius group and other important staphylococcal pathogens of humans and dogs.
Until recently, Staphylococcus intermedius was considered responsible for most cases of canine pyoderma, a major skin disease of dogs (8). However, using a multilocus sequencing approach, independent research groups have demonstrated that isolates phenotypically identified as Staphylococcus intermedius consist of three distinct species, including S. intermedius, Staphylococcus pseudintermedius, and Staphylococcus delphini, which together represent the S. intermedius group (SIG) (1a, 4). Importantly, it was discovered that S. pseudintermedius, not S. intermedius, is the common canine pyoderma pathogen and that S. delphini, isolated from a variety of different animals, may be more clinically important than was previously thought (1a, 4). The recently identified S. pseudintermedius (5) is occasionally isolated from serious human infections, and the emergence and spread of methicillin-resistant S. pseudintermedius strains are major veterinary and public health issues (1a, 3, 4, 7, 11-13). Sasaki et al. could biochemically differentiate S. intermedius from the other SIG species but was unable to identify phenotypic markers to discriminate S. pseudintermedius, the most clinically relevant species, from S. delphini (12), and DNA sequencing is currently required to identify S. pseudintermedius (1a, 12). In order to address the need for a method of discriminating clinical isolates of S. pseudintermedius, we have developed a rapid, simple, and robust PCR-restriction fragment length polymorphism (RFLP) approach which has been validated independently in laboratories in separate countries.
Our previous population genetic study of SIG isolates examined sequence diversity at five gene loci among 104 isolates (1a). In the current study, sequence analysis of one of the loci, pta, which encodes the enzyme phosphoacetyltransferase, revealed the presence of an MboI restriction site in all S. pseudintermedius isolates, which was absent in all S. intermedius and S. delphini isolates examined. Based on this discovery we have designed a simple, robust, and inexpensive PCR-RFLP diagnostic test for the identification of S. pseudintermedius. Staphylococcal genomic DNA isolation was carried out as previously described (1a). PCR amplification of a 320-bp fragment of the pta gene was carried out in a 50-μl volume with a 0.2 μM concentration of each oligonucleotide primer (pta_f1, AAA GAC AAA CTT TCA GGT AA, and pta_r1, GCA TAA ACA AGC ATT GTA CCG), a 0.2 mM concentration of the deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.5 U Taq DNA polymerase, and 50 ng DNA template, in a 1× reaction buffer. Thermocycling conditions included a 95°C incubation for 2 min followed by 30 cycles of 95°C for 1 min, 53°C for 1 min, and 72°C for 1 min, with a final incubation of 72°C for 7 min. Twenty-five-microliter samples of the PCR mixtures were incubated with 5 U of MboI and 5 μl of 5× digestion buffer for 2 h, and the digestion products were resolved in 2% (wt/vol) agarose by electrophoresis. We investigated the efficacy of the technique by the analysis of well-characterized isolates of S. pseudintermedius, S. intermedius, and S. delphini that represented the breadth of diversity identified within the SIG and that included the type strains (1). In addition, representative isolates of Staphylococcus aureus, Staphylococcus schleiferi subsp. coagulans, and Staphylococcus schleiferi subsp. schleiferi, which are associated with human and canine infections, were included (Table 1). A pta PCR product of 320 bp was successfully amplified from all strains examined. S. pseudintermedius PCR products all contained a single MboI site, resulting in two restriction fragments of 213 bp and 107 bp, respectively (Fig. 1). In contrast, SIG species S. delphini and S. intermedius and the S. schleiferi strains did not contain an MboI restriction site (Fig. 1). S. aureus isolates contained a unique MboI site, resulting in restriction fragments of 156 bp and 164 bp that appeared as a single band after agarose electrophoresis; this band was readily distinguishable from the S. pseudintermedius restriction fragment profile (Fig. 1). Our data indicate that the PCR-RFLP assay is an effective approach to S. pseudintermedius identification, allowing discrimination from the other SIG species and important staphylococcal pathogens of dogs (6, 9). In order to test the reproducibility of the method, independent investigators applied the approach in a diagnostic laboratory in Italy to a panel of 112 coagulase-positive staphylococcal field isolates which had been isolated from dogs, cats, cows, horses, wild foxes, and a captive bear and identified as either SIG or S. aureus (2). Of the 86 isolates from dogs, 85 were identified as S. pseudintermedius by the PCR-RFLP approach, consistent with our previous findings that S. pseudintermedius is a common commensal of dogs and the major canine pyoderma pathogen. The remaining isolate resulted in a pta restriction profile which indicated an S. aureus identity. All but 1 of the 14 isolates from cats, all 3 isolates from wild foxes, and the single isolate from a captive bear were identified as S. pseudintermedius. The remaining isolate of feline origin had an S. aureus-specific restriction profile. A previous study reported the isolation of S. intermedius from different species of the canoidea, including bears and foxes (1). Our data suggest that S. pseudintermedius may be the most common SIG species associated with these animals. All five isolates from equid genital swabs did not contain a pta MboI site, consistent with our previous findings that S. delphini is the SIG species commonly identified among horses, and three isolates from equine respiratory tracts and a skin lesion were identified as S. aureus. Finally, of the seven isolates from cows, two were S. pseudintermedius and five were identified as S. aureus. To our knowledge, this represents the first reported identification of S. pseudintermedius in association with cows.
TABLE 1.
Staphylococcal isolates previously identified to the species level by a DNA sequencing approacha
| Strain | Species | Host origin | MboI restriction siteb |
|---|---|---|---|
| ED99 | S. pseudintermedius | Dog | + |
| AV8001 | S. pseudintermedius | Dog | + |
| 95072195 | S. pseudintermedius | Dog | + |
| 94062394 | S. pseudintermedius | Dog | + |
| N900260 | S. pseudintermedius | Human | + |
| HT20030686 | S. pseudintermedius | Dog | + |
| M72199 | S. pseudintermedius | Dog | + |
| 3279 (MRSP) | S. pseudintermedius | Dog | + |
| 3414 | S. pseudintermedius | Cat | + |
| 9075 | S. pseudintermedius | Dog | + |
| 690 | S. pseudintermedius | Cat | + |
| 8478 | S. pseudintermedius | Dog | + |
| HH1 | S. pseudintermedius | Dog | + |
| Can6 | S. pseudintermedius | Dog | + |
| Can10 | S. pseudintermedius | Human | + |
| BH47 | S. pseudintermedius | Dog | + |
| AV8024 | S. pseudintermedius | Dog | + |
| 8016 | S. pseudintermedius | Dog | + |
| LMG22219 | S. pseudintermedius | Cat | + |
| LMG22220 | S. pseudintermedius | Horse | + |
| ATCC 49171 | S. delphini | Dolphin | − |
| HT20030680 | S. delphini | Camel | − |
| 9106 | S. delphini | Horse | − |
| 8485 | S. delphini | Horse | − |
| AV8051 | S. delphini | Pigeon | − |
| HT20030677 | S. delphini | Camel | − |
| AV8047 | S. delphini | Pigeon | − |
| HT20030676 | S. delphini | Camel | − |
| HT20030674 | S. delphini | Camel | − |
| 8086 | S. delphini | Horse | − |
| HT20030679 | S. delphini | Camel | − |
| AV8061 | S. intermedius | Pigeon | − |
| NCTC11048 | S. intermedius | Pigeon | − |
| AV8063 | S. intermedius | Pigeon | − |
| AV8063 | S. intermedius | Pigeon | − |
| ATCC 43808 | S. schleiferi subsp. schleiferi | Human | − |
| CCUG37248 | S. schleiferi subsp. coagulans | Dog | − |
| Newman | S. aureus | Human | + |
| N315 | S. aureus | Human | + |
The isolates were previously identified by Bannoehr et al. (1). +, present; −, absent.
S. pseudintermedius pta MboI restriction fragments were 107 bp and 213 bp, and S. aureus pta MboI restriction fragments were 156 bp and 164 bp.
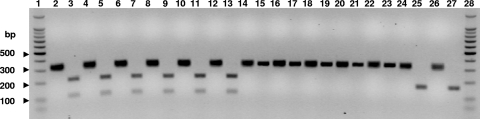
FIG. 1.
Agarose gel electrophoresis of MboI restriction digest pta PCR products. Lane 1, 100-bp ladder; lane 2, S. pseudintermedius ED99; lane 3, S. pseudintermedius ED99, MboI digested; lane 4, S. pseudintermedius HH1; lane 5, S. pseudintermedius HH1, MboI digested; lane 6, S. pseudintermedius Can6; lane 7, S. pseudintermedius Can6, MboI digested; lane 8, S. pseudintermedius Can10; lane 9, S. pseudintermedius Can10, MboI digested; lane 10, S. pseudintermedius LMG22219; lane 11, S. pseudintermedius LMG22219, MboI digested; lane 12, S. pseudintermedius 3414; lane 13, S. pseudintermedius 3414, MboI digested; lane 14, S. delphini ATCC 49171; lane 15, S. delphini ATCC 49171, MboI digested; lane 16, S. delphini 9106; lane 17, S. delphini 9106, MboI digested; lane 18, S. delphini HT20030680; lane 19, S. delphini HT20030680, MboI digested; lane 20, S. intermedius NCTC11048; lane 21, S. intermedius NCTC11048, MboI digested; lane 22, S. intermedius AV8061; lane 23, S. intermedius AV8061, MboI digested; lane 24, S. aureus Newman; lane 25, S. aureus Newman, MboI digested; lane 26, S. aureus N315; lane 27, S. aureus N315, MboI digested; lane 28, 100-bp ladder.
Until now, the lack of unique phenotypic markers for S. pseudintermedius in comparison to the other SIG members has precluded its identification without DNA sequencing. Importantly, due to the presence of common phenotypic markers, S. pseudintermedius is occasionally misidentified as S. aureus in human clinical diagnostic laboratories (10). The simple PCR-RFLP approach presented here represents the first reported diagnostic method which is effective for the identification of S. pseudintermedius and for its discrimination from other SIG species and several other important staphylococcal pathogens. As such, the method should prove extremely useful in routine veterinary diagnostic and clinical microbiology laboratories.
Acknowledgments
We thank Fabiola Feltrin, Angela Ianzano, and Cinzia Onorati for technical support.
Footnotes
Published ahead of print on 17 December 2008.
REFERENCES
- 1.Aarestrup, F. M. 2001. Comparative ribotyping of Staphylococcus intermedius isolated from members of the Canoidea gives possible evidence for host-specificity and co-evolution of bacteria and hosts. Int. J. Syst. Evol. Microbiol. 511343-1347. [DOI] [PubMed] [Google Scholar]
- 1a.Bannoehr, J., N. L. Ben Zakour, A. S. Waller, L. Guardabassi, K. L. Thoday, A. H. van den Broek, and J. R. Fitzgerald. 2007. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 1898685-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, F., M. F. Cochet, J. L. Pellerin, N. Ben Zakour, A. Lebon, A. Navarro, I. Proudy, Y. Le Loir, and M. Gautier. 2004. Development of a PCR test to differentiate between Staphylococcus aureus and Staphylococcus intermedius. J. Food Prot. 672302-2305. [DOI] [PubMed] [Google Scholar]
- 3.Descloux, S., A. Rossano, and V. Perreten. 2008. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius. J. Clin. Microbiol. 461818-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devriese, L. A., K. Hermans, M. Baele, and F. Haesebrouck. 2008. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet. Microbiol. 133206-207. [DOI] [PubMed] [Google Scholar]
- 5.Devriese, L. A., M. Vancanneyt, M. Baele, M. Vaneechoutte, E. De Graef, C. Snauwaert, I. Cleenwerck, P. Dawyndt, J. Swings, A. Decostere, and F. Haesebrouck. 2005. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 551569-1573. [DOI] [PubMed] [Google Scholar]
- 6.Frank, L. A., S. A. Kania, K. A. Hnilica, R. P. Wilkes, and D. A. Bemis. 2003. Isolation of Staphylococcus schleiferi from dogs with pyoderma. J. Am. Vet. Med. Assoc. 222451-454. [DOI] [PubMed] [Google Scholar]
- 7.Guardabassi, L., S. Schwarz, and D. H. Lloyd. 2004. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54321-332. [DOI] [PubMed] [Google Scholar]
- 8.Hajek, V. 1976. Staphylococcus intermedius, a new species isolated from animals. Int. J. Syst. Bacteriol. 26401-408. [Google Scholar]
- 9.Morgan, M. 26 September 2008, posting date. Methicillin-resistant Staphylococcus aureus and animals: zoonosis or humanosis? J. Antimicrob. Chemother. [Epub ahead of print.] [DOI] [PubMed]
- 10.Pottumarthy, S., J. M. Schapiro, J. L. Prentice, Y. B. Houze, S. R. Swanzy, F. C. Fang, and B. T. Cookson. 2004. Clinical isolates of Staphylococcus intermedius masquerading as methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 425881-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki, T., K. Kikuchi, Y. Tanaka, N. Takahashi, S. Kamata, and K. Hiramatsu. 2007. Methicillin-resistant Staphylococcus pseudintermedius in a veterinary teaching hospital. J. Clin. Microbiol. 451118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki, T., K. Kikuchi, Y. Tanaka, N. Takahashi, S. Kamata, and K. Hiramatsu. 2007. Reclassification of phenotypically identified Staphylococcus intermedius strains. J. Clin. Microbiol. 452770-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Hoovels, L., A. Vankeerberghen, A. Boel, K. Van Vaerenbergh, and H. De Beenhouwer. 2006. First case of Staphylococcus pseudintermedius infection in a human. J. Clin. Microbiol. 444609-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]