Abstract
The clinical value of viral load and integration testing for human papillomavirus (HPV) remains unclear. Data on HPV type 18 (HPV18) is limited. We examined the HPV18 viral load and integration status of 78 women with normal cervix or neoplasia. While the crude viral load appeared to increase with lesion severity, the association was not significant after normalization with sample cellularity. Unlike reports for HPV16, the amino-terminal 1 region of HPV18 E2 was most frequently (71.0%) disrupted, representing the best marker for integration. A substantial proportion (57.1%) of invasive cancers harbored only the episomal genome, thus jeopardizing the clinical value of integration testing. A large proportion (41.7%) of normal/low-grade lesions showed viral integration, suggesting that integration of HPV18 starts early and is unlikely to be a sole determinant for progression. Interpretation of viral load should take into account the form of HPV infection as single infections had significantly higher viral loads than coinfections (P = 0.046). More data generated from routinely collected samples are warranted to verify the clinical value of viral load and integration testing. Viral load quantitation for HPV18 is premature for clinical use at this stage.
There is no doubt that infection with human papillomavirus (HPV) may lead to the establishment of cervical cancer (3, 33). Though infection with HPV is common, not all HPV types will lead to cancer. There are 15 high-risk HPV types that have a higher propensity for the development of cervical cancer, with HPV type 16 (HPV16) and HPV18 being the most prevalent high-risk types worldwide (8, 25).
Women who have tested positive for high-risk HPV tend to harbor abnormal cervical cytology, and some studies have reported a correlation between viral load and disease severity (19, 20, 24, 28, 31). However, the consistency of these findings has been questioned, and it is now realized that the relationship is a lot more complex (5-7, 35). As the severity of disease increases, HPV is often found integrated into the host genome, making the interpretation of viral load even more complex. Furthermore, the available viral load data have been generated using different methodologies that may not be directly comparable (13, 17, 28, 29).
The HPV genome can exist in two physical forms: a closed circular episomal form or linearized and integrated into the host genome. Integration of the viral genome is often associated with the disruption of the E2 gene, which has regulatory control of the oncogenes E6 and E7. Nevertheless, this concept has been questioned by a recent study where the integration of the HPV16 genome was not necessarily associated with the overexpression of E6 and E7 (14). HPV18 has been found to be strongly associated with adenocarcinoma of the cervix. Studies carried out with HPV18 have shown that the viral genome is almost invariably found to be integrated in high-grade cervical intraepithelial lesions and invasive cancers (2, 9, 22, 27, 32), and these findings potentially allow the use of integration status as a marker to triage HPV18 infections with different clinical implications. However, such applications should be supported by data generated from a large series of clinical samples. In this study, we have used refined methods to determine the viral load, the physical status of the viral genome, and the type of HPV18 infection (whether single infection or coinfection) in a cohort of Chinese women with different grades of cervical lesions in an attempt to provide some information about the usefulness of these viral parameters in clinical setting.
MATERIALS AND METHODS
Study subjects.
A cross-sectional study was conducted in Hong Kong, where cervical cancer ranks ninth in female cancers, with an age-standardized incidence of 7.8 per 100,000 women (16). During the course of our previous study, a group of women with cervical disease status confirmed by histology and with HPV18 detected from cervical cytology samples was indentified. These samples were used for the current study. The study was approved by the local institutional ethics committee.
HPV detection and genotyping.
Total DNA was extracted from cervical scrapes using a commercially available extraction kit (QIAamp DNA Mini Kit; Qiagen GmbH, Hilden, Germany). HPV genotypes were determined using a Linear Array HPV Genotyping Test (Roche, Molecular Systems, Inc., CA) which can detect 37 HPV types. Samples found to be positive for HPV18 were subjected to viral load and integration status determination.
Viral load, E2 physical status, and integration.
Real-time quantitative PCR (qPCR) targeting the HPV18 E7 gene was used to determine the total crude (episomal and integrated) viral load. This region was selected as it is retained in both the episomal and integrated forms. To account for variation in the number of cells collected in each cervical scrape sample, levels of the housekeeping gene, beta-actin, were also determined using qPCR. Crude viral load measurements were normalized with the beta-actin gene levels using the following formula: (E7 gene copy number/beta-actin gene copy number) × 2. Values are expressed as the number of viral copies/cell equivalent.
Based on the assumption that HPV integration disrupts the E2 gene, the integrity of E2 was taken as a marker that HPV had integrated into the host genome. To account for the possibility that disruption might have involved only part of the E2 gene as has been shown in other HPV types (5-7), four nonoverlapping PCRs were designed to span the entire E2 gene, covering the amino (N)-terminal, hinge, and carboxyl (C)-terminal regions. Primer sets used for qPCR are listed in Table 1.
TABLE 1.
Primer sets for real-time PCRs
| Primer sequence | Primer position (nt)a | HPV 18 amplification region | Primer concn (μM) | Amplicon size (bp) |
|---|---|---|---|---|
| 5′-AGACACCGAAGGAAACCCTTT-3′ | 2821-2841 | E2 (amino terminus 1) | 0.25 | 223 |
| 5′-GCTTTATGTGCTTTACTTTTTGA-3′ | 3201-3043 | |||
| 5′-TGCAAGACACATGCGAGGAA-3′ | 3109-3129 | E2 (amino terminus 2) | 0.25 | 157 |
| 5′-CATGTTCCTGCATCAGTCATAT-3′ | 3244-3265 | |||
| 5′-AAAATATGGGAACACAGGTACG-3′ | 3359-3380 | E2 (hinge) | 0.25 | 192 |
| 5′-GCCGACGTCTGGCCGTAGGTCT-3′ | 3529-3550 | |||
| 5′-TACAGGCAACAACAAAAGACG-3′ | 3632-3652 | E2 (carboxyl terminus 1) | 0.25 | 177 |
| 5′-CCTGTTTTTTCATTGCCTGC-3′ | 3789-3808 | |||
| 5′-GTCACGAGCAATTAAGCGAC-3′ | 669-688 | E7 | 0.25 | 212 |
| 5′-CACAAAGGACAGGGTGTTCA-3′ | 861-880 | |||
| 5′-GCACGGCATCGTCACCAACT-3′ | 1283-1303 | Beta-actin | 0.125 | 142 |
| 5′-CATCTTCTCGCGGTTGGCCT-3′ | 1404-1424 |
nt, nucleotide. Position numbering is according to the HPV18 reference strain (GenBank accession no. NC_001357).
Using the E2/E7 gene copy number ratio, the physical status of the HPV18 genome for each sample was determined. Viral genomes were regarded as “purely integrated” when the E2/E7 gene copy number ratio was 0. When the E2/E7 ratio was 1, the sample was regarded to be “purely episomal”. Concomitant forms were defined as samples having an E2/E7 ratio between 0 and 1.
In total, six sets of real-time PCRs were performed for each sample using a Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) with a 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA). A standard curve for E7 and for each of the E2 gene targets was generated by plotting threshold cycle values against 10-fold serial dilutions of plasmids containing the full genome of HPV18 (ATCC, Rockville, MD). The standards used for quantitation of the beta-actin gene were purchased commercially (Applied Biosystems, Foster City, CA).
A 200-μl aliquot of cervical sample was used for DNA extraction. Five microliters of the extracted preparation was amplified in a 25-μl reaction mixture containing 0.125 to 0.25 μmol of primers (Table 1). The cycling conditions were 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 58°C for 15 s (except for beta-actin, which required 56°C for 15 s), and 72°C for 30 s. All real-time PCR assays showed a wide linear range that covered at least 10 to 10,000,000 copies/μl, and all showed high amplification efficiency. The specificity of each amplification was confirmed by checking the dissociation curve against the expected melting temperature of the amplification product.
Statistical analysis.
The differences in viral load levels between groups were compared by the Mann-Whitney U test or the Kruskal-Wallis test using SPSS software (version 15.0; SPSS Inc., IL). The distribution of categorical variables including integration status and location of disruption among groups was assessed by the chi-square test or the Fisher's test, as appropriate. This statistical analysis was performed using the Statcalc program (Epi Info; Centres for Disease Control and Prevention, Atlanta, GA). Two-sided P values less than or equal to 0.05 were regarded as significant.
RESULTS
Seventy-eight Chinese women aged 20 to 76 (mean age, 44.3 years; standard deviation, 10.8) years, all infected with HPV18, were examined in this study. Two women had a normal cervix, 4 had low-grade squamous intraepithelial lesions (LGSIL), 6 had grade 1 cervical intraepithelial neoplasia (CIN 1), 9 had CIN 2, 15 had CIN 3, and 42 had invasive cervical carcinoma (ICC). Of the ICC, 21 were of squamous cell carcinoma, 17 were adenocarcinoma, and 4 were adenosquamous carcinoma. For the purpose of analysis in this study, the cervical grades were grouped into three categories: normal/LGSIL/CIN 1, CIN 2/CIN 3, and ICC.
Viral load.
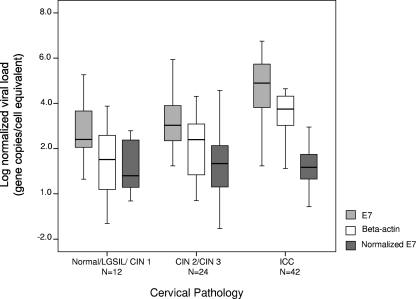
The crude E7 gene copy numbers were distributed over a wide range from 4.4 to 21,358,178.0 (median, 6,622.4; interquartile range, 245.0 to 185,546.7) gene copies/μl. The gene copy numbers for beta-actin ranged from 0.05 to 44,125.1 (median, 1,237.2; interquartile range, 106.3 to 8,123.4) gene copies/μl. The viral load after normalization with beta-actin ranged from <0.01 to 37,850.3 (median, 12.8; interquartile range, 3.3 to 96.2) gene copies/cell equivalent. The distributions of crude and normalized viral load and beta-actin gene levels among the different grades of cervical lesion are shown in Fig. 1. The crude total viral load (episomal and integrated) appeared to increase with disease severity. However, when normalized with the beta-actin gene levels, this association was no longer observed (P = 0.830 by a Kruskal-Wallis test). Study subjects were arbitrarily divided into three age groups (20 to 34 years, 35 to 54 years, and 55 to 76 years) to analyze for a possible effect between age and viral load. No significant association between viral load and age was observed (P = 0.591 by a Kruskal-Wallis test). The lack of association was further confirmed by dividing subjects into two age groups, i.e., 20 to 40 years and 41 to 76 years (P = 0.402 by Mann-Whitney U test).
FIG. 1.
Distribution of viral load among different grades of cervical lesions. The middle line indicates the median; the box represents the interquartile range. Lines extending from each box are the upper and lower limits. Viral load was determined as described in Materials and Methods.
Viral genome physical status.
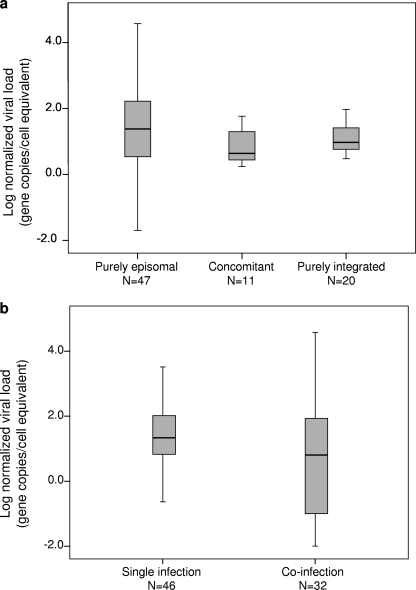
Of the 78 study samples, 20 (25.6%) showed no detectable amplification for any of the E2 regions investigated and, hence, were regarded as harboring pure integrated forms. Forty-seven samples (60.3%) did not show any decrease in gene copy numbers for the four E2 gene regions compared to E7 gene levels; hence, these were regarded as harboring the pure episomal form. The remaining 11 (14.1%) samples contained both the episomal and integrated forms, which are thus regarded as concomitant forms. The distribution of viral load among samples with different physical forms of the HPV18 genome is shown in Fig. 2a. The normalized viral load was spread over a wide range. The median (interquartile range) values were as follows: for episomal forms, 24.0 (3.4 to 166.6) gene copies/cell equivalent; for concomitant forms, 4.3 (2.8 to 20.1) gene copies/cell equivalent; and for the integrated form, 8.8 (3.7 to 22.1) gene copies/cell equivalent. No significant association between viral load and the physical status of the genome was found (P = 0.212 by the Kruskal-Wallis test).
FIG. 2.
Distribution of viral load according to the physical status of the genome (a) and type of HPV18 infection (single or coinfection) (b). In both panels, the middle line indicates median, the box represents interquartile range, and lines extending from each box are the upper and lower limits. Viral load was determined as described in Materials and Methods.
Using the two age stratifications described above, the physical status of the viral genome had no significant association with age (P = 0.578 by the Kruskal-Wallis test and P = 0.670 by the Mann-Whitney U test, respectively, for the two age stratification methods).
E2 disruption status.
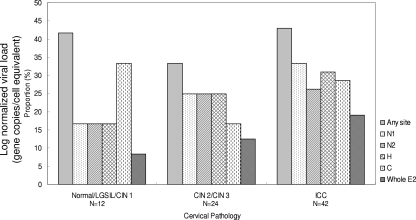
Altogether, 31/78 (39.8%) samples were found to have E2 disruptions. The most common disruption occurred at the N1 (nucleotides 2821 to 3043) region in 22/31 (71.0%) samples, followed by the hinge (67.7%), the C-terminal (64.5%), and then the N2 (nucleotides 3109 to 3265) (61.3%) regions. The preferential site of disruption did not change with the severity of lesion (Fig. 3). The proportion of samples with E2 disruption (any site combined) did not show a significant association with the degree of cervical lesion (E2 disruption proportion in normal/LGSIL/CIN 1 cases, 41.7%; in CIN 2/CIN 3 cases, 33.3%; and in ICC cases, 42.9%; PTrend = 0.737, by the chi-square test for trend) (Fig. 3).
FIG. 3.
Proportion and location of E2 disruption according to the type of cervical lesions. Specimens with disruption involving more than one E2 region are counted more than once. H, hinge region.
Of the 31 samples that contained an E2 disruption, 12 had a complete loss of the whole E2 gene (identified by no decrease in gene copy numbers in any of the four regions of E2 investigated). The proportion of complete loss of the E2 gene increased with the severity of cervical lesion, but it did not reach a significant level (8.3% in normal/LGSIL/CIN 1, 12.5% in CIN 2/CIN 3, and 19.0% in ICC cases; PTrend = 0.314 by the chi-square test for trend) (Fig. 3).
Infection form.
Of the 78 subjects, 46 (59.0%) had single infections of HPV18 alone. The other 32 (41.0%) subjects had coinfections of HPV18 with other HPV types (Table 2). The most common coinfecting HPV type was HPV16 (50.0%), followed by HPV52 (15.6%); HPV types 31, 59, 61, and 81 (6.3% each); and HPV types 51, 53, and 84 (3.1% each). Figure 2b shows the distribution of viral load among samples with HPV18 single infections and coinfections. The viral loads obtained for HPV18 single infections were significantly higher than the load in HPV18 coinfections (median [interquartile range] = 21.8 [6.75 to 104.8] versus 4.5 [0.05 to 72.25] viral copies/cell equivalent; P = 0.046 by the Mann-Whitney U test). When the form of the infection (single or coinfection) was analyzed with respect to the severity of cervical lesion, a significant trend of increase in the proportion of single infections was found (normal/LGSIL/CIN 1, 33.3%; CIN 2/3, 58.3%; ICC, 66.7%; PTrend = 0.050 by the chi-square test for trend).
TABLE 2.
Viral load and viral genome physical status of HPV18 infection according to the degree of cervical lesion
| Cervical lesion type (n) | Viral load measurement (mean [interquartile range])
|
Form of the viral genome (no. of samples [%])
|
Type of HPV infection (no. of samples [%])
|
|||||
|---|---|---|---|---|---|---|---|---|
| Crude total viral load (copies/μl) | Beta-actin level (copies/μl) | Normalized viral load (copies/cell equivalent) | Pure episomal | Pure integrated | Concomitant | Single | Coinfectiona | |
| Normal/LGSIL/CIN 1 (12) | 255.1 (118.3-6,949.7) | 36.6 (1.9-392.1) | 6.8 (2.0-245.0) | 7 (58.3) | 3 (25.0) | 2 (16.7) | 4 (33.3) | 8 (66.7) |
| CIN 2/CIN 3 (24) | 1,069.8 (225.2-8,784.5) | 251.5 (7.1-1,237.2) | 21.9 (2.3-140.3) | 16 (66.7) | 6 (25.0) | 2 (8.3) | 14 (58.3) | 10 (41.7) |
| ICC (42) | 58,985.0 (3,215.0-477,860.7) | 5,578.5 (1,452.7-20,193.6) | 11.7 (3.5-53.4) | 24 (57.1) | 11 (26.2) | 7 (16.7) | 28 (66.7) | 14 (33.3) |
Coinfection with other HPV type(s).
No significant association was found between infection form and the physical status of the HPV18 genome (P = 0.122 by a chi square test). Using the same age stratifications as described above, no significant association between the form of infection and age was observed (P = 0.253 by Kruskal-Wallis test and P = 0.094 by Mann-Whitney U test, respectively, for the two age stratification methods).
Adeno/adenosquamous carcinoma versus squamous cell carcinoma.
When the viral load results were analyzed with respect to the histological group of adeno/adenosquamous carcinoma (n = 21) and squamous cell carcinoma (n = 21), no significant difference was found (P = 0.400 by the Mann-Whitney U test) though a wider range of viral load was seen for the squamous cell carcinoma (median, 8.5 copies/cell equivalent; interquartile range, 0.05 to 53.4 copies/cell equivalent) than with adeno/adenosquamous carcinoma (median, 16.3 copies/cell equivalent; interquartile range, 7.0 to 36.4 copies/cell equivalent). The physical status of the viral genome also did not differ between the two histological groups; the pure episomal form was found in 10 (47.6%) cases of squamous cell carcinoma and in 14 (66.7%) cases of adeno/adenosquamous carcinoma (P = 0.212 by the chi-square test).
When the infection form (single or coinfection) of the two histological groups was analyzed, it was found that the proportion of HPV18 single infections was significantly higher for adeno/adenosquamous carcinoma cases (85.7% versus 14.3%; P = 0.009 by the chi-square test).
DISCUSSION
Many previous studies have found a positive association between viral load and disease severity, which has led to the idea of using viral load as a biomarker to determine the status of cervical disease or to predict its progression (4, 11, 13, 19, 23, 24, 28, 30, 31). However, at the same time, contradictory data have also been generated (1, 15, 26, 34). The explanation for the observation from some studies that the viral load increases with disease severity remains obscure. As the disease worsens, the virus becomes integrated, which is a genome form not capable of self-replicating, and therefore the mechanism for maintaining a high viral load is uncertain. Many studies that attempted to examine viral load associations did not analyze the physical status of the viral genome (4, 12, 13, 18, 28, 29). In addition, some studies have used a method that measured the total viral load of a mixture of HPV types (13, 18, 28, 29). It is now known that the relationship between viral load, integration, and disease severity varies with different HPV types (5-7).
In this study, we found that normalization with a human gene to adjust for variation in the degree of cellularity of the specimen is important for an accurate interpretation of the association between viral load and disease severity. Our results showed that as the disease worsened, the crude viral load appeared to increase. However, when the viral load was normalized, such an association was no longer seen. The observation of a higher crude viral load in high-grade lesions may be explained by the fact that abnormal cells express fewer intracellular adhesion molecules than the normal cells and thus are more readily exfoliated (28). Although we were able to normalize the viral load to minimize biases due to variation in the cellular content among different specimens, it was not possible to distinguish the HPV-infected and noninfected portion of cells collected in the cervical scrape sample. While cervical scrape is the most convenient and feasible sample type, it is inevitable that the sample will contain a variable mixture of relevant (HPV infected) and irrelevant (noninfected) cells.
We found that the interpretation of viral load can be complicated by its association with the physical state of the viral genome and with the form of HPV infection (single or coinfection). Though not reaching statistical significance, specimens harboring the episomal viral genome form seemed to have a higher viral load. This observation is in line with the fact that viral replication can occur only in the episomal form. While coinfection with multiple HPV types is common, the biological interaction between HPV types in such circumstances is unknown. Our finding of a significantly lower viral load in coinfections is in line with the hypothesis that the coinfecting types are competing with each other for the limited microelements required for viral replication. Overall, our results indicated that measuring viral load to assess HPV18 infection is premature for clinical use at the present stage.
Many previous studies have found that almost all HPV18 infections exist in an integrated form in women with invasive cancer (2, 9, 15, 22, 32). In the current study, we found a rather high proportion of ICC (57.1%) harboring only the episomal form of the virus. We believe one explanation for this discrepancy could be the sensitivity of the method used. Methods such as Southern blotting or two-dimensional gel electrophoresis were often employed to document the physical state of the HPV genome in previous studies (2, 9, 10, 22, 27). One disadvantage of these methods is that they often require a large amount of DNA to produce a positive signal. In contrast, the method employed in the current study, real-time PCR, is able to detect trace amounts of DNA. A recent study by Huang et al. (17) also reported that the episomal form of HPV18 was seen in all lesion grades ranging from CIN 1 to cervical cancer, with the highest proportion of the episomal form seen in cervical cancer stages II to IV (66.9%). We also obtained a similar proportion of the episomal form (57.1%) for ICC cases. A well-established notion is that disruption of the E2 gene results in the overexpression of E6 and E7, which is the principal oncogenic mechanism conferred by high-risk HPV. Our findings together with those from Huang et al. (17), suggest that there might be alternative oncogenic pathways that bypass E2 disruption.
To date, only a few studies have examined the integration state of the HPV18 genome in normal or low-grade lesions (17). We found that integration was not uncommon (41.7%) in the normal/LGSIL/CIN 1 group. This indicates that integration occurs early in the disease progression process, and probably integration alone cannot drive the full progress to high-grade or invasive cancer.
We observed that the most common site of disruption, regardless of lesion grade, was the N1 region. The available information on the location of disruption is mainly derived from HPV16, where the hinge region was identified as the most common site of disruption (1, 21). However, if the hinge region had been selected for our study, we could potentially have missed 32.3% of the E2 disruptions. These data emphasize the importance of selecting the right target of the E2 gene as a surrogate marker for determining the state of genome integration.
One should note that integration of the HPV genome could also lead to disruption of gene regions other than the E2. The current study might have missed integrations that disrupt only viral gene regions outside the E2. Nevertheless, the oncogenic consequence of such non-E2-disrupting integrations is unknown, and there is no evidence to suggest that these integrations could lead to an overexpression of E6 and E7. Thus, the approach of this study was to perform a more comprehensive analysis on four nonoverlapping regions of the most important gene, E2.
Adenocarcinoma of the cervix has a strong association with HPV18 (35). Here, we have made some subgroup analyses according to the histological type. Although no significant differences in viral load or the physical state of the genome between adeno/adenosquamous carcinomas and squamous cell carcinoma were seen, we found that the proportion of single infections was significantly higher among adeno/adenosquamous carcinomas than in squamous cell carcinomas. Further studies are required to explore the implication of this finding.
Our study has provided further information on the complex relationship between viral load, integration of the viral genome, and disease severity. These data, which can be generated by a widely available technique, real-time PCR, on a routinely feasible specimen, cervical scrape, should inform researchers about the potential advantages and drawbacks of applying viral load and integration testing in a clinical setting.
Acknowledgments
The work described in this paper was fully supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, People's Republic of China (project number CUHK4429/03 M).
Footnotes
Published ahead of print on 26 November 2008.
REFERENCES
- 1.Andersson, S., H. Safari, M. Mints, I. Lewensohn-Fuchs, U. Gyllensten, and B. Johansson. 2005. Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN). Br. J. Cancer. 922195-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berumen, J., L. Casas, E. Segura, J. L. Amezcua, and A. Garcia-Carranca. 1994. Genome amplification of human papillomavirus types 16 and 18 in cervical carcinomas is related to the retention of E1/E2 genes. Int. J. Cancer 56640-645. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87796-802. [DOI] [PubMed] [Google Scholar]
- 4.Carcopino, X., M. Henry, D. Benmoura, A. S. Fallabregues, H. Richet, L. Boubli, and C. Tamalet. 2006. Determination of HPV type 16 and 18 viral load in cervical smears of women referred to colposcopy. J. Med. Virol. 781131-1140. [DOI] [PubMed] [Google Scholar]
- 5.Chan, P. K., J. L. Cheung, T. H. Cheung, K. W. Lo, S. F. Yim, S. S. Siu, and J. W. Tang. 2007. Profile of viral load, integration, and E2 gene disruption of HPV58 in normal cervix and cervical neoplasia. J. Infect. Dis. 196868-875. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, J. L., T. H. Cheung, J. W. Tang, and P. K. Chan. 2008. Increase of integration events and infection loads of human papillomavirus type 52 with lesion severity from low-grade cervical lesion to invasive cancer. J. Clin. Microbiol. 461356-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, J. L., K. W. Lo, T. H. Cheung, J. W. Tang, and P. K. Chan. 2006. Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J. Infect. Dis. 1941706-1712. [DOI] [PubMed] [Google Scholar]
- 8.Clifford, G. M., J. S. Smith, M. Plummer, N. Munoz, and S. Franceschi. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer. 8863-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corden, S. A., L. J. Sant-Cassia, A. J. Easton, and A. G. Morris. 1999. The integration of HPV-18 DNA in cervical carcinoma. Mol. Pathol. 52275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, A. P., R. Reid, M. Campion, and A. T. Lörincz. 1991. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J. Virol. 65606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuzick, J., G. Terry, L. Ho, T. Hollingworth, and M. Anderson. 1994. Type-specific human papillomavirus DNA in abnormal smears as a predictor of high-grade cervical intraepithelial neoplasia. Br. J. Cancer 69167-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalstein, V., D. Riethmuller, J. L. Pretet, K. Le Bail Carval, J. L. Sautiere, J. P. Carbillet, B. Kantelip, J. P. Schaal, and C. Mougin. 2003. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal French cohort study. Int. J. Cancer 106396-403. [DOI] [PubMed] [Google Scholar]
- 13.Gravitt, P. E., R. D. Burk, A. Lorincz, R. Herrero, A. Hildesheim, M. E. Sherman, M. C. Bratti, A. C. Rodriguez, K. J. Helzlsouer, and M. Schiffman. 2003. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol. Biomarkers Prev. 12477-484. [PubMed] [Google Scholar]
- 14.Häfner, N., C. Driesch, M. Gajda, L. Jansen, R. Kirchmayr, I. B. Runnebaum, and M. Dürst. 2008. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene 271610-1617. [DOI] [PubMed] [Google Scholar]
- 15.Ho, C. M., T. Y. Chien, S. H. Huang, B. H. Lee, and S. F. Chang. 2006. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol. Oncol. 10254-60. [DOI] [PubMed] [Google Scholar]
- 16.Hong Kong Cancer Registry, Hospital Authority. 2005. Facts on cervical cancer. Hong Kong Cancer Registry, Hospital Authority, Hong Kong. http://www3.ha.org.hk/cancereg/eng/cx.pdf.
- 17.Huang, L. W., S. L. Chao, and B. H. Lee. 2008. Integration of human papillomavirus type-16 and type-18 is a very early event in cervical carcinogenesis. J. Clin. Pathol. 61627-631. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y., M. N. Huang, N. Li, X. G. Li, N. Li, and L. Y. Wu. 16 November 2007, posting date. Association between human papillomavirus DNA load and development of cervical intraepithelial neoplasia and cervical cancer. Int. J. Gynecol. Cancer 18755-760. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 19.Hudelist, G., M. Manavi, K. I. Pischinger, T. Watkins-Riedel, C. F. Singer, E. Kubista, and K. F. Czerwenka. 2004. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol. Oncol. 92873-880. [DOI] [PubMed] [Google Scholar]
- 20.Josefsson, A. M., P. K. Magnusson, N. Ylitalo, P. Sorensen, P. Qwarforth-Tubbin, P. K. Andersen, M. Melbye, H. O. Adami, and U. B. Gyllensten. 2000. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 3552189-2193. [DOI] [PubMed] [Google Scholar]
- 21.Kalantari, M., F. Karlsen, G. Kristensen, R. Holm, B. Hagmar, and B. Johansson. 1998. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int. J. Gynecol. Pathol. 17146-153. [DOI] [PubMed] [Google Scholar]
- 22.Manavi, M., G. Hudelist, A. Fink-Retter, D. Gschwantler-Kaulich, K. Pischinger, and K. Czerwenka. 2008. Human papillomavirus DNA integration and messenger RNA transcription in cervical low- and high-risk squamous intraepithelial lesions in Austrian women. Int. J. Gynecol. Cancer 18285-294. [DOI] [PubMed] [Google Scholar]
- 23.Mansell, M. E., L. Ho, G. Terry, A. Singer, and J. Cuzick. 1994. Semi-quantitative human papillomavirus DNA detection in the management of women with minor cytological abnormality. Br. J. Obstet. Gynaecol. 101807-809. [DOI] [PubMed] [Google Scholar]
- 24.Moberg, M., I. Gustavsson, E. Wilander, and U. Gyllensten. 2005. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br. J. Cancer 92891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muñoz, N., F. X. Bosch, S. de Sanjosé, R. Herrero, X. Castellsagué, K. V. Shah, P. J. Snijders, C. J. Meijer, and the International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348518-527. [DOI] [PubMed] [Google Scholar]
- 26.Onan, M. A., C. Taskiran, G. Bozdayi, A. Biri, O. Erdem, A. Acar, G. Gunaydin, S. Rota, O. Ataoglu, and H. Guner. 2005. Assessment of human papilloma viral load of archival cervical intraepithelial neoplasia by real-time polymerase chain reaction in a Turkish population. Eur. J. Gynaecol. Oncol. 26632-635. [PubMed] [Google Scholar]
- 27.Pirami, L., V. Giache, and A. Becciolini. 1997. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive and invasive lesions of the uterine cervix. J. Clin. Pathol. 50600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlecht, N. F., A. Trevisan, E. Duarte-Franco, T. E. Rohan, A. Ferenczy, L. L. Villa, and E. L. Franco. 2003. Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int. J. Cancer 103519-524. [DOI] [PubMed] [Google Scholar]
- 29.Snijders, P. J., C. J. Hogewoning, A. T. Hesselink, J. Berkhof, F. J. Voorhorst, M. C. Bleeker, and C. J. Meijer. 2006. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int. J. Cancer 1191102-1107. [DOI] [PubMed] [Google Scholar]
- 30.Swan, D. C., R. A. Tucker, G. Tortolero-Luna, M. F. Mitchell, L. Wideroff, E. R. Unger, R. A. Nisenbaum, W. C. Reeves, and J. P. Icenogle. 1999. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J. Clin. Microbiol. 371030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Duin, M., P. J. Snijders, H. F. Schrijnemakers, F. J. Voorhorst, L. Rozendaal, M. A. Nobbenhuis, A. J. van den Brule, R. H. Verheijen, T. J. Helmerhorst, and C. J. Meijer. 2002. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int. J. Cancer 98590-595. [DOI] [PubMed] [Google Scholar]
- 32.Vinokurova, S., N. Wentzensen, I. Kraus, R. Klaes, C. Driesch, P. Melsheimer, F. Kisseljov, M. Dürst, A. Schneider, and M. von Knebel Doeberitz. 2008. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 68307-313. [DOI] [PubMed] [Google Scholar]
- 33.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Muñoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 18912-19. [DOI] [PubMed] [Google Scholar]
- 34.Wensveen, C. W., M. J. Kagie, N. J. Nagelkerke, R. W. Veldhuizen, and J. B. Trimbos. 2005. Can. viral load, semi-quantitatively evaluated, of human papillomavirus predict cytological or histological outcome in women with atypical squamous or glandular cells of undetermined significance cytology? Eur. J. Gynaecol. Oncol. 26393-397. [PubMed] [Google Scholar]
- 35.Woodman, C. B., S. I. Collins, and L. S. Young. 2007. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer. 711-22. [DOI] [PubMed] [Google Scholar]