Abstract
Orientia tsutsugamushi, the cause of scrub typhus, is a major pathogen in the Asia-Pacific region. The severity of infection ranges from mild features to multiorgan failure and death. The aim of this prospective study was to define the O. tsutsugamushi loads in the blood samples of patients with scrub typhus on the day of hospital admission and to determine whether this was associated with disease severity. Quantitation was performed using a real-time PCR assay targeting the 16S rRNA gene of O. tsutsugamushi. A total of 155 patients with a confirmed diagnosis of scrub typhus had a median (interquartile range [IQR], range) O. tsutsugamushi DNA load in blood of 13 (0 to 334, 0 to 310,253) copies/ml. This included 74 patients who had undetectable bacterial loads. An analysis of bacterial load versus clinical features for all 155 patents demonstrated that duration of illness (P < 0.001), presence of eschar (P = 0.004), and concentrations of aspartate aminotransferase, alanine aminotransferase, and alkaline phosphatase (P < 0.001 for all three) were positively correlated with bacterial load. Patients who died had a significantly higher bacterial load than those who survived (mean [standard deviation] values: 17,154 [12.7] versus 281 [5.2] copies/ml; P < 0.001). This study has demonstrated a relationship between bacterial load and disease severity in adults with scrub typhus.
Orientia tsutsugamushi, the cause of scrub typhus, is a major pathogen in the Asia-Pacific region, where it accounts for up to 23% of all febrile episodes (4). Infection commonly presents as an acute febrile illness 7 to 10 days after the bite of an infected larval trombiculid mite (chigger). The major presenting features are fever, severe headache, and myalgia. Other signs and symptoms include rash, lymphadenopathy, hepatosplenomegaly, cough, sore throat, abdominal pain, and central nervous system involvement. The severity of infection ranges from mild features to multiorgan failure and death, which occurs for around 4% of patients presenting to a hospital (10, 21). Severity of illness has been reported to show geographical variability (10), although the basis for this is unknown and has multiple explanations, including differences in host genetic susceptibility or response to infection and variability in the virulence of the organism. A relationship between magnitude of bacteremia and severity of clinical picture has been reported for a range of bacterial infections in both adults and children, based on quantitative bacterial culture (25) and, more recently, molecular techniques (5, 8). A previous study reported the O. tsutsugamushi DNA loads in 7 patients with scrub typhus, as defined by quantitative real-time PCR targeting a gene encoding a 47-kDa protein (20). This ranged from 1,076 to 28,812 DNA copies/μl in blood, but clinical features and outcomes were not described (20).
The aim of this study was to define the O. tsutsugamushi loads in blood samples taken on the day of hospital admission from a large, unselected population of patients with scrub typhus in northeast Thailand and to compare this with disease severity.
MATERIALS AND METHODS
Patients with scrub typhus were identified during a prospective study of acute febrile illness in Udon Thani hospital, northeast Thailand, between October 2000 and December 2001, as described previously (21). In brief, the inclusion criteria were an age of ≥15 years, the presence of a fever (>37.8°C) of unknown cause, and agreements to participate and to attend an outpatient follow-up for a convalescent-phase serum sample at 2 weeks. The exclusion criterion was a blood smear positive for malaria parasites or the presence of other definable infections, such as pneumonia or urinary tract infection. Blood was drawn on admission for aerobic blood culture, serological testing, and molecular diagnostics. The duration of symptoms prior to admission, antibiotic treatment at presentation, and clinical features, including the presence of an eschar, the results of laboratory tests, and in-hospital outcome (survival or death), were recorded using a standardized data collection form. Paired sera were tested using the indirect immunofluorescence antibody (IFA) test (3). EDTA samples were evaluated by PCR when the IFA titer of immunoglobulin M against O. tsutsugamushi showed a fourfold or greater rise between paired samples or a single titer of ≥1:400. A total of 156 patients were positive by serological criteria; one EDTA sample was not available, and so, 155 samples were evaluated by real-time PCR. The study was approved by the Ethical Review Subcommittee of the Ministry of Public Health, Thailand.
DNA from laboratory cultures of O. tsutsugamushi was extracted using a Wizard SV Genomic DNA purification kit (Promega). A plasmid standard was constructed by cloning a 221-bp 16S ribosomal DNA fragment amplified from O. tsutsugamushi strain Kato genomic DNA into pGEMT easy Vector Systems (Promega). The fragment was amplified using primers OT1F and OT1R (21). The purified DNA fragment was ligated and transformed into Escherichia coli (DH5α), as described by the manufacturer. The presence of the cloned DNA fragment in plasmid pG16S was verified by sequencing. Plasmid was linearized with PstI, and DNA concentrations were determined using a Quant-it DNA assay kit with high sensitivity (Molecular Probes) and a Rotor-Gene 3000 (Corbett Research, Australia) in the DNA concentration measurement mode.
DNA was extracted from admission EDTA samples as previously described (21) and maintained at −80°C prior to use. A quantitative real-time PCR assay was developed based on the 16S rRNA gene of O. tsutsugamushi strain Kato. Dual-labeled probe (TaqMan) and primers were designed and modified using Primer3 software (17). The primers amplified a 151-bp product (positions 106 to 124 and 235 to 256 of GenBank accession no. D38624). Reaction mixtures (20 μl) contained 5 μl DNA, 10 μl QUANTIPROBES (QuantiMix Easy Probes kit; Biotools), 0.2 μM probe (5′ [6-carboxyfluorescein]-TAA GTG CTA ATA CCG TAT GCC CTC TA-[BHQ1] 3′), and 0.1 μM each of primers OT1-R (5′ CTC TCA GAC CAG CTA CAG ATC ACA 3′) and OT3-F (5′ CCC ATC AGT ACG GAA TAA CA 3′). Tenfold dilutions of linearized plasmid pG16S ranging from 4.2 × 104 to 0.042 copies/μl reaction mixture were used to perform a standard curve. Real-time PCR was performed using the Rotor-Gene 3000. The cycling conditions were 95°C for 10 min (1 cycle), followed by 45 cycles at 95°C for 10 s and 58°C for 45 s.
A standard curve was constructed by plotting the logarithmic values of a known number of plasmid pG16S copies versus the cycle threshold value. The standard curve determined using plasmid DNA revealed a linear assay over 6 orders of magnitude (4 × 104 to 0.04 copies/μl). The lower limit of detection was 0.04 DNA copies/μl. Clinical DNA samples were evaluated and the copy number was calculated based on the standard curve. Intra- and interassay analyses were performed to define the reproducibility of the assays. Intra-assay variability was determined for five replicate values within each run. Interassay variability was calculated from mean values for samples run in five separate assays run on different days, using freshly prepared standard DNA. The mean (range) coefficient of variation values for intra-assay and interassay variation were 1.18% (1.0 to 1.2%) and 4.1% (3.0 to 7.3%), respectively. Assay specificity was assessed by testing with DNA extracted from laboratory cultures of Rickettsia typhi, Rickettsia conorii, and Leptospira interrogans serovar Autumnalis (the last being selected because leptospirosis is a common cause of febrile illness in northeast Thailand and presents with clinical features that are similar to those of scrub typhus), together with DNA extracted from EDTA blood samples from 60 patients randomly selected from the febrile illness study who had alternative diagnoses and did not have scrub typhus based on the IFA test. An initial nonparametric analysis comparing the quantitative PCR results was performed on all 155 patients with scrub typhus, using Spearman's rho for analysis of continuous variables and the Kruskal-Wallis test for analysis of nominal outcome variables. A further parametric analysis was carried out on the 81 patients with detectable quantitative PCR levels. Continuous variables were transformed toward normality, and Pearson's correlation and Student's t test were used as appropriate. Associations between groups were divided according to bacterial load, and clinical characteristics and outcomes were analyzed using Fisher's exact test and the Kruskal-Wallis test as appropriate. All analyses were carried out using Stata/SE 10.0 (StataCorp LP, College Station, TX).
RESULTS
A total of 155 patients with a confirmed diagnosis of scrub typhus had a median (interquartile range [IQR], range) O. tsutsugamushi DNA load in blood of 13 (0 to 334, 0 to 310,253) copies/ml. This included 74 patients who had undetectable bacterial loads. Assay specificity was high, with no signal detected in DNA samples extracted from the blood samples of 60 patients with negative IFA test results or from DNA extracted from laboratory cultures of R. typhi, R. conorii, or L. interrogans (data not shown).
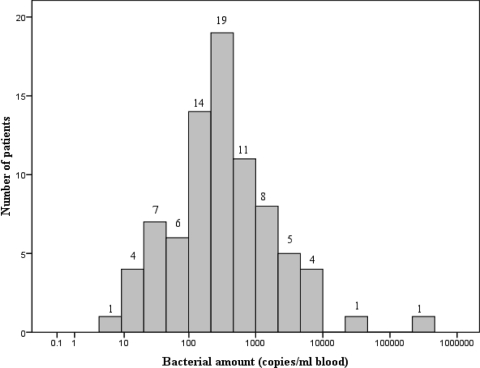
A nonparametric analysis comparing the quantitative PCR results with clinical features and outcomes for all 155 patients is shown in Table 1. Duration of illness, presence of eschar, and all three hepatic enzymes were positively correlated with bacterial load (Table 1). A second analysis was performed on the 81 patients (52%) who had positive quantitative real-time PCR results. The median (IQR) O. tsutsugamushi load in blood for these 81 patients was 284 (124 to 943) DNA copies/ml, with a minimum detectable level of 5 copies/ml, as shown in Fig. 1. Patients were divided into three groups according to bacterial load: <100 DNA copies/ml (n = 18), 100 to 1,000 DNA copies/ml (n = 44), and >1,000 DNA copies/ml (n = 19). A comparison of the clinical features and outcomes for the three groups is shown in Table 2. Factors correlated with bacterial concentration included serum bicarbonate, serum creatinine, bilirubin, hepatic enzyme levels, and APACHE II score (Table 2). Patients who died had significantly higher bacterial loads than those who survived (mean [standard deviation] values: 17,154 [12.7] versus 281 [5.2] copies/ml; P < 0.001). This result should be regarded as a strong trend, as a nonparametric analysis performed on all 155 patients (including all those with undetectable bacterial loads) was not statistically significant; it is unsurprising, with only three deaths overall, that this association is not statistically robust.
TABLE 1.
Relationship between bacterial load and clinical features in 155 patients with scrub typhusa
| Variable | Valueb | Association with bacterial loada
|
|
|---|---|---|---|
| ρ | P | ||
| Admission characteristics | |||
| No. of males (%) | 126 (66) | NS* | |
| Age (yr) | 44 (31-56) | 0.14 | NS |
| No. of patients with eschars (%) | 14 (7.3) | 0.004* | |
| Duration of symptoms (days) | 6 (4-8) | 0.34 | <0.001 |
| Hemoglobin concn (mg/dl) | 12.4 (10.6-13.8) | 0.10 | NS |
| White blood cell count (103/ml) | 11.0 (8.5-13.9) | 0.02 | NS |
| Platelet count (109/ml) | 156 (95-229) | 0.09 | NS |
| Serum bicarbonate concn (mmol/liter) | 23 (21-26) | 0.06 | NS |
| Serum creatinine concn (mg/dl) | 1.3 (1.0-2.4) | −0.16 | NS |
| Blood urea nitrogen concn (mg/dl) | 16 (11-36) | −0.20 | 0.01 |
| Total serum bilirubin concn (μmol/liter) | 1.1 (0.6-2.7) | −0.14 | NS |
| Direct serum bilirubin concn (μmol/liter) | 0.6 (0.4-1.7) | −0.15 | NS |
| Aspartate aminotransferase concn (U/liter) | 97 (55-163) | 0.41 | <0.001 |
| Alanine aminotransferase concn (U/liter) | 70 (43-113) | 0.40 | <0.001 |
| Alkaline phosphatase concn (U/liter) | 170 (106-262) | 0.29 | <0.001 |
| APACHE II score | 7 (4-10) | −0.04 | NS |
| Outcome | |||
| No. of deaths (%) | 5 (2.6) | NS* | |
| Fever clearance time (h) | 41 (16-84) | 0.06 | NS |
*, P by Kruskal-Wallis test; ρ, Spearman's rho; NS, not significant.
Values with ranges in parentheses are medians (IQRs), and other values are quantities (percentages).
FIG. 1.
Histogram of the calculated O. tsutsugamushi loads (numbers of DNA copies/ml blood) in 81 patients with PCR-confirmed scrub typhus.
TABLE 2.
Relationship between bacterial load and clinical features in 81 patients with PCR-confirmed scrub typhusa
| Variable | Association with bacterial load
|
Valueb for indicated no. of DNA copies/ml
|
P | |||
|---|---|---|---|---|---|---|
| ρc | P | <100 (n = 18) | 100-1,000 (n = 44) | >1,000 (n = 19) | ||
| Admission characteristics | ||||||
| No. of males (%) | NS† | 10 (55.6) | 26 (59.1) | 14 (73.7) | NS¶ | |
| Age (yr) | 0.22§ | 0.05 | 35 (33-47) | 42 (28-54) | 48 (37-58) | NS |
| No. of patients with eschars (%) | NS† | 0 | 10 (22.7) | 1 (5.3) | 0.03¶ | |
| Duration of symptoms (days) | 0.16§ | NS | 5.5 (4.5-8.0) | 7.0 (4.5-10.0) | 8.0 (7.0-11.0) | NS |
| Hemoglobin concn (mg/dl) | −0.03§ | NS | 11.8 (11.1-13.6) | 13.5 (11.3-14.3) | 12.5 (10.2-13.8) | NS |
| Platelet count (109/ml) | 0.24§ | 0.03 | 168 (147-239) | 184 (135-243) | 145 (121-213) | NS |
| White cell count (103/ml) | −0.17 | NS | 10.1 (7.1-12.9) | 11.1 (8.5-14.3) | 11.7 (9.9-13.5) | NS |
| Serum bicarbonate concn (mmol/liter) | −0.30 | 0.01 | 26 (24-29) | 23 (21-26) | 22 (15-25) | 0.01 |
| Blood urea nitrogen concn (mg/dl) | 0.16§ | NS | 13 (12-19) | 15 (9-19) | 18 (9-48) | NS |
| Serum creatinine concn (mg/dl) | −0.37§ | <0.001 | 1.0 (1.0-1.3) | 1.1 (1.0-1.3) | 1.5 (1.1-3.4) | 0.002 |
| Total serum bilirubin concn (μmol/liter) | 0.31 | 0.01 | 0.7 (0.5-1.0) | 0.7 (0.6-1.5) | 1.4 (0.7-3.7) | 0.02 |
| Direct serum bilirubin concn (μmol/liter) | 0.29 | 0.01 | 0.4 (0.3-0.7) | 0.4 (0.3-0.8) | 0.9 (0.5-2.7) | 0.02 |
| Aspartate aminotransferase concn (U/liter) | 0.36§ | 0.001 | 106 (73-145) | 129 (89-172) | 182 (97-226) | 0.02 |
| Alanine aminotransferase concn (U/liter) | 0.29 | 0.01 | 66 (49-99) | 98 (74-136) | 142 (76-182) | 0.04 |
| Alkaline phosphatase concn (U/liter) | 0.27§ | 0.02 | 165 (97-275) | 199 (135-317) | 324 (196-396) | 0.04 |
| APACHE II score | 0.28§ | 0.01 | 4 (3-7) | 6 (4-9) | 10 (3-16) | 0.04 |
| Outcome | ||||||
| No. of deaths (%) | <0.001† | 0 | 0 | 3 (15.8) | 0.02¶ | |
| Fever clearance time (h) | 0.06 | NS | 48 (19-74) | 48 (20-145) | 65 (30-136) | NS |
The default tests for columns 3 and 7 are Spearman's rho (ρ) and the Kruskal-Wallis test, respectively, unless otherwise indicated by superscript symbols as follows: †, t test; ¶, Fisher's exact test. NS, not significant.
Values with ranges in parentheses are medians (IQRs), and other values are quantities (percentages).
§, Pearson's correlation coefficient (R).
DISCUSSION
This is the first report of a positive association between O. tsutsugamushi load in admission blood and disease severity in patients with scrub typhus. The bacterial loads in 81 patients ranged between 100 and 1,000 DNA copies/ml in just over half of this group, with approximately one-quarter each having fewer than 100 or more than 1,000 DNA copies/ml. The finding that 74 patients had no detectable O. tsutsugamushi in blood is consistent with a previous study that reported a diagnostic sensitivity for scrub typhus of 44.8% for a 16S rRNA PCR assay (21). Variation in bacterial load could relate to efficiency in host immune response. O. tsutsugamushi is an obligate intracellular pathogen, and the ability to mount an effective host cellular immune response is likely to be crucial for the control of infection but may be variable within the population. The efficiency with which O. tsutsugamushi can invade and multiply within cells of the human host, disseminate to other sites, and seed into the bloodstream could also be influenced by characteristics of the infecting strain. This possibility assumes that the genetic complements of O. tsutsugamushi vary between strains. A phylogenetic analysis of 23 O. tsutsugamushi isolates cultured from patients with scrub typhus presenting to our study site in Udon Thani (n = 19) or in Tak province, northwest Thailand (n = 4), demonstrated that the majority had genotypes related to the Karp-type strain (74%), a lower proportion were related to the Gilliam-type strain (21%), and the remainder clustered with other strains identified previously in Thailand (2). If these results are representative of the larger population, they suggest that the majority of strains infecting our patient group were probably related to a single serotype (Karp), but this does not preclude the possibility of significant variation in genomic content involving genes involved in host-pathogen interactions. The whole-genome sequence of O. tsutsugamushi strain Ikeda (Gilliam serotype) has revealed features that are consistent with a highly plastic genome, including the presence of a large number of foreign genes (15). It is plausible, therefore, that there is considerable variability in genomic content even within a given serotype and that this is associated with variable disease severity in humans. Finally, it is possible that differences in load in the host could relate to the chigger. The main vectors of disease in Thailand are reported to be Leptotrombidium imphalum and Leptotrombidium deliense, with a third species (Leptotrombidium chiangraiensis) having been identified in Chiang Rai, northern Thailand (13). Individual chiggers can be infected by more than one strain of O. tsutsugamushi (18), and it is not clear whether disease in humans differs following a bite from a chigger positive for a single versus multiple strains of O. tsutsugamushi. It is not clear whether the sizes of the infecting inocula vary between the different vector species. During experimental human infection, two volunteers bitten by Leptotrombidium fletcheri developed more-severe clinical disease than two volunteers bitten by Leptotrombidium arenicola (19). Similarly, mice experimentally infected with tissue suspensions prepared from L. arenicola-infected mice showed higher mortality rates and longer durations of illness than mice infected with tissue suspensions prepared from L. fletcheri-infected mice (18). Although these studies had mite-related differences, there were several other variables in these assays, including the possibility of a nonidentical O. tsutsugamushi population present in the different Leptotrombidium species. The presence of an eschar was associated with bacterial load in blood in our study, and we speculate as to whether there is a link between the development of an eschar and the size of initial inoculum or between the presence of an eschar and the virulence potential of the infecting strain.
Our quantitative findings contrast with those of a previous report of seven patients with scrub typhus in which the numbers of DNA copies (originally reported in copies/μl but converted here to copies/ml) ranged from 1.0 × 106 to 2.88 × 107/ml (20). These patients were from Chiang Rai and represented a subset of 126 patients with laboratory-confirmed scrub typhus, recruited into a treatment trial published in the year 2000 (24). The basis for the selection of the seven patients was that they had previously been positive by real-time PCR, and it is possible that they were not representative of the larger group from which they were drawn (20). It is also possible that the ethnicities of these seven patients differed from those of the patients in our study, since the Chiang Rai population in the north includes hill tribe ethnic groups, whereas our study population in northeast Thailand was predominantly Thai. It is also possible that the infecting strains were genetically different. The majority of our patient group would be predicted to have been infected with strains related to the Karp or Gilliam serotype (2). A study of O. tsutsugamushi strains isolated from patients in Vientiane, Laos, on the opposite side of the Mekong river from Udon Thani reported a comparable pattern of genotypes (16). Although the O. tsutsugamushi strains circulating in Chiang Rai may be similar to those found in Thailand and Laos, it is also conceivable that they differ. This would be consistent with a report that strains of O. tsutsugamushi in Chiang Rai are more antibiotic resistant than those elsewhere in Thailand (23).
O. tsutsugamushi becomes disseminated in the human host during scrub typhus. In fatal cases, postmortem histopathological examination has demonstrated bacteria in endothelial cells of the heart, lung, brain, kidney, pancreas, and skin, within cardiac muscle cells, and in macrophages located in the liver and spleen (14). The three patients who died from multiorgan failure in our study had higher bacterial loads in admission blood than survivors. A link between bacterial load and disease severity was also suggested by the positive association with APACHE II score. Other prominent findings were associations between bacterial load and liver and kidney function. Scrub typhus is well documented in the literature as a cause of hepatic dysfunction (1, 6, 7, 9, 12, 22, 26). Renal impairment together with rare cases of renal failure is also recognized to occur during scrub typhus (27), and an elevated creatinine level has been reported to be an independent predictor of mortality (22). Histopathological examination of liver and kidney tissues from individuals with organ impairment during scrub typhus has demonstrated cellular damage that appears to be directly related to cellular invasion by O. tsutsugamushi (11).
In conclusion, scrub typhus is similar to many other bacterial infections in that bacterial burden in the host is related to disease severity. Further study is required to determine whether this is a function of the host, the infecting strain, or a combination of the two.
Acknowledgments
We thank our colleagues at Udon Thani Hospital and the Mahidol-Oxford Tropical Medicine Research Unit for support. We thank Yupin Suputtamongkol, Department of Medicine, Faculty of Medicine Siriraj Hospital, Mahidol University, and Mongkol Chenchittikul from the NIH, Ministry of Public Health of Thailand, for IFA testing; Stuart Blacksell for provision of laboratory cultures of O. tsutsugamushi and Rickettsia spp.; and Vanaporn Wuthiekanun, who provided a laboratory culture of L. interrogans.
This study was funded by a grant awarded to P.S. by the Federation of Infection Society, United Kingdom, and by the Wellcome Trust of Great Britain.
Footnotes
Published ahead of print on 17 December 2008.
REFERENCES
- 1.Aung, T., W. Supanaranond, W. Phumiratanaprapin, B. Phonrat, S. Chinprasatsak, and N. Ratanajaratroj. 2004. Gastrointestinal manifestations of septic patients with scrub typhus in Maharat Nakhon Ratchasima Hospital. Southeast Asian J. Trop. Med. Public Health 35845-851. [PubMed] [Google Scholar]
- 2.Blacksell, S. D., R. Luksameetanasan, T. Kalambaheti, N. Aukkanit, D. H. Paris, R. McGready, F. Nosten, S. J. Peacock, and N. P. Day. 2008. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol. Med. Microbiol. 52335-342. [DOI] [PubMed] [Google Scholar]
- 3.Bozeman, F. M., and B. L. Elisberg. 1963. Serological diagnosis of scrub typhus by indirect immunofluorescence. Proc. Soc. Exp. Biol. Med. 112568-573. [DOI] [PubMed] [Google Scholar]
- 4.Brown, G. W., D. M. Robinson, D. L. Huxsoll, T. S. Ng, and K. J. Lim. 1977. Scrub typhus: a common cause of illness in indigenous populations. Trans. R. Soc. Trop. Med. Hyg. 70444-448. [DOI] [PubMed] [Google Scholar]
- 5.Carrol, E. D., M. Guiver, S. Nkhoma, L. A. Mankhambo, J. Marsh, P. Balmer, D. L. Banda, G. Jeffers, S. A. White, E. M. Molyneux, M. E. Molyneux, R. L. Smyth, and C. A. Hart. 2007. High pneumococcal DNA loads are associated with mortality in Malawian children with invasive pneumococcal disease. Pediatr. Infect. Dis. J. 26416-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanta, C., K. Triratanapa, P. Ratanasirichup, and W. Mahaprom. 2007. Hepatic dysfunction in pediatric scrub typhus: role of liver function test in diagnosis and marker of disease severity. J. Med. Assoc. Thai. 902366-2369. [PubMed] [Google Scholar]
- 7.Deepak, N. A., and N. D. Patel. 2006. Differential diagnosis of acute liver failure in India. Ann. Hepatol. 5150-156. [PubMed] [Google Scholar]
- 8.Hackett, S. J., M. Guiver, J. Marsh, J. A. Sills, A. P. J. Thomson, E. B. Kaczmarski, and C. A. Hart. 2002. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch. Dis. Child. 8644-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, M. L., J. W. Liu, K. L. Wu, S. N. Lu, S. S. Chiou, C. H. Kuo, S. K. Chuah, J. H. Wang, T. H. Hu, K. W. Chiu, C. M. Lee, and C. S. Changchien. 2005. Short report: abnormal liver function in scrub typhus. Am. J. Trop. Med. Hyg. 73667-668. [PubMed] [Google Scholar]
- 10.Kawamura, A., Jr. 1995. Tsutsugamushi disease: an overview, p. 1-34. In A. Kawamura, Jr., H. Tanaka, and A. Tamura (ed.), Tsutsugamushi disease. University of Tokyo Press, Tokyo, Japan.
- 11.Kim, D.-M., D. W. Kang, J. O. Kim, J. H. Chung, H. L. Kim, C. Y. Park, and S.-C. Lim. 2008. Acute renal failure due to acute tubular necrosis caused by direct invasion of Orientia tsutsugamushi. J. Clin. Microbiol. 461548-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, D.-M., K. D. Yu, J. H. Lee, H. K. Kim, and S.-H. Lee. 2007. Controlled trial of a 5-day course of telithromycin versus doxycycline for treatment of mild to moderate scrub typhus. Antimicrob. Agents Chemother. 512011-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerdthusnee, K., B. Khuntirat, W. Leepitakrat, P. Tanskul, T. Monkanna, N. Khlaimanee, I. Inlao, A. Kengluecha, S. Mungviriya, K. Chandranoi, P. Krairojananan, D. Bodhidatta, W. Rodkwamthook, D. Phulsuksombati, N. Sangjun, P. Watcharapichat, J. W. Jones, and R. E. Coleman. 2003. Scrub typhus: vector competence of Leptotrombidium chiangraiensis chiggers and transmission efficacy and isolation of Orientia tsutsugamushi. Ann. N. Y. Acad. Sci. 99025-35. [DOI] [PubMed] [Google Scholar]
- 14.Moron, C. G., V. L. Popov, H. M. Feng, D. Wear, and D. H. Walker. 2001. Identification of the target cells of Orientia tsutsugamushi in human cases of scrub typhus. Mod. Pathol. 14752-759. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama, K., A. Yamashita, K. Kurokawa, T. Morimoto, M. Ogawa, M. Fukuhara, H. Urakami, M. Ohnishi, I. Uchiyama, Y. Ogura, T. Ooka, K. Oshima, A. Tamura, M. Hattori, and T. Hayashi. 2008. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 15185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parola, P., S. D. Blacksell, R. Phetsouvanh, S. Phongmany, J. M. Rolain, N. P. Day, P. N. Newton, and D. Raoult. 2008. Genotyping of Orientia tsutsugamushi from humans with scrub typhus, Laos. Emerg. Infect. Dis. 141483-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 18.Shirai, A., D. L. Huxsoll, A. L. Dohany, R. D. Montrey, R. M. Werner, and E. Gan. 1982. Characterization of Rickettsia tsutsugamushi strains in two species of naturally infected, laboratory-reared chiggers. Am. J. Trop. Med. Hyg. 31395-402. [DOI] [PubMed] [Google Scholar]
- 19.Shirai, A., J. P. Saunders, A. L. Dohany, D. L. Huxsoll, and M. G. Groves. 1982. Transmission of scrub typhus to human volunteers by laboratory-reared chiggers. Jpn. J. Med. Sci. Biol. 359-16. [DOI] [PubMed] [Google Scholar]
- 20.Singhsilarak, T., W. Leowattana, S. Looareesuwan, V. Wongchotigul, J. Jiang, A. L. Richards, and G. Watt. 2005. Short report: detection of Orientia tsutsugamushi in clinical samples by quantitative real-time polymerase chain reaction. Am. J. Trop. Med. Hyg. 72640-641. [PubMed] [Google Scholar]
- 21.Sonthayanon, P., W. Chierakul, V. Wuthiekanun, S. D. Blacksell, K. Pimda, Y. Suputtamongkol, S. Pukrittayakamee, N. J. White, N. P. Day, and S. J. Peacock. 2006. Rapid diagnosis of scrub typhus in rural Thailand using polymerase chain reaction. Am. J. Trop. Med. Hyg. 751099-1102. [PubMed] [Google Scholar]
- 22.Varghese, G. M., O. C. Abraham, D. Mathai, K. Thomas, R. Aaron, M. L. Kavitha, and E. Mathai. 2006. Scrub typhus among hospitalised patients with febrile illness in South India: magnitude and clinical predictors. J. Infect. 5256-60. [DOI] [PubMed] [Google Scholar]
- 23.Watt, G., C. Chouriyagune, R. Ruangweerayud, P. Watcharapichat, D. Phulsuksombati, K. Jongsakul, P. Teja-Isavadharm, D. Bhodhidatta, K. D. Corcoran, G. A. Dasch, and D. Strickman. 1996. Scrub typhus infections poorly responsive to antibiotics in northern Thailand. Lancet 34886-89. [DOI] [PubMed] [Google Scholar]
- 24.Watt, G., P. Kantipong, K. Jongsakul, P. Watcharapichat, D. Phulsuksombati, and D. Strickman. 2000. Doxycycline and rifampicin for mild scrub-typhus infections in northern Thailand: a randomised trial. Lancet 3561057-1061. [DOI] [PubMed] [Google Scholar]
- 25.Yagupsky, P., and F. S. Nolte. 1990. Quantitative aspects of septicemia. Clin. Microbiol. Rev. 3269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, C. H., G. J. Hsu, M. Y. Peng, and T. G. Young. 1995. Hepatic dysfunction in scrub typhus. J. Formos. Med. Assoc. 94101-105. [PubMed] [Google Scholar]
- 27.Yen, T. H., C. T. Chang, J. L. Lin, J. R. Jiang, and K. F. Lee. 2003. Scrub typhus: a frequently overlooked cause of acute renal failure. Ren. Fail. 25397-410. [DOI] [PubMed] [Google Scholar]