Abstract
We report on the use of an electronic microarray to simultaneously type influenza A and B viruses and to distinguish influenza A virus subtypes H1N1 and H3N2 from the potentially pandemic avian virus subtype H5N1. The assay targets seven genes: the H1, H3, H5, N1, and N2 genes of influenza A virus; the matrix protein M1 gene of influenza A virus; and the nonstructural protein (NS) gene of influenza B virus. By combining a two-step reverse transcription-multiplex PCR with typing and subtyping on the electronic microarray, the assay achieved an analytical sensitivity of 102 to 103 copies of transcripts per reaction for each of the genes. The assay correctly typed and subtyped 15 different influenza virus isolates, including two influenza B virus, five A/H1N1, six A/H3N2, and two A/H5N1 isolates. In addition, the assay correctly identified 8 out of 10 diluted, archived avian influenza virus specimens with complete typing and subtyping information and 2 specimens with partial subtyping information. In a study of 146 human clinical specimens that had previously been shown to be positive for influenza virus or another respiratory virus, the assay showed a clinical sensitivity of 96% and a clinical specificity of 100%. The assay is a rapid, accurate, user-friendly method for simultaneously typing and subtyping influenza viruses.
The influenza viruses, members of the family Orthomyxoviridae, have genomes consisting of either seven or eight single-stranded RNA segments (11). Based on differences in the matrix protein and the nucleoprotein, the influenza viruses have been divided into three types: A, B, and C. Type C viruses cause mild respiratory illness in children and young adults, whereas types A and B cause more-severe respiratory illness. The type A viruses are further divided into subtypes on the basis of two proteins on the surface of the virus: hemagglutinin (HA) and neuraminidase (NA). Although 16 different HA subtypes (H1 to H16) and 9 different NA subtypes (N1 to N9) have been identified, only three combinations (H1N1, H2N2, and H3N2) have circulated widely in the human population. Currently, influenza A/H1N1, A/H3N2, and B viruses are responsible for seasonal influenza epidemics (6, 18, 23, 25). Infections with these influenza viruses have a significant social and economic impact. Each year in the United States, influenza virus infections are responsible for more than 200,000 hospitalizations and 36,000 deaths (http://www.cdc.gov/flu/about/disease.htm). Recently, variants of avian influenza virus H5N1 that are highly pathogenic in poultry have been found to infect humans and to be highly virulent (20). Of the 383 people reported to have been infected by this virus to date, 241 have died (http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_05_28/en/index.html). Although human-to-human transmission of H5N1 has been suspected in very rare cases, the possibility of viral mutations that could increase the rate of human transmission has raised concerns that this subtype could pose an important public health threat (http://www.cdc.gov/flu/avian/outbreaks/current.htm).
To enable a quick response to a potential outbreak, it is desirable to have a fast, accurate, and comprehensive diagnostic method capable of simultaneously typing and subtyping influenza viruses. Currently, the diagnostic methods available for identifying influenza viruses include viral culture, direct fluorescent antibody testing, rapid point-of-care immunoassays, real-time reverse transcription-PCR (RT-PCR), sequencing, and multiplex RT-PCR. Although viral culture is the “gold standard” for typing and subtyping of influenza viruses, it usually takes 3 to 7 days to culture the virus (2). Both rapid point-of-care immunoassays (24) and real-time RT-PCR (17, 18) can provide results within 30 min to 1 h, but they do not provide subtype information. Multiplex RT-PCR, which uses multiple primer pairs to amplify the influenza virus genome, can provide an approach to the screening of influenza virus variants. However, analysis of different amplification products from multiplex RT-PCR using traditional agarose gel electrophoresis can be problematic and slow (26). Recently, different types of microarrays in combination with multiplex amplification have been reported for the typing and subtyping of influenza viruses (9, 13, 14, 19, 21, 22, 27).
We report on the development of an influenza virus genotyping assay using an electronic microarray (eMA). This assay distinguishes influenza virus types A and B, and it identifies the common influenza virus A subtypes H1N1 and H3N2 as well as the potentially pandemic avian influenza virus subtype H5N1. The performance of the assay was evaluated using 15 different influenza virus isolates, 10 avian influenza virus H5N1 specimens, and 146 human clinical respiratory specimens.
MATERIALS AND METHODS
Cloned RNA transcripts.
Influenza virus gene transcripts of H1, H3, H5, and N1 from an H5N1 isolate [this N1 transcript is hereafter designated N1(H5)] were made by cloning synthetic constructs. Transcripts of M1, NS, N2, and N1 from an H1N1 isolate [this N1 transcript is hereafter designated N1(H1)] were made by cloning RT-PCR products into pPCR-Script (Stratagene, San Diego, CA), pBluescript (Stratagene), or pET28 (Novagen brand; EMD, Madison, WI). The sequences or viruses used for cloning were A/NewCaledonia/20/99 (H1N1), A/New York/206/2006 (H3N2), A/California/7/2004 (H3N2), A/Vietnam/1203/2004 (H5N1), A/Vietnam/Bl-014/2005 (H5N1), and B/Shanghai/361/2002. RNA transcripts were synthesized using T7 RNA polymerase (MEGAscript kits; Ambion, Austin, TX), purified on NucAway columns (Ambion, Austin, TX), analyzed on polyacrylamide gels, and quantitated by optical density.
Assay design.
The eMA influenza virus genotyping assay was designed to detect eight targets: the matrix gene segment (M1) of influenza A virus; the nonstructural gene segment (NS) of influenza B virus; the HA genes H1, H3, and H5; and the NA genes N1(H1), N2, and N1(H5). Sequences were obtained from GenBank and the Influenza Sequence Database (15) and were then aligned. Highly conserved regions were selected for primers and for capture and discriminator oligonucleotides. The Influenza Primer Design Resource website, developed at the Medical College of Wisconsin (MCW) (www.ipdr.mcw.edu) (1), was used for additional in silico evaluation of the selected primers and capture oligonucleotides. The analysis indicated that coverage for the isolates sequenced from 2002 through 2007 ranged from 93 to 100%.
Multiple sets of primers, capture oligonucleotides, and discriminator oligonucleotides were tested for each of the targets. Primers were initially evaluated in singleplex and analyzed by microcapillary electrophoresis. Primer pairs capable of amplifying approximately 10 copies were subsequently tested by the multiplex assay, initially with single RNA transcripts. Sensitivities of approximately 10 copies per reaction were observed The multiplex PCR was then run with three RNA transcripts that together corresponded to a subtype (such as M1 with H3 and N2). This format resulted in an approximately 10-fold loss in analytical sensitivity and uneven amounts of the three amplicons. PCR parameters and primer concentrations were adjusted to produce more-balanced amounts of the three amplicons.
Capture and discriminator oligonucleotides were screened using the eMA. Discriminators are bifunctional oligonucleotides that are complementary to an amplicon and to one of the universal reporters. The criteria for selection were a high signal level and no detectable cross-reactivity with the other targets. The final assay was composed of 16 PCR primers, 10 capture oligonucleotides, 13 bifunctional discriminator oligonucleotides, and 2 fluorescently labeled universal reporter probes (Tables 1 and 2).
TABLE 1.
PCR primers used for the multiplex influenza assay
| Target | Gene | Primer | Sequence | Amplicon size (bp) |
|---|---|---|---|---|
| FA | M1 | FA forward | 5′-CTT CTA ACC GAG GTC GAA ACG TA-3′ | 233 |
| FA reverse | 5′-ACA AAG CGT CTA CGC TGC AGT CC-3′ | |||
| FB | NS | FB forward | 5′-ATG GCC ATC GGA TCC TCA ACT CAC TC-3′ | 244 |
| FB reverse | 5′-TCA TGT CAG CTA TTA TGG AGC TGT T-3′ | |||
| H1 | HA | H1 forward | 5′-ACA ATA ATA TTT GAG GCA AAT GGA AAT CTA ATA-3′ | 264 |
| H1 reverse | 5′-GAT TGA ATG GAT GGG ATG TTC CT-3′ | |||
| H3 | HA | H3 forward | 5′-CAT GCA GTA CCA AAC GGA AC-3′ | 180 |
| H3 reverse | 5′-CAT CAC ACT GAG GGT CTC CCA A-3′ | |||
| H5 | HA | H5 forward | 5′-GCA TTG GTT ACC ATG CAA ACA A-3′ | 177 |
| H5 reverse | 5′-CGA GGA GCC ATC CAG CTA CAC TAC A-3′ | |||
| N1(H1) | NA | N1(H1) forward | 5′-GAT GGG CTA TAT ACA CAA AAG ACA ACA-3′ | 257 |
| N1(H1) reverse | 5′-TGC TGA CCA TGC AAC TGA TT-3′ | |||
| N1(H5) | NA | N1(H5) forward | 5′-ATA CGG CAA TGG TGT CTG GAT-3′ | 327 |
| N1(H5) reverse | 5′-GCA CCG TCT GGC CAA GA-3′ | |||
| N2 | NA | N2 forward | 5′-AGC ATG GTC CAG CTC AAG TT-3′ | 183 |
| N2 reverse | 5′-CAA GTT CCA TTG ATA CAA ACG CAT TC-3′ |
TABLE 2.
Capture, discriminator, and universal reporter oligonucleotide sequences used for the multiplex influenza virus assaya
| Target | Name | Sequence |
|---|---|---|
| FA | FA capture | 5′-Biotin-TCA GGC CCC CTC AAA GCC GAR ATC GC-3′ |
| FA discriminator1 | 5′-gca gta tat cgc ttg aca AAC CGA GGT CGA AAC GTA-3′ | |
| FA discriminator2 | 5′-gca gta tat cgc ttg aca GTG CCC AGT GAG CGA GGA CT-3′ | |
| FA discriminator3 | 5′-gca gta tat cgc ttg aca TTT GTG TTC ACG CTC ACC G-3′ | |
| FB | FB capture | 5′-Biotin-TAT CCC AAT TTG GTC AAG AGC ACC GAT TAT CAC CAG-3′ |
| FB discriminator | 5′-ctg agt ccg aac att gag ATG GCC ATC GGA TCC TCA ACT CAC TC-3′ | |
| H1 | H1 capture | 5′-Biotin-GGA GCT ATA AAC AGC AGT CTT CCT TTC CAG A-3′ |
| H1 discriminator1 | 5′-ctg agt ccg aac att gag TTG AGG CAA ATG GAA ATC TAA-3′ | |
| H1 discriminator2 | 5′-ctg agt ccg aac att gag TGT CCA AAG TAT GTC AGG AGT-3′ | |
| H3 | H3 capture1 | 5′-Biotin-ATG ACC AAA TTG AAG TTA CTA ATG CTA CTG AGC-3′ |
| H3 capture2 | 5′-Biotin-ATG ACC AAA TTG AAG TCA CTA ATG CTA CTG AGC-3′ | |
| H3 discriminator | 5′-ctg agt ccg aac att gag CAT GCA GTA CCA AAC GGA AC-3′ | |
| H5 | H5 capture | 5′-Biotin-ACG TTC TTT TCC ATT ATT GTG TCA ACC TGC TCT GT-3′ |
| H5 discriminator | 5′-ctg agt ccg aac att gag TTG TTT GCA TGG TAA CCA ATG-3′ | |
| N1(H1) | N1(H1) capture | 5′-Biotin-TAT AGG GCC TTA ATG AGC TGT CCT CTA GG-3′ |
| N1(H1) discriminator1 | 5′-gca gta tat cgc ttg aca GCT CTA TTA AAT GAC AAA CAT TCA-3′ | |
| N1(H1) discriminator2 | 5′-gca gta tat cgc ttg aca TCC GTC CCC ATA CAA TTC AAA G-3′ | |
| N1(H5) | N1(H5) capture1 | 5′-Biotin-TTT GAA ATG ATT TGG GAT CCA AAT GGG TGG AC-3′ |
| N1(H5) capture2 | 5′-Biotin-TTT GAA ATG ATT TGG GAT CCA AAT GGA TGG AC-3′ | |
| N1(H5) discriminator | 5′-gca gta tat cgc ttg aca ttt ttt gca gta tat cgc ttg aca ATC CAG ACA CCA TTG CCG T-3′ | |
| N2 | N2 capture | 5′-Biotin-GAT GGA AAA GCA TGG CTG CAT GTT TGT GT-3′ |
| N2 discriminator1 | 5′-gca gta tat cgc ttg aca AGC ATG GTC CAG CTC AAG TT-3′ | |
| N2 discriminator2 | 5′-gca gta tat cgc ttg aca GAT GAT AAA AAT GCA ACT GCT AG-3′ | |
| Red universal reporters | 5′-TGT CAA GGG ATA TAC TGC-red fluorescent dye-3′ | |
| Green universal reporters | 5′-CTC AAT GTT CGG ACT CAG-green fluorescent dye-3′ | |
| I19 | Nonspecific capture | 5′-Biotin-CAG ATG GAA GAC TCT TGT AAT TAT TTT TCA TTA CCT-3′ |
Lowercase letters represent sequences complementary to the red universal reporter sequence; lowercase underlined letters represent sequences complementary to the green universal reporter sequence.
Multiplex RT-PCR.
A two-step RT-PCR was used for multiplex amplification. Three microliters of RNA was added to 17 μl of RT mixture, containing 1× PCR buffer II (Applied Biosystems, Foster City, CA), 2.5 μM random hexamer, 50 U of murine leukemia virus reverse transcriptase, and 20 U of RNase inhibitor (Applied Biosystems), and was incubated at 22°C for 5 min, 42°C for 14 min, and 95°C for 1 min. The 20-μl RT reaction mixture was combined with 30 μl of a PCR mixture. The final 50-μl PCR mixture contained the primers (synthesized by Integrated DNA Technologies, Coralville, IA) for all eight targets, 1× PCR buffer II, 3.5 mM MgCl2, 2.4 mM deoxynucleoside triphosphates, and 2.5 U of FastStart Taq DNA polymerase (Roche Applied Science, Indianapolis, IN). The primer concentrations were 0.2 μM, except for the H3 and H5 primers, whose concentrations were reduced to 0.1 μM, and the N1(H5) primer, whose concentration was increased to 0.45 μM. The thermal profile used was 95°C for 10 min, followed by 40 cycles of PCR at 95°C for 5 s, 58°C for 15 s, and 72°C for 30 s. The RT-PCR amplification (both RT and PCR) was achieved within 100 min on a GeneAmp PCR system, model 9700 (Applied Biosystems). Initially, PCR products were analyzed on a LabChip 90 microcapillary electrophoresis system (Caliper Life Sciences, Hopkinton, MA). During the latter part of development, products were analyzed on the eMA using the Nanochip 400 (NC400) instrument (Nanogen, San Diego, CA).
Typing and subtyping on the Nanochip 400.
The Nanochip 400 has been described previously (4, 7). The specific addressing and reporting protocols used for this assay are as follows.
(i) Capture oligonucleotide addressing.
Five capture mixes, each containing 100 nM concentrations of two or three biotinylated capture oligonucleotides complementary to two different amplicons, were sequentially addressed for 15 s at 350 nA to a number of electrode sites equal to the number of samples being analyzed.
(ii) Amplicon addressing and hybridization.
RT-PCR products were diluted 1 to 8 in CAPdown sample buffer A (Nanogen, San Diego, CA), consisting of 114 mM histidine and 142.5 mM 1-thioglycerol. The diluted material was addressed at 995 nA for 2 min to one set of five electrode sites.
(iii) Reporting.
A mix of discriminator (500 nM each) and reporter (2.5 μM green reporter, 4.5 μM red reporter) oligonucleotides was loaded onto the eMA and hybridized for 1 min. The eMA was washed at 47°C in a high-salt buffer (Nanogen), cooled to 24°C, and imaged with the instrument's charge-coupled device camera at 553 nm and 668 nm for green and red fluorescent signals, respectively. The relative fluorescence units (RFU) were determined on each test electrode and on a background electrode containing a nonspecific capture oligonucleotide, and the ratio of the signal on the test electrode to that on the background electrode (signal-to-noise ratio [SNR]) was calculated. The positive cutoff was set to a minimum SNR of 3.0 based on previous experience with the eMA for assays with multiple targets (5, 10, 12).
Analytical sensitivity.
Tenfold serial dilutions of RNA transcripts were used. A single NS transcript was used for influenza B virus, and equal concentrations of three transcripts (an M1, an HA, and an NA transcript) were used for influenza A viruses. At each concentration, three replicates were run. Analytical sensitivity was defined as the lowest copy number at which complete type and subtype information was successfully detected on the eMA for all three replicates.
Analytical specificity.
Analytical specificity was evaluated using 15 different influenza virus isolates, including 2 influenza B viruses (B/Hong Kong/330/01; B/Shanghai/361/02) and 13 influenza A viruses, of which 5 were H1N1 (A/New Caledonia/20/99, A/Beijing/95, and specimens 120905A, 011303A, and 011503A), 6 were H3N2 (A/Wyoming/03/3, A/California/7/04, A/Fujian/411/02, A/Sydney/97, A/Panama/99, and specimen 121905A-3 M5), and 2 were H5N1 (A/Vietnam/1194/04 and a culture of a human H5N1 specimen). These virus isolates had been tested previously by culture and were obtained from different sources. Nucleic acids were prepared by processing 100 to 500 μl of virus isolates using the MagNA Pure compact nucleic acid isolation kit 1 system (Roche Applied Science, Indianapolis, IN) or the QIAamp MinElute virus spin kit (Qiagen, Valencia, CA) according to the manuals and were eluted in 50 μl of elution buffer.
Human nasopharyngeal specimens and avian specimens.
The majority of the human nasopharyngeal specimens were collected from 2004 through 2006 at the MCW; 15 of the influenza B virus specimens were collected from 1996 though 1998. Nucleic acids from the specimens were prepared from either 400 μl (human nasopharyngeal specimens) or 200 μl (avian specimens) of sample by using Total Nucleic Acid spin columns (Roche Applied Science) and were eluted into 50 μl. The specimens had been analyzed previously at the MCW by using several different reference methods. Influenza virus, respiratory syncytial virus, human parainfluenza virus type 1 (HPIV-1), and HPIV-3 were analyzed using multiplex RT-PCR followed by an enzyme hybridization assay (EHA) (3, 5) and/or by tissue culture. Human adenovirus, enterovirus, and rhinovirus were analyzed by real-time PCR and multiplex RT-PCR followed by an EHA (3, 5).
RESULTS
Analytical sensitivity and specificity.
The analytical sensitivity of the eMA influenza virus genotyping assay, as shown in Table 3, was 102 copies per reaction for influenza B virus and for influenza A virus subtypes H1N1 and H3N2. For influenza A virus subtype H5N1, the analytical sensitivity was 103 copies per reaction, because only one of three replicates was detected with complete typing (M1) and subtyping (H5 and N1) at 102 copies per reaction.
TABLE 3.
Analytical sensitivity of the assay with RNA transcripts
| Genotype | Gene | No. of positive calls/no. of samples tested with the following no. of copies per RT reaction:
|
Analytical sensitivity (no. of copies/RT reaction) | |||
|---|---|---|---|---|---|---|
| 105 | 104 | 103 | 102 | |||
| B | NS1 | 3/3 | 3/3 | 3/3 | 3/3 | 102 |
| A/H1N1 | M1 | 3/3 | 3/3 | 3/3 | 3/3 | 102 |
| H1 | 3/3 | 3/3 | 3/3 | 3/3 | 102 | |
| N1 | 3/3 | 3/3 | 3/3 | 3/3 | 102 | |
| A/H3N2 | M1 | 3/3 | 3/3 | 3/3 | 3/3 | 102 |
| H3 | 3/3 | 3/3 | 3/3 | 3/3 | 102 | |
| N2 | 3/3 | 3/3 | 3/3 | 3/3 | 102 | |
| A/H5N1 | M1 | 3/3 | 3/3 | 3/3 | 3/3 | 102 |
| H5 | 3/3 | 3/3 | 3/3 | 3/3 | 102 | |
| N1 | 3/3 | 3/3 | 3/3 | 1/3 | 103 | |
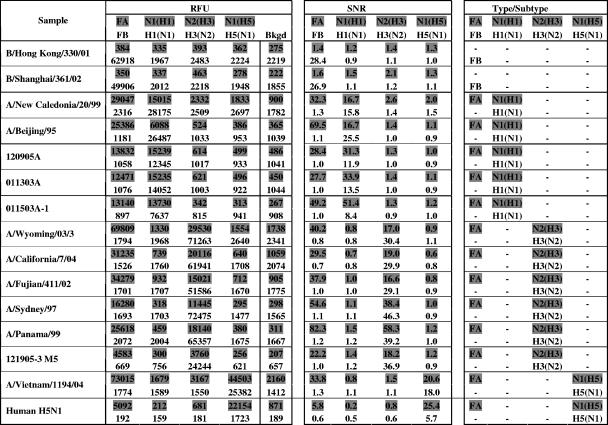
The analytical specificity of the eMA influenza virus genotyping assay was evaluated by testing 15 cultured virus isolates: 2 influenza B viruses and 13 influenza A viruses (5 H1N1, 6 H3N2, and 2 H5N1). The results (Fig. 1) indicated no cross-reaction between the different gene targets; no wrong types or subtypes were observed across the 15 virus isolates. In addition, the eMA influenza virus genotyping assay has been demonstrated to correctly type and subtype 14 cultured A/H5N1 virus isolates representing multiple clades from 2002 to 2006 (8).
FIG. 1.
Representative NC400 data from the eMA influenza virus genotyping assay, showing correct detection of 15 influenza virus isolates at 1/10 dilutions of RNA, with no wrong types or subtypes across different virus isolates. Red fluorescent signals are represented by shading. Green fluorescent signals are represented by unshaded numbers. The SNR is the ratio of the RFU on a test site to the RFU on a background site (Bkgd). Typing and subtyping are called when the SNR is ≥3.0.
Clinical evaluation.
The clinical performance of the assay was first evaluated using 146 human clinical nasopharyngeal samples collected at the MCW (Table 4). Previously, influenza A virus had been detected in 59 specimens and influenza B virus in 45 specimens. Forty-two of the specimens were reported to be negative for influenza virus but positive for other respiratory viruses (4 for HPIV-1, 9 for HPIV-3, 5 for respiratory syncytial virus, 12 for rhinovirus, 5 for enterovirus, and 7 for adenovirus). In comparison to the reference methods (culture, real-time PCR, multiplex PCR, and EHA), the eMA influenza virus genotyping assay correctly identified 55 out of 59 (93%) samples as influenza virus A and 41 out of 45 (91%) samples as influenza virus B. Among the 55 influenza A virus-positive samples, 7 (13%) were subtyped as H1N1 and 48 (87%) were subtyped as H3N2. None of the influenza virus-negative specimens tested positive by the eMA influenza virus genotyping assay. Based on these results, the clinical sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated to be 93.2%, 100%, 100%, and 95.6%, respectively, for influenza A virus and 91.1%, 100%, 100%, and 96.2%, respectively, for influenza B virus. For influenza A and B viruses combined, the clinical sensitivity, specificity, PPV, and NPV were 92.3%, 100%, 100%, and 84.0%, respectively (Table 4).
TABLE 4.
Performance of eMA influenza virus genotyping assay compared with the reference methods for 146 human clinical samples
| Target and result by the eMA influenza virus genotyping assay | No. of samples with the following result by the reference methodsa:
|
Performance characteristic (%)
|
|||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | Sensitivity | Specificity | PPV | NPV | |
| FA | 93.2 | 100 | 100 | 95.6 | |||
| Positive | 55 | 0 | 55 | ||||
| Negative | 4 | 87 | 91 | ||||
| Total | 59 | 87 | 146 | ||||
| FB | 91.1 | 100 | 100 | 96.2 | |||
| Positive | 41 | 0 | 41 | ||||
| Negative | 4 | 101 | 105 | ||||
| Total | 45 | 101 | 146 | ||||
| FA + FB | 92.3 | 100 | 100 | 84.0 | |||
| Positive | 96 | 0 | 96 | ||||
| Negative | 8 | 42 | 50 | ||||
| Total | 104 | 42 | 146 | ||||
Results by the reference methods are the combined results of culture, real-time PCR, multiplex RT-PCR, and EHA.
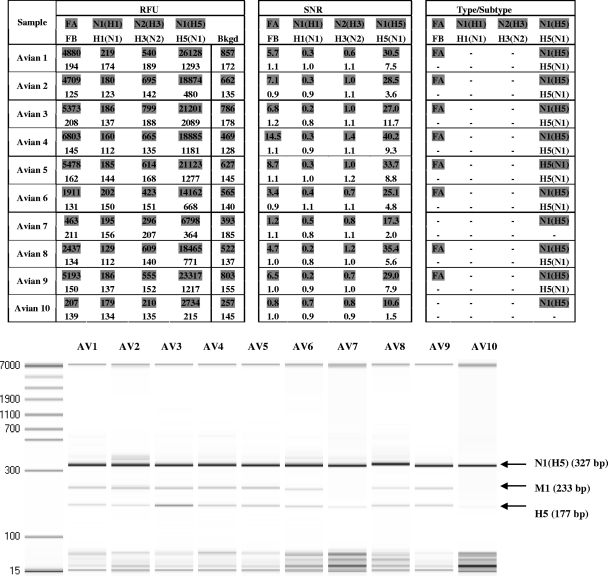
The clinical performance of the assay was further evaluated using 1/102 dilutions of 10 purified avian specimens (Fig. 2). The eMA influenza virus genotyping assay correctly typed and subtyped the viruses in 8 of these 10 (80%) samples as influenza virus A H5N1 compared to culture and multiplex PCR, used as the reference methods. Two samples (Avian 7 and Avian 10) were positive for N1(H5) but negative for M1 and H5.
FIG. 2.
NC400 data from the eMA influenza virus genotyping assay and microcapillary electrophoresis gel, showing the detection of influenza virus in 10 avian specimens at 1/102 dilutions of RNA. Red fluorescent signals are represented by shading. Green fluorescent signals are represented by unshaded numbers. The SNR is the ratio of the RFU on a test site to the RFU on a background site (Bkgd). Typing and subtyping are called when the SNR is ≥3.0.
Analysis of discrepant specimens.
The eMA influenza virus genotyping assay failed to detect 8 of 104 (7.7%) human clinical nasopharyngeal samples that were initially influenza virus positive by the reference methods (culture, real-time PCR, multiplex PCR, and EHA). Gel analysis of the amplicons of these eight discrepant samples revealed no PCR bands for any genes (data not shown). These eight samples were retested by both an EHA and the eMA assay. Of the four influenza A virus-discrepant samples, two retested positive and two retested negative by EHA. All four retested negative with the eMA. Similar retesting results were obtained for the four influenza B virus-discrepant samples: two were positive and two negative by EHA, and all four were negative with the eMA. It is possible that degradation of the RNA occurred in the four samples that were negative by both methods at the time of retesting. When these four specimens were removed from the analysis, the recalculated clinical sensitivity and NPV were 96.5% and 97.8%, respectively, for influenza A virus and 95.3% and 98.1%, respectively, for influenza B virus. For influenza A and B viruses combined, the clinical sensitivity and NPV were 96.0% and 92.0%, respectively. The specificity and PPV remained at 100%.
For the two avian samples (Avian 7 and Avian 10) that were positive for N1(H5) and negative for M1 and H5, no retest could be conducted by either EHA or eMA due to the limited availability of the original samples. Microcapillary electrophoresis gel analysis of the PCR products of these 10 avian specimens confirmed that little or none of the M1 and H5 amplicons were produced from the Avian 7 and Avian 10 specimens (Fig. 2).
DISCUSSION
We have described the development of an eMA influenza virus genotyping assay for detecting and identifying the types and subtypes of human influenza A (H1N1 and H3N2) and influenza B viruses and avian H5N1 viruses. The assay has been validated with 15 influenza virus isolates, 146 human clinical nasopharyngeal specimens, and 10 avian specimens.
One concern regarding nucleic acid assays for influenza A virus subtyping is that they may be short-lived due to constant viral mutations (18, 21) The oligonucleotide set for the eMA influenza virus genotyping assay was designed based on recent sequences (2002 to 2007) with in silico coverage rates of 93 to 100% for the eight targets. However, in silico analysis suggests that the selected oligonucleotide set would have performed well at detecting H3 and H1 viruses isolated 10 years ago. More importantly, in silico analyses of the sequences of the 2008-to-2009 season influenza virus vaccine strains (for H1N1, A/Brisbane/59/2007; for H3N2, A/Brisbane/10/2007; B/Florida/4/2006) suggest that the eMA influenza virus genotyping assay described here would detect these newer virus strains.
The specific detection of the H5N1 subtype is more problematic, due to the close relationship of the H5N1 HA and NA to H5 from avian H5N2 viruses and N1 from avian H1N1, H6N1, and H7N1 viruses. The H5 oligonucleotide assay set would be expected to detect a subset of H5 genes from avian H5N2 viruses, and the N1 oligonucleotide set would be expected to detect N1 genes associated with other avian virus subtypes, such as H1N1, H6N1, and H7N1. Thus, for identification of H5N1 viruses, it is necessary that both the H5 and the associated N1 be positive.
The eMA influenza virus genotyping assay demonstrated good analytical and clinical specificity. The analytical sensitivity and clinical sensitivity of the assay are comparable to those reported for other microarray-based influenza virus typing and subtyping methods (9, 16, 21, 27). In addition, the eMA influenza virus genotyping assay provided subtype information for 55 influenza A virus-positive specimens—7 identified as H1N1 and 48 as H3N2—although the results were not validated using culture, the gold standard, at this time; such testing is planned in the future. The eMA influenza virus genotyping assay identified the viruses in 80% (8 of 10) of the avian specimens as A/H5N1 when 1/102-diluted RNA was used. The other two samples tested positive for N1(H5) and negative for M1 and H5. The analytical sensitivity suggested that M1 and H5 were more sensitive than N1(H5). The negative M1 and H5 results for the two avian specimens could be caused by deterioration of diluted RNA during shipping and handling or by mutations occurring in the regions of the M1 and H5 primers. The latter situation could not be verified without the availability of the sequences for these specimens.
Among the several microarrays reported for the typing and subtyping of influenza viruses (9, 13, 14, 19, 21, 22, 27), a majority used at least 55 oligonucleotides as primers and detecting probes. On the other hand, the eMA influenza virus genotyping assay requires a total of 41 oligonucleotides: 16 PCR primers, 10 capture oligonucleotides, 13 bifunctional discriminator oligonucleotides, and 2 fluorescently labeled universal reporter oligonucleotides. The work flow of the assay is simplified by combining a two-step multiplex RT-PCR assay with automated detection on an eMA. The NC400 instrument can be programmed to load the capture oligonucleotides, dilute the PCR products, perform the hybridizations, and process the data. Hands-on post-PCR procedures such as dilutions and hybridization, required by many microarray-based assays (9, 13, 14, 21, 27), can be accomplished automatically by the NC400 instrument. Although this feature of the instrument was not employed during the experiments reported here, it has been used in other assays (10). This desirable feature helps to minimize potential carryover contamination in clinical testing.
The assay can be performed rapidly: the entire procedure from sample extraction (30 min) to multiplex RT-PCR (95 min) to detection on the eMA (30 min) can be achieved in less than 3.5 h for 8 samples and approximately 6.5 h for 64 samples. This speed is comparable to or better than those of other microarray-based assays (21, 27). Although real-time RT-PCR can provide the results within 1 h (17, 18), it is typically designed for detecting one to three targets per reaction. For the detection of multiplex (e.g., eight) targets in the eMA influenza virus genotyping assay, the samples have to be divided into multiple reaction tubes. This may be an issue when the amount of sample is small.
In summary, we have developed a rapid, accurate, user-friendly multiplex assay for simultaneously detecting influenza A and B viruses and for identifying the two major subtypes currently circulating in humans, A/H1N1 and A/H3N2, as well as the avian virus subtype A/H5N1. This assay could fill a niche between simple typing of influenza A and B viruses and the more complex, technically challenging microarrays. The assay could provide a method for influenza surveillance, where early detection and rapid subtype information can be important.
Acknowledgments
We thank the New Mexico State Health Laboratory and Guy Boivin of Laval University for providing cultured viruses.
This work was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI070428 and AI066584), and HX Diagnostics.
Footnotes
Published ahead of print on 10 December 2008.
REFERENCES
- 1.Bose, M. E., J. C. Littrell, A. D. Patzer, A. J. Kraft, J. A. Metallo, J. Fan, and K. J. Henrickson. 2008. The Influenza Primer Design Resource: a new tool for translating influenza sequence data into effective diagnostics. Influenza Other Respir. Viruses 223-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis, J. S., and M. C. Zambon. 2002. Molecular diagnosis of influenza. Rev. Med. Virol. 12375-389. [DOI] [PubMed] [Google Scholar]
- 3.Fan, J., K. J. Henrickson, and L. L. Savatski. 1998. Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (hexaplex). Clin. Infect. Dis. 261397-1402. [DOI] [PubMed] [Google Scholar]
- 4.Heller, M. J. 1996. An active microelectronics device for multiplex DNA analysis. Eng. Med. Biol. 15100-104. doi: 10.1109/51.486725. [DOI] [Google Scholar]
- 5.Henrickson, K. J., A. Kraft, D. Canter, and J. Shaw. 2007. Comparison of electronic microarray to enzyme hybridization assay for multiplex reverse-transcriptase PCR detection of common respiratory viruses in children. Clin. Microbiol. Newsl. 29113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes, E. C., J. K. Taubenberger, and B. T. Grenfell. 2005. Heading off an influenza pandemic. Science 309989. [DOI] [PubMed] [Google Scholar]
- 7.Huang, Y., J. Shirajian, A. Schroder, Z. Yao, T. Summers, D. Hodko, and R. Sosnowski. 2004. Multiple sample amplification and genotyping integrated on a single electronic microarray. Electrophoresis 253106-3116. [DOI] [PubMed] [Google Scholar]
- 8.Huang, Y., H. Tang, S. Duffy, M. Ghosh, S. Norman, K. Henrickson, A. Kraft, W. G. Weisburg, and E. L. Mather. 2007. Abstr. 23rd Ann. Clin. Virol. Symp., abstr. Tp-4.
- 9.Kessler, N., O. Ferraris, K. Palmer, W. Marsh, and A. Steel. 2004. Use of the DNA flow-through chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J. Clin. Microbiol. 422173-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar, S., L. Wang, J. Fan, A. Kraft, M. E. Bose, S. Tiwari, M. Van Dyke, R. Haigis, T. Luo, M. Ghosh, H. Tang, M. Haghnia, E. L. Mather, W. G. Weisburg, and K. J. Henrickson. 2008. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J. Clin. Microbiol. 463063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb, R. A. 1989. Genes and proteins of the influenza viruses, p. 1-88. In R. M. Krug, H. Fraenkel-Conrat, and R. R. Wagner (ed.), The influenza viruses. Plenum Press, New York, NY.
- 12.Li, H., M. A. McCormac, R. W. Estes, S. E. Sefers, R. K. Dare, J. D. Chappell, D. D. Erdman, P. F. Wright, and Y. W. Tang. 2007. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J. Clin. Microbiol. 452105-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodes, M. J., D. Suciu, M. Elliott, A. G. Stover, M. Ross, M. Caraballo, K. Dix, J. Crye, R. J. Webby, W. J. Lyon, D. L. Danley, and A. McShea. 2006. Use of semiconductor-based oligonucleotide microarrays for influenza A virus subtype identification and sequencing. J. Clin. Microbiol. 441209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. D. M. E. Osterhaus, N. Cox, and A. W. Hampson (ed.), Options for the control of influenza IV. Elsevier Science, Amsterdam, The Netherlands.
- 16.Mehlmann, M., A. B. Bonner, J. V. Williams, D. M. Dankbar, C. L. Moore, R. D. Kuchta, A. B. Podsiad, J. D. Tamerius, E. D. Dawson, and K. L. Rowlen. 2007. Comparison of the MChip to viral culture, reverse transcription-PCR, and the QuickVue influenza A+B test for rapid diagnosis of influenza. J. Clin. Microbiol. 451234-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng, E. K. O., P. K. C. Cheng, A. Y. Y. Ng, T. L. Hoang, and W. W. L. Lim. 2005. Influenza A H5N1 detection. Emerg. Infect. Dis. 111303-1305. www.cdc.gov/eid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petric, M., L. Comanor, and C. A. Petti. 2006. Role of the laboratory in diagnosis of influenza during seasonal epidemics and potential pandemics. J. Infect. Dis. 194S98-S110. [DOI] [PubMed] [Google Scholar]
- 19.Sengupta, S., K. Onodera, A. Lai, and U. Melcher. 2003. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 414542-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shortridge, K. F., J. S. M. Peiris, and Y. Guan. 2003. The next influenza pandemic: lessons from Hong Kong. J. Appl. Microbiol. 9470S-79S. [DOI] [PubMed] [Google Scholar]
- 21.Townsend, M. B., E. D. Dawson, M. Mehlmann, J. A. Smagala, D. M. Dankbar, C. L. Moore, C. B. Smith, N. J. Cox, R. D. Kuchta, and K. L. Rowlen. 2006. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J. Clin. Microbiol. 442863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Z., L. T. Daum, G. J. Vora, D. Metzgar, E. A. Walter, L. C. Canas, A. P. Malanoski, B. Lin, and D. A. Stenger. 2006. Identifying influenza viruses with resequencing microarrays. Emerg. Infect. Dis. 12638-646. www.cdc.gov/eid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg, A., and M. L. Walker. 2005. Evaluation of three immunoassay kits for rapid detection of influenza virus A and B. Clin. Diagn. Lab. Immunol. 12367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegler, T., and N. J. Cox. 1995. Influenza viruses, p. 918-919. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, DC.
- 26.Zou, S. 1997. A practical approach to genetic screening for influenza virus variants. J. Clin. Microbiol. 352623-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou, S., J. Han, L. Wen, Y. Liu, K. Cronin, S. H. Lum, L. Gao, J. Dong, Y. Zhang, Y. Guo, and Y. Shu. 2007. Human influenza A virus (H5N1) detection by a novel multiplex PCR typing method. J. Clin. Microbiol. 451889-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]