Luong et al. (7) recently described the first report and characterization of Aurantimonas altamirensis recovered from human clinical specimens. According to that report, the isolates were described as being “possibly associated” with infection. In April 2008, a 50-year-old male patient with a 10-day history of scrotal swelling and redness was admitted to Methodist Hospital in Indianapolis, IN. An A. altamirensis (002-2918A) bacteremia isolate recovered on the third hospital day (HD) was presumptively associated with the scrotum infection, which we describe below.
The patient had numerous prior hospitalizations with a complicated clinical history of diabetes mellitus, hypertension, congestive heart failure (CHF), peripheral neuropathy, hyperthyroidism, hyperlipidemia, and chronic renal failure (stage IV). In addition, both legs had been amputated below the knees due to complications of diabetes mellitus. On admission, the patient presented with fever and dysuria and complained of urinary frequency for 14 days prior to hospitalization. Significant findings on physical examination included swelling and pitting edema of the scrotum and swelling of the lower abdominal skin and extremities with suprapubic pain. Prominent redness and tenderness of the scrotum and adjacent area along with swelling of the thighs were also observed.
The scrotal swelling was presumptively diagnosed as secondary to CHF or infection. Intravenous Lasix was administered due to exacerbation of CHF during hospitalization along with clindamycin, vancomycin, and ciprofloxacin and continuation of his current home medications. Two sets of blood cultures were drawn. Swelling of the scrotum decreased at the second HD, and Lasix doses were decreased, following control of the swelling of the scrotum and extremities. At the third HD, both sets of aerobic blood culture bottles yielded a gram-negative coccobacillus. The organism was initially identified using Vitek 2 (bioMerieux, Hazelwood, MO) and conventional biochemical tests (8), which provided identification as Brucella spp. and CDC group EO-3, respectively. The 002-2918A isolate was susceptible to ciprofloxacin (Etest; AB Biodisk, Solna, Sweden); thus, clindamycin and vancomycin were discontinued and the patient was maintained only on ciprofloxacin (250 mg; twice a day). The patient was discharged on oral ciprofloxacin (250 mg; twice a day for a week).
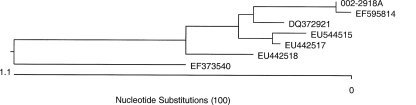
The 002-2918A isolate was forwarded to JMI Laboratories (North Liberty, IA) as part of the SENTRY Antimicrobial Surveillance Program (5), where bacterial identification was performed by 16S rRNA sequencing. The sequence was compared with a DNA library using BIBI (Bioinformatics Bacterial Identification) available through the Internet (http://pbil.univ-lyon1.fr/bibi) (4). The isolate identification was confirmed as A. altamirensis. The 16S rRNA sequence displayed greatest identity with that of A. altamirensis under accession number EF595814 (99.9%) (7), followed by DQ372921 (99.6%) (6) and EU442517 and EU442518 (99.5%) (7) (Fig. 1).
FIG. 1.
Phylogenetic relationships obtained by the ClustalW method using the 16S rRNA gene sequences from Aurantimonas spp. The accession numbers EF595814 (7), DQ372921 (6), EU544515 and EU442517 (7), and EU442518 (7) represent A. altamirensis species. The accession number EF373540 represents Aurantimonas kwangyangensis used as the outlier species.
The isolate was tested for susceptibility using the broth microdilution method as described by the Clinical and Laboratory Standards Institute (CLSI; M7-A7, 2006) (1). MICs were interpreted according to the breakpoints for Pseudomonas aeruginosa and other non-Enterobacteriaceae published in the M100-S18 document (CLSI, 2008) (2). The 002-2918A isolate was fully susceptible to 32 antimicrobial agents, except for a modestly elevated MIC for aztreonam (8 to 16 μg/ml); however, previous publications have reported isolates displaying a phenotype of resistance to fluoroquinolones, trimethoprim-sulfamethoxazole, and nitrofurantoin (3, 7).
The clinical relevance of the genus Aurantimonas has remained uncertain, since few reports have demonstrated an etiological role for the species, and it has usually been implicated as a contaminant derived from environmental and/or water sources (7). However, we describe a case of A. altamirensis bacteremia possibly associated with a scrotum infection. The presence of clinical symptoms of infection and the clinical response to appropriate antimicrobial therapy suggest an etiological role for this organism. Additionally, this report emphasizes the major diagnostic challenges when treating immunocompromised patients with complicated and unusual infectious processes and the limitations of contemporary automated identification systems.
Footnotes
Published ahead of print on 26 November 2008.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2006. M7-A7. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 2.Clinical and Laboratory Standards Institute. 2008. M100-S18. Performance standards for antimicrobial susceptibility testing, 18th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Daley, D., S. Neville, and K. Kociuba. 1997. Peritonitis associated with a CDC group EO-3 organism. J. Clin. Microbiol. 353338-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devulder, G., G. Perrière, F. Baty, and J. P. Flandrois. 2003. BIBI, a bioinformatics bacterial identification tool. J. Clin. Microbiol. 411785-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones, R. N. 2003. Global epidemiology of antimicrobial resistance among community-acquired and nosocomial pathogens: a five-year summary from the SENTRY Antimicrobial Surveillance Program (1997-2001). Semin. Respir. Crit. Care Med. 24121-134. [DOI] [PubMed] [Google Scholar]
- 6.Jurado, V., J. M. Gonzalez, L. Laiz, and C. Saiz-Jimenez. 2006. Aurantimonas altamirensis sp. nov., a member of the order Rhizobiales isolated from Altamira Cave. Int. J. Syst. Evol Microbiol. 562583-2585. [DOI] [PubMed] [Google Scholar]
- 7.Luong, M. L., S. Bekal, D. C. Vinh, D. Lauzon, V. Leung, G. N. Al-Rawahi, B. Ng, T. Burdz, and K. Bernard. 2008. First report of isolation and characterization of Aurantimonas altamirensis from clinical samples. J. Clin. Microbiol. 462435-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreckenberger, P. C., and D. Lindquist. 2007. Algorithms for identification of aerobic gram-negative bacteria, p. 371-376. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.