Abstract
We studied the epidemiology of the recently described serotype 6C of Streptococcus pneumoniae among a collection of carriage isolates recovered between 1996 and 2007 in Portugal. Of 4,064 isolates, 106 (2.6%) were of serotype 6C, 17.9% of which were multidrug resistant. The strains were genetically diverse.
The polysaccharide capsule of Streptococcus pneumoniae is considered to be the most important virulence factor in this species. It is antigenically diverse, and its identification by serology (Quellung reaction) has been used for decades as a primary criterion for the classification of pneumococci (18). Up to now, 91 serotypes have been described, with serotype 6C being the most recent one (14). The latter is indistinguishable from serotype 6A by the Quellung reaction, although it contains a galactose molecule instead of a glucose in the capsular repeating oligosaccharide unit due to the presence of a different glycosyl transferase (wciN) gene (14). Serotypes 6A and 6C, together with serotype 6B, constitute serogroup 6, which has related capsular structures (1, 10). The putative serotype 6D that would result from the introduction of wciN6C into the serotype 6B operon has been proposed, although it remains unidentified (7).
The 7-valent pneumococcal conjugate vaccine (PCV7, targeting serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F) was introduced in the United States in 2000 and became commercially available in several European countries during 2001. PCV7 confers protection, within serogroup 6, against serotype 6B. The most recent data available suggest that it provides cross-protection against disease caused by serotype 6A but not that caused by serotype 6C (12).
Due to its recent identification, data on the epidemiology of serotype 6C are still very scarce (13). A recent study from South Africa found that serotype 6C strains had a higher propensity to cause meningitis than did serotype 6A or 6B strains (5). A study from the CDC in the United States reported that in 2006, the rate of invasive disease caused by serotype 6C was significantly higher than that in 1999 but lower for serotype 6A (3). Another very recent publication from the United States also documented the increasing prevalence of this serotype after 2001 (8).
In this study, we describe the epidemiology of serotype 6C strains colonizing healthy Portuguese preschool children in studies conducted between 1996 and 2007.
We screened 4,064 S. pneumoniae strains isolated from a total of 6,559 nasopharyngeal samples obtained from children attending day care centers in the Lisbon and Oeiras areas of Portugal, in studies conducted in nine different years between 1996 and 2007: 1996 to 1999, 2001 to 2003, and 2006 to 2007. All samples were collected between January and March of each year. Identification of pneumococci was done as previously described (17). The serotypes of drug-resistant isolates (n = 1,659) were reported previously (9, 11, 16, 17).
Antimicrobial susceptibility testing was performed using the Kirby-Bauer technique according to Clinical and Laboratory Standards Institute recommendations and definitions (4) for chloramphenicol, erythromycin, clindamycin, tetracycline, and sulfamethoxazole-trimethoprim. MICs of penicillin and ceftriaxone were determined by use of an Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations and were interpreted according to CLSI guidelines (4). Multidrug resistance was defined as resistance to three or more antibiotics tested.
To identify strains of serogroup 6, all isolates lacking serotype assignment were screened by PCR for the presence of a specific region of the wzy gene as described previously (2). Specifically, two primer pairs were used, 6Bwzy-f (5′-CGA CGT AAC AAA GAA CTA GGT GCT GAA AC-3′) and 6Bwzy-r (5′-AAG TAT ATA ACC CTG TAA AAC TCT GAC-3′), generating a product of 200 bp for all serogroup 6 strains, and cpsA-f (5′-GGT GTT CTC TAT CCT TGT CAG CTC TGT GTC GCT C-3′) and cpsA-r (5′-GTG TGA ATG GTC GAA TCA ACT CTA TAA ATG CC-3′), generating a product of 657 bp. The highly conserved cpsA gene exists in all but two capsular loci and was used as an internal control (2). To assign serotypes 6A, 6B, and 6C (and the putative serotype 6D), all serogroup 6 isolates were (i) typed by the Quellung reaction (18) and (ii) screened by PCR for a region of wciN using previously described primers (5′-TACCATGCAGGGTGGAATGT-3′ and 5′-CCATCCTTCGAGTATTGC-3′) that result in product sizes of 2.0 kb for serotypes 6A and 6B and 1.8 kb for serotype 6C (13). The PCRs were done using a 10-μl volume containing 1× GoTaq Flexi buffer (Promega, Madison, WI), 2.5 mM of MgCl2, 0.08 mM of deoxynucleoside triphosphates, 1.0 μM of each primer, and 0.1 U μl−1 of GoTaq Flexi DNA polymerase (Promega, Madison, WI). DNA was isolated from freshly grown bacterial cultures, picked with a sterile tip, and briefly immersed into the PCR mix. Thermocycling was performed using a My Cycler thermal cycler (Bio-Rad Laboratories, Hercules, CA) under the following conditions: 94°C for 4 min; 35 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 45 s (for wzy) or 2 min (for wciN); and a final extension step at 72°C for 5 min. PCR analysis was done by electrophoresis on 1% or 2% Seakem LE agarose gels in 1× Tris-acetate-EDTA buffer. Gels were stained in a 0.1-μg ml−1 ethidium bromide solution.
Preparation of chromosomal DNA, restriction with SmaI endonuclease, pulsed-field gel electrophoresis (PFGE), and analysis of patterns with Bionumerics software (version 5.10; Applied Maths, Gent, Belgium) were done as previously described (15, 17). Multilocus sequence typing was performed, as previously described (6), on selected strains of each PFGE cluster, choosing at least one representative of each year within each group.
Out of 4,064 pneumococcal strains recovered from colonization samples in studies conducted between 1996 and 2007, 809 (19.9%) were of serogroup 6: 7.3% were of serotype 6A, 9.6% were of serotype 6B, and 2.6% were of serotype 6C. Temporal fluctuations were observed for all serotypes (Table 1). Serotype 6B was dominant among serogroup 6 isolates from 1996 until 2002, when it started to decline, being almost absent by 2006. For serotype 6A, a sharp decrease was observed in 1998, and no significant fluctuations were observed with the availability of PCV7 (since June 2001). The prevalence of serotype 6C ranged from 0.2% to 5.8%, reaching the highest value in 2007. There were no strains of the hypothetical serotype 6D.
TABLE 1.
Distribution of serogroup 6 pneumococci over timea
| Yr | Total no. of Pn isolates | No. of isolates (%) of:
|
|||
|---|---|---|---|---|---|
| SG 6 | Serotype 6A | Serotype 6B | Serotype 6C | ||
| 1996 | 277 | 77 (27.8) | 33 (11.9) | 40 (14.4) | 4 (1.4) |
| 1997 | 353 | 84 (23.8) | 26 (7.4) | 46 (13.0) | 12 (3.4) |
| 1998 | 465 | 66 (14.2) | 5 (1.1) | 60 (12.9) | 1 (0.2) |
| 1999 | 596 | 137 (23.0) | 45 (7.6) | 68 (11.4) | 24 (4.0) |
| 2001 | 466 | 135 (29.0) | 57 (12.2) | 75 (16.1) | 3 (0.6) |
| 2002 | 559 | 97 (17.4) | 30 (5.4) | 61 (10.9) | 6 (1.1) |
| 2003 | 559 | 94 (16.8) | 41 (7.3) | 36 (6.4) | 17 (3.0) |
| 2006 | 392 | 64 (16.3) | 46 (11.7) | 2 (0.5) | 16 (4.1) |
| 2007 | 397 | 55 (13.8) | 31 (7.8) | 1 (0.3) | 23 (5.8) |
| Total | 4,064 | 809 (19.9) | 314 (7.3) | 389 (9.6) | 106 (2.6) |
Pn, pneumococcus; SG, serogroup.
Of the 106 serotype 6C strains identified, 35.8% were resistant to at least one of the antibiotics tested. The proportions of penicillin-resistant isolates according to parenteral meningeal and nonmeningeal criteria were 30.2% and 0%, respectively, as 29.2% of the 106 isolates had an MIC of 0.12 μg ml−1, and a single isolate (1%) had an MIC of 0.25 μg ml−1 (4). Resistance levels were 21.7% for erythromycin, 18.9% for clindamycin, 18.9% for tetracycline, and 1.9% for sulfamethoxazole-trimethoprim. Multidrug resistance was detected in 17.9% of the isolates and was observed in 2006 and 2007 only.
PFGE fingerprinting clustered the 106 serotype 6C strains into 11 groups (Table 2). By multilocus sequence typing, the 27 strains representing all PFGE groups yielded 11 sequence types (STs) that, using eBURST and the complete database of S. pneumoniae (available at http://spneumoniae.mlst.net/), fell into four clonal complexes (CCs) based on a minimum similarity of five identical loci (numbered by ST of the predicted founder as CC138, CC395, CC386, and CC3034) and two singletons (ST3671 and ST2789) (Table 2). Three novel STs were found in this study (STs 3671, 3673, and 3711).
TABLE 2.
Characteristics of serotype 6C strainsa
| PFGE group (no. of isolates) | ST (no. of isolates)b | CC | Resistance pattern (no. of isolates) | No. of DCCs |
|---|---|---|---|---|
| 6C-1 (52) | ST395 (5) | 395 | Susceptible | 12 |
| ST1714 (1) | ||||
| ST1692 (1) | ||||
| ST3711 (1) | ||||
| 6C-2 (18) | ST1150 (4) | 138 | P (11) | 3 |
| Susceptible (7) | ||||
| 6C-3 (1) | ST1150 (1) | 138 | P | 1 |
| 6C-4 (4) | ST3673 (2) | 138 | Ery | 2 |
| 6C-5 (1) | ST2789 (1) | Singleton | Susceptible | 1 |
| 6C-6 (2) | ST3671 (1) | Singleton | Susceptible | 1 |
| 6C-7 (6) | ST1150 (1) | 138 | P (3) | 3 |
| ST2689 (2) | Susceptible (3) | |||
| 6C-8 (2) | ST2185 (1) | 3034 | Ery, Clin, Tet, SXT | 1 |
| 6C-9 (3) | ST395 (2) | 395 | Susceptible | 1 |
| 6C-10 (11) | ST3396 (1) | 386 | P, Ery, Clin, Tet (8) | 1 |
| Ery, Clin, Tet (3) | ||||
| 6C-11 (6) | ST3396 (1) | 386 | P, Ery, Clin, Tet | 1 |
P, penicillin G MIC of 0.12 μg ml−1, with the exception of a single isolate that had an MIC of 0.25 μg ml−1; Ery, erythromycin; Clin, clindamycin; Tet, tetracycline; SXT, sulfamethoxazole-trimethoprim; No. of DCCs, number of day care centers where the PFGE group was found.
Number of representative strains tested.
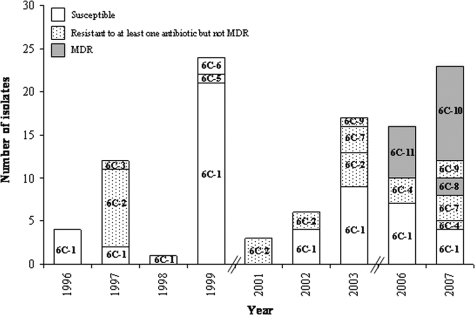
The dominant group 6C-1 (STs 395, 1692, 1714, and 3711) included 52 strains susceptible to all antibiotics that were recovered from several day care centers and were detected in all but one sampling period (Fig. 1 and Table 2). The next two most frequently found groups, 6C-2 (ST1150, with 18 isolates) and 6C-10 (ST3396, with 11 isolates), contained strains that were resistant to penicillin (according to meningeal criteria). In addition, strains of group 6C-10 were also resistant to erythromycin, clindamycin, and tetracycline (Table 2). While group 6C-2 was found in 1997 and between 2001 and 2003, group 6C-10 was identified in 2007 only (Fig. 1). The remaining eight groups were sporadic, representing 0.9 to 5.7% of the serotype 6C collection.
FIG. 1.
Distribution of serotype 6C PFGE groups through the nine study periods. MDR, multidrug resistant.
Our study shows that the recently described serotype 6C is frequently carried by healthy young children in Portugal. Overall, of all isolates that would have been conventionally typed by the Quellung reaction as belonging to serotype 6A, close to one-quarter (25.2%) were identified as belonging to serotype 6C; this value ranged between 5.0% and 42.6% depending on the sampling period.
Serotype 6C strains circulating in Portugal are genetically diverse. Similar observations were recently reported in a study from the United States (8), and, of interest, there were no common STs among serotype 6C isolates from both studies. This observation implies that if the introduction of wciN6C into the capsular operon of serotype 6A strains occurred only once, as suggested previously by Park et al., it must have occurred a sufficiently long time ago to yield such lineage diversity (13). In our collection, one fully antibiotic-susceptible lineage (6C-1, with STs 395, 1692, 1714, and 3711) persisted over one decade and accounted for close to half of all serotype 6C isolates, suggesting that it is well adapted even when considering the natural selection driven by antibiotic pressure. Still, strains of serotype 6C circulating in Portugal are often drug resistant, and in particular, multidrug resistance has been detected in 2006 and 2007. These data are in contrast with recent observations from South Africa, where most invasive strains of this serotype were fully susceptible or resistant to macrolides only, but are comparable to resistance rates reported in a recent study from the United States (8).
In summary, serotype 6C has been circulating in Portugal since at least 1996. It is genetically diverse and often antibiotic resistant. Continued surveillance of this serotype is important since it is not targeted by PCV7, and it is not known if PCV13 (which targets not only serotype 6B but also serotype 6A) will confer cross-protection to it. Additional data from other countries and estimates of its invasive disease potential will help us understand the epidemiology of this serotype.
Acknowledgments
This work was supported by projects PREVIS (LSHM-CT-2003-503413) and GRACE (LSHM-CT-2005-518226) from the European Commission and PTDC/SAU-ESA/65048/2006 from the Fundação para a Ciência e Tecnologia (FCT), Portugal. S.N. was supported by grant SFRH/BD/40706/2007 from the FCT.
We acknowledge the use of the pneumococcal MLST database, which is located at the Imperial College of London and is funded by the Wellcome Trust.
Footnotes
Published ahead of print on 10 December 2008.
REFERENCES
- 1.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brito, D. A., M. Ramirez, and H. de Lencastre. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 412378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho, M. G., C. Henderson, A. Trujillo, H. H. Joshi, I. H. Park, S. Hollingshead, C. Whitney, M. Nahm, B. Beall, and the TABC ABCs Team. 2008. Emergence of genetically diverse invasive pneumococcal serotype 6C, abstr. S01-O1, p. 35. Abstr. 6th Int. Symp. Pneumococci Pneumococcal Dis., Reykjavik, Iceland.
- 4.CLSI. 2008. Performance standards for antimicrobial susceptibility testing. Eighteenth informational supplement, vol. 28, no. 1. Approved standard M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.du Plessis, M., A. von Gottberg, S. A. Madhi, O. Hattingh, L. de Gouveia, and K. P. Klugman. 2008. Serotype 6C is associated with penicillin-susceptible meningeal infections in human immunodeficiency virus (HIV)-infected adults among invasive pneumococcal isolates previously identified as serotype 6A in South Africa. Int. J. Antimicrob. Agents 32S66-S70. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 1443049-3060. [DOI] [PubMed] [Google Scholar]
- 7.Hermans, P. W., M. Blommaart, I. H. Park, M. H. Nahm, and D. Bogaert. 2008. Low prevalence of recently discovered pneumococcal serotype 6C isolates among healthy Dutch children in the pre-vaccination era. Vaccine 26449-450. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs, M. R., S. Bajaksouzian, R. A. Bonomo, C. E. Good, A. R. Windau, A. M. Hujer, C. Massire, R. Melton, L. B. Blyn, D. J. Ecker, and R. Sampath. 29 October 2008. Occurrence, distribution, and origins of serotype 6C Streptococcus pneumoniae, a recently recognized serotype. J. Clin. Microbiol. doi: 10.1128/JCM.01524-08. [DOI] [PMC free article] [PubMed]
- 9.Mato, R., I. S. Sanches, C. Simas, S. Nunes, J. A. Carriço, N. G. Sousa, N. Frazão, J. Saldanha, A. Brito-Avô, J. S. Almeida, and H. D. Lencastre. 2005. Natural history of drug-resistant clones of Streptococcus pneumoniae colonizing healthy children in Portugal. Microb. Drug Resist. 11309-322. [DOI] [PubMed] [Google Scholar]
- 10.Mavroidi, A., D. Godoy, D. M. Aanensen, D. A. Robinson, S. K. Hollingshead, and B. G. Spratt. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J. Bacteriol. 1868181-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunes, S., R. Sá-Leão, J. Carriço, C. R. Alves, R. Mato, A. Brito-Avô, J. Saldanha, J. S. Almeida, I. S. Sanches, and H. de Lencastre. 2005. Trends in drug resistance, serotypes, and molecular types of Streptococcus pneumoniae colonizing preschool-age children attending day care centers in Lisbon, Portugal: a summary of 4 years of annual surveillance. J. Clin. Microbiol. 431285-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, I. H., M. R. Moore, J. J. Treanor, S. Pelton, T. Pilishvili, B. Beall, M. Shelly, G. Gallagher, B. Mahon, and M. H. Nahm. 2008. Reduction in serotype 6A invasive pneumococcal disease after accounting for effect of serotype 6C, abstr. P1-006, p. 88. Abstr. 6th Int. Symp. Pneumococci Pneumococcal Dis., Reykjavik, Iceland.
- 13.Park, I. H., S. Park, S. K. Hollingshead, and M. H. Nahm. 2007. Genetic basis for the new pneumococcal serotype, 6C. Infect. Immun. 754482-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park, I. H., D. G. Pritchard, R. Cartee, A. Brandao, M. C. Brandileone, and M. H. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 451225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sá-Leão, R., A. S. Simões, S. Nunes, N. G. Sousa, N. Frazão, and H. de Lencastre. 2006. Identification, prevalence and population structure of non-typable Streptococcus pneumoniae in carriage samples isolated from preschoolers attending day-care centres. Microbiology 152367-376. [DOI] [PubMed] [Google Scholar]
- 16.Sá-Leão, R., A. Tomasz, I. S. Sanches, A. Brito-Avô, S. E. Vilhelmsson, K. G. Kristinsson, and H. de Lencastre. 2000. Carriage of internationally spread clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J. Infect. Dis. 1821153-1160. [DOI] [PubMed] [Google Scholar]
- 17.Sá-Leão, R., A. Tomasz, I. S. Sanches, S. Nunes, C. R. Alves, A. Brito-Avô, J. Saldanha, K. G. Kristinsson, and H. de Lencastre. 2000. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J. Clin. Microbiol. 384137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorensen, U. B. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 312097-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]