Abstract
Human noroviruses (NoVs) cause epidemic and endemic acute gastroenteritis in children and adults. To study the prevalence and genetic diversity of NoV in children in Tunisia, a total of 788 fecal samples were collected during a 4-year period in the region of Monastir, from children 12 years of age or younger, hospitalized or presenting in dispensaries with symptoms of acute gastroenteritis. NoV was detected by reverse transcription-PCR and confirmed by sequence analysis. This is the first report that describes the molecular epidemiology of NoV in Tunisian children: NoVs were characterized as the causative agent in 128 (16.2%) of the samples. Fourteen samples contained a mixture of two NoVs, and 33 samples were coinfected with additional enteric viruses. Eight distinct NoV genotypes were detected (GGI.2, GGI.4, GGII.1, GGII.4, GGII.8, GGII.14, GGIIb/GGII.2, and GGIIb/GGII.3). GGII.4 was the most prevalent genotype, accounting for 83 (64.8%) cases. Interestingly the GGII.4 variant Hunter, described as spreading all over the world in 2004, was found in Tunisia as early as January 2003. The delay of 1 year between the isolation in Tunisia and the worldwide emergence is somewhat surprising, considering the importance of the contacts between North Africa and Europe particularly. Nevertheless, this illustrates the idea that sporadic gastroenteritis cases may be a reservoir for emerging epidemic NoV strains.
Human noroviruses (NoVs) are now recognized as an important cause of epidemic and sporadic diarrheal disease in humans of all ages worldwide (16, 20). They are members of the genus Norovirus in the family Caliciviridae. They are positive-sense, single-stranded RNA viruses. Their genome is organized into three open reading frames (ORFs). ORF1 is the largest and encodes a polyprotein precursor for several nonstructural proteins, ORF2 encodes the capsid protein, and ORF3 encodes a small protein that plays a role in the stability of viral capsid protein VP1 (4, 42). According to nucleotide sequence analysis of the capsid region, NoVs are subdivided into several genogroups that can be further divided into several clusters or genotypes. Genogroups GI, GII, and GIV have been found in humans, though GII seems to be the predominant strain around the world (1, 8, 27, 38). Genogroup I with 14 clusters and genogroup II with 17 clusters contain most of the strains infecting humans (16, 46), but only one NoV genotype (GGII.4) is the predominant circulating virus associated with global epidemics of gastroenteritis (9, 36).
In many countries, numerous works have studied the incidence and the molecular epidemiology of NoVs causing gastroenteritis. Indeed, each year, NoVs may cause more than 1.1 million hospitalizations and 200,000 deaths in children <5 years of age in developing countries (28). However, in Tunisia, as in many other African countries, while diarrhea is a major cause of illness among children, little is known about the molecular epidemiology of NoVs in gastroenteritis. A first study conducted by our laboratory between January 2003 and June 2005 showed NoVs as the second most common agent responsible for sporadic cases of viral gastroenteritis in Tunisian children but equal to rotaviruses in terms of necessity of hospitalization and severity of the clinical symptoms (32). This present study completes the previous survey by focusing on the molecular epidemiology of NoV strains circulating in Tunisia. Indeed, we screened 788 stool specimens collected from Tunisian children under 12 years of age who had acute diarrhea between January 2003 and April 2007. Using reverse transcription-PCR (RT-PCR) with GI- and GII-specific primer sets and sequencing, we were able to phylogenetically analyze the strains. The genetic characterization of the NoVs detected in our study showed a diversity of strains circulating in Tunisia but with a high predominance of the GGII.4 genotype. Surprisingly, our results show the presence of the GGII.4 variant Hunter from the beginning of our study in January 2003, whereas its first isolation was reported in the literature to be in February 2004, which is 1 year later.
MATERIALS AND METHODS
Patients.
Seven hundred eighty-eight patients consulting for acute gastroenteritis at a pediatric hospital or dispensaries in Monastir, Tunisia, between January 2003 and April 2007 were enrolled in this prospective study. The mean age of the study population was 20.08 ± 20.607 months with an age range of 14 days to 12 years. The majority of patients (n = 588, 74.6%) were <2 years old, and eight (1%) were newborns. Cases were identified by reviewing hospital admission logs for demographic characteristics of the patients (name, age, sex, etc.) and symptoms (fever, vomiting, abdominal pain, dehydration, etc.), and then parents were interviewed for confirmation of medical history and study consent.
Specimens.
Fecal specimens were collected within 24 h of admission. Four hundred eight (51.8%) specimens were collected from hospitalized children (inpatients) throughout the study, and 380 (48.2%) were collected from outpatient children presenting in dispensaries from January 2003 to May 2004. They were all negative for bacteria and parasites. The specimens were frozen, sent to the laboratory, and then stored at −40°C until being tested for NoVs.
RNA extraction.
Viral RNA was extracted from 10% stool suspensions in phosphate-buffered saline (pH 7.5) with a QIAamp viral RNA kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions, and was stored at −40°C.
RT-PCR and sequencing.
NoVs were detected by RT-PCR using several sets of primers in separate reactions. The primer set JV12/JV13 (40) was used to amplify a fragment of the RNA polymerase gene. The primers sets G1SKF/G1SKR and G2SKF/G2SKR (17) were used to detect a fragment of the capsid gene of NoV genogroups I and II, respectively. RT-PCR was performed with the One Step RT-PCR kit (Qiagen) according to the manufacturer's instructions and to the cycles of amplification given by the authors of each primer set.
All the amplified cDNA samples were systematically purified from the gel by using the QIAquick gel extraction kit (Qiagen). Sequencing of the PCR products was performed with the same primers as those for amplification by using the ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applera Corporation, Foster City, CA) and an ABI 3100 automated sequencer (PE Biosystems). The entire sequences of ORF2 were amplified for seven samples (five from 2003, one from 2004, and one from 2007) using primer pair FW1/RT5 and the Titan one-tube RT-PCR kit (Roche Applied Biosystems) (A. H. Kamel, M. A. Ali, P. Pothier, and G. Belliot, submitted for publication).
The ORF1/ORF2 junction of the viral genome was sequenced for all positive samples (n = 14) whose genotype in the capsid gene differs from the genotype in the polymerase gene. The PCR products obtained with the primer set JV12/G2SKR (1,112 bp) were cloned into the pGEM-T Easy vector system (Promega Corporation, Madison, WI), according to the manufacturer's instructions, and then sequenced using the same primers.
Characterization and phylogenetic analysis.
The nucleotide sequences of the amplicons were compared to corresponding sequences of NoV strains available in the GenBank database using Fasta program version 3, available from the European Bioinformatics Institute (http://www.ebi.ac.uk). For the RNA polymerase, analyses were also performed with reference strains available in the database of the European Food-Borne Viruses Network (https://hypocrates.rivm.nl/bnwww/Divine-Event/index.html). Sequence alignments were carried out by using Clustal W software. RNA polymerase nucleotide sequence identities within a genotype range from 87 to 99% and 91 to 100% for GGI and GGII NoVs, respectively. Capsid nucleotide sequence identities range from 88 to 99% within a genotype for both genogroups (41).
For the phylogenetic analyses, sequence alignment and clustering were performed by the UPGMA (unweighted-pair group method using average linkages) using Bionumerics software (Applied Maths).
Nucleotide sequence accession numbers.
The sequences obtained in this study have been submitted to GenBank under the following accession numbers (isolate numbers and dates are given in parentheses): EU650205 (Monastir 18/2003), EU650206 (Monastir 294/2003), EU650207 (Monastir 3/2003), EU650208 (Monastir 273/2003), EU650209 (Monastir 430/2003), EU650210 (Monastir 17285/2004), EU650211 (Monastir 127/2003), EU650212 (Monastir 474/2003), EU650213 (Monastir 1795/2004), EU650214 (Monastir 578/2003), EU650215 (Monastir 17291/2006), EU650216 (Monastir 519/2003), EU650217 (Monastir 862/2003), EU650218 (Monastir 39/2004), EU650219 (Monastir 249/2003), EU650220 (Monastir 310/2003), EU650221 (Monastir 375/2003), EU650223 (Monastir 8655/2007), EU650224 (Monastir 389/2003), EU650225 (Monastir 17736/2005), EU650226 (Monastir 13562/2004), and EU650227 (Monastir 4456/2005). For the entire ORF2 sequences, accession numbers are as follows: EU916955 (Monastir 3968/2004), EU916956 (Monastir 8655/2007), EU916957 (Monastir 127/2003), EU916958 (Monastir 113/2003), EU916959 (Monastir 529/2003), EU916960 (Monastir 493/2003), and EU916961 (Monastir 715/2003).
RESULTS
NoV prevalence.
Of the 788 fecal specimens from patients with gastrointestinal symptoms, 128 (16.2%) contained NoVs. Of these, 24 (18.7%), 7 (5.5%), 1 (0.8%), and 1 (0.8%) were also positive for rotavirus, astrovirus, adenovirus, and Aichi virus, respectively.
The distribution of the sampling during the years of our study is indicated in Table 1: 76.6% (n = 604) of the samples were collected during 2003 and 2004, and 83.6% of NoV-positive samples were detected in these years. However, the ratio between the number of NoV-positive samples and the number of samples tested decreased over time.
TABLE 1.
Annual distribution of NoV-positive samples detected in Monastir, Tunisia, between January 2003 and April 2007
| Yr | No. of samples
|
No. of strains by genotypea
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tested | Positive for:
|
GGII.1 | GGIIb/GGII.2 | GGIIb/GGII.3 | GGII.4 | GGII.8 | GGII.14 | GGI.2 | GGI.4 | |||
| NoV | GGI | GGII | ||||||||||
| 2003 | 358 | 64 | 9 | 55 | 4 | 10 | 3 | 49 | 2 | 0 | 8 | 1 |
| 2004 | 246 | 43 | 2 | 41 | 1 | 1 | 21 | 17 | 2 | 0 | 2 | 0 |
| 2005 | 106 | 14 | 14 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | |
| 2006 | 64 | 6 | 6 | 0 | 0 | 0 | 2 | 0 | 4 | 0 | 0 | |
| 2007 | 14 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Total | 788 | 128 | 11 | 117 | 5 | 11 | 24 | 83 | 4 | 4 | 10 | 1 |
The total is more than 128 because mixed infections with two different genotypes were detected for several samples.
Nucleotide sequence and phylogenetic analysis.
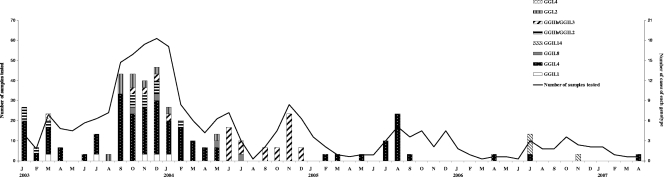
Of the 128 NoV-positive samples, a total of 142 different strains were found, 11 in samples with a single GGI strain, 103 in samples with a single GGII strain, and 28 in samples with mixtures of two different GGII strains. It has to be noted that no mixed infection with GGI and GGII strains was observed. All strains detected in this study were grouped into eight different genotypes, with GGII genotypes accounting for six of the eight genotypes: GGII.4 (n = 83) was the most predominant genotype, followed by GGI.2 (n = 10), GGI.4 (n = 1), GGII.1 (n = 5), GGII.8 (n = 4), and GGII.14 (n = 4); in addition, the recombinant strains designated GGIIb/GGII.2 and GGIIb/GGII.3 were identified in 11 and 24 samples, respectively (Table 1). The analysis of the genetic diversity of the NoV strains during our study showed an extensive cocirculation of various genotypes from January 2003 to May 2004, with all but GGII.14 being identified, as shown in Fig. 1, whereas in the second half of 2004, from June to December, GGIIb/GGII.3 was almost the only genotype represented, with the exception of one sample positive for a GGII.8 strain. It has to be noted that the GGII.4 strain was not detected in this period but became the only genotype detected in 2005 and was also present in 2006 and 2007, at the same time as strain M7 (GGII.14), which emerged in 2006, with three cases occurring in July and one case in November.
FIG. 1.
Monthly distribution of NoV genotypes and numbers of samples tested in Tunisian children between January 2003 and April 2007. J through D, January through December, respectively.
The existence of mixed infections in 14 samples was revealed by sequencing of the ORF1/ORF2 junction; this was realized for all positive samples whose genotype in the capsid gene differs from the genotype in the polymerase gene, in order to distinguish between the potential presence of recombinant strains and mixed infections. It has to be noted that no recombinant NoV strain other than GGIIb was identified. The coinfections found could be classified into three groups: 11 coinfections between GGII.4 and GGIIb/GGII.2, two cases of coinfection between GGII.4 and GGII.8, and one case between GGII.4 and GGIIb/GGII.3.
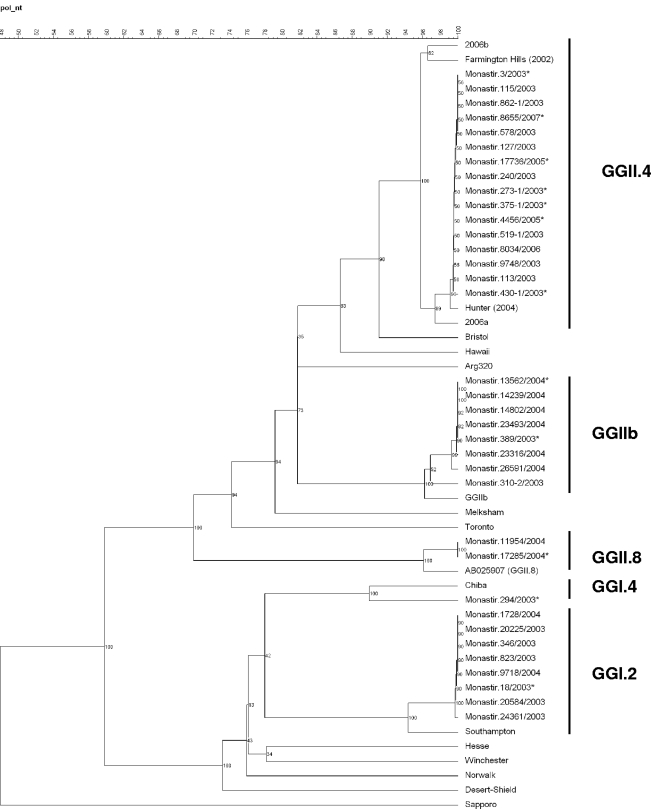
Based on the nucleotide sequences, representative phylogenetic trees of the polymerase and capsid regions were constructed by the UPGMA method using Bionumerics software (Fig. 2 and 3). Phylogenetic analysis of both regions showed that the GGII.4 strains identified in this study from 2003 to 2007 were all closely related to the GGII.4 2004 variant Hunter (GenBank accession numbers DQ078801 and DQ078794), first described in Australia in February 2004 (9). The sequence analysis based on 285 bp from the polymerase region, which is the most conserved region of the genome and the most widely analyzed in the literature, showed that our strains differ from Hunter by 1 bp. The analysis of 302-bp sequences of the capsid gene showed 100% identity with Hunter.
FIG. 2.
Phylogenetic analysis based on the partial nucleotide sequences (285 bp) of the RNA-dependent RNA polymerase coding gene of the NoV strains associated with pediatric gastroenteritis in Monastir, Tunisia, between January 2003 and April 2007. The tree was constructed using UPGMA clustering. GenBank accession numbers for reference strains in this figure are as follows: Hunter virus, DQ078794; Southampton virus, L07418; Chiba virus, AB042808; Amsterdam virus, AB025907; GGIIb virus, AJ487794; Arg 320 virus, AF190817; Farmington Hills virus, AY502023. For more clarity, the sequences that present 100% nucleotide identity are not all represented on the tree. Only some of them, representative of each genotype, are indicated. In the same way, only some representative sequences were submitted to GenBank. Their GenBank accession numbers are indicated in Materials and Methods, and they are marked with asterisks in the tree. The percent bootstrap values in which the major groupings were observed among 100 replicates are indicated in each branch.
FIG. 3.
Phylogenetic analysis based on the partial nucleotide sequences (302 bp) of the capsid coding gene of NoV isolates detected in Tunisia. Reference strains of NoV were selected from the GenBank database under the following accession numbers: Farmington Hills virus, AY502023; Hunter virus, DQ078794; Amsterdam virus, AF195848; Hawaii virus, U07611; Melksham virus, X81879; Arg 320 virus, AF190817; M7/1999/US virus, AY130761. The GenBank accession numbers of strains of this study are indicated in the text, and they are marked with asterisks in the tree. The numbers in the branches indicate the bootstrap values.
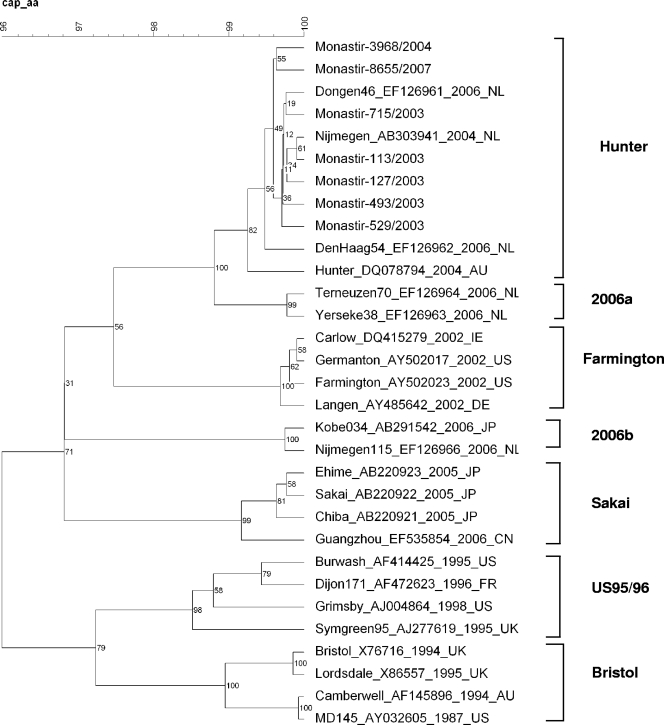
To confirm that the Hunter variant was effectively found throughout the study period, and especially since January 2003, we sequenced the complete capsid gene of seven samples collected at different times: two samples from January 2003, one from April 2003, one from September 2003, one from October 2003, one from February 2004, and one from April 2007. The alignment of the amino acid sequences of the complete ORF2 with reference strains representing each GGII.4 variant confirms that the GGII.4 strains found in this study, in 2003 as well as in 2007, cluster with the 2004 Hunter variant (Fig. 4).
FIG. 4.
Phylogenetic tree constructed from complete amino acid sequences of ORF2 of the seven Tunisian isolates and the GGII.4 representative strains from GenBank. For the samples in this study, accession numbers are as follows: EU916955 (Monastir-3968/2004), EU916956 (Monastir-8655/2007), EU916957 (Monastir-127/2003), EU916958 (Monastir-113/2003), EU916959 (Monastir-529/2003), EU916960 (Monastir-493/2003), and EU916961 (Monastir-715/2003). The numbers on each branch indicate the bootstrap values for the clusters supported by that branch.
Concerning the other genotypes, the alignment of the sequences showed in-group nucleotide identities of 100%, except for GGII.1 and GGIIb/GGII.2 strains, which can be divided into two subgroups.
The 10 GGI.2 strains showed 94.7% nucleotide identity in the polymerase region with the reference strain Southampton/1991/UK (GenBank accession number L07418), and the strain GGI.4 presented 90.8% nucleotide identity in the polymerase gene with Chiba/1987/JP (accession number AB042808). The capsid gene of these strains failed to amplify by our method. Among the four GGII.8 strains, two were amplified in the polymerase gene and displayed 99 to 100% nucleotide identity with strains referenced in the database of the European Food-Borne Viruses Network. The two other GGII.8 strains were amplified in the capsid gene and showed 96% nucleotide identity with Amsterdam/1998/NL (accession number AF195848). The four strains of GGII.14 displayed 95.7% nucleotide identity in the capsid gene with the strain designated M7/1999/US (accession number AY130761). The 24 GGIIb/GGII.3 strains found in our study were all sequenced in the polymerase and capsid genes. They displayed 97.5% nucleotide identity in the polymerase gene with the GGIIb strain described by Buesa et al. (6) (accession number AJ487794). Their capsid region showed 97.4% nucleotide identity with Mexico/1995 (accession number U22498). The 11 GGIIb/GGII.2 strains were divided into two groups that differed by five nucleotides according to the alignment of 267-bp sequences of the capsid gene; these two groups of six and five strains displayed 97.4% and 98.5% nucleotide identity with Melksham/1994/UK (accession number X81879), respectively. Their polymerase gene showed 97.9% nucleotide identity with the GGIIb strain (accession number AJ487794). All the GGII.1 strains shared 95.3% nucleotide identity in the capsid gene to the Hawaii strain (accession number U07611), except for one that differed by six nucleotides from the others but presented also 95.3% nucleotide identity with the Hawaii strain. The polymerase gene of these strains could not be amplified by our method.
DISCUSSION
We investigated the molecular epidemiology of human NoVs for more than 4 years in children under 12 years of age, who represent more than 28% of the total population of the urban region of Monastir in Tunisia. Our study supports the increasing recognition that NoV strains of genogroup II are commonly responsible for sporadic cases of gastroenteritis in children (1, 3, 5, 11, 12, 14, 27, 30, 47).
Our results have shown the presence of eight distinct genotypes of NoVs. This is consistent with the genetic diversity found in other studies (1, 3, 6, 14, 24, 31) and indicates that gastroenteritis can be due to genetically different NoVs in the same area. However, the prevalence of the different genotypes changes over time, with a period of cocirculation of numerous strains, followed by periods of circulation of almost one unique genotype. Indeed, we observed a cocirculation of seven distinct strains until May 2004 (including GGIIb/GGII.3 and GGII.4), and then strains classified as GGIIb/GGII.3 were almost the only ones detected in the second half of 2004. Again in 2005 the pattern of circulation changed, with the disappearance of these strains and their almost-exclusive replacement by the GGII.4 genotype. These changes in the predominance of a particular genotype have already been observed in several countries; the GGII.3 strain, predominant in Japan in the winter season 2001-2002 (26), and the GGII.1 strain, predominant in the Italian region of Parma and in Hungary in 2000 (24, 31), were all replaced the next season by the emerging strain GGII.4. In our study the end of cocirculation of diverse strains in May 2004 coincides with the end of collecting stools from children presenting in dispensaries in addition to hospitalized children. However, no correlation was found between the two events (data not shown), challenging the hypothesis that some genotypes could be responsible for more severe symptoms and therefore be more often detected in the hospitalized population than in the community.
The natural emergent recombinant GGIIb strains were detected for the first time in France in August 2000 (2) and then spread throughout France and Europe in the following seasons, with outbreaks and sporadic cases reported in Spain, Sweden, and Hungary (6, 20, 31). They were also reported in other parts of the world, such as in Japan in the 2003-2004 season (29). Our study showed that the recombinant GGIIb strains were the second most frequently detected NoVs in Tunisian children, as previously found in Hungary between 2001 and 2003 (31). However, it is interesting that these strains were observed in our study only in 2003 and 2004 and have not diffused since. The same pattern has been observed in France, with a high prevalence until 2004 (5) and then only episodic gastroenteritis cases due to the GGIIb strains occurring (personal data).
Except between June and December 2004 when GGIIb/GGII.3 was the predominant circulating genotype, GGII.4 was the most prevalent cluster isolated during the whole study. This genotype is considered the predominant genotype worldwide, responsible for the majority of outbreaks of gastroenteritis since 1995 (9, 19, 22, 25, 35), and is also shown in recent molecular epidemiological studies as the most prevalent genotype in sporadic community cases in recent years (1, 3, 10, 45). The emergence and global spread of this virus coincided with a worldwide increase in the number of gastroenteritis outbreaks, suggesting that this strain could be more virulent or better adapted for survival in the environment or may be more contagious than other NoVs (43). It is interesting that this overall increase in the number of outbreaks is punctuated with epidemic seasons due to GGII.4, followed by postepidemic years with non-GGII.4 strains being more commonly found in outbreaks (34). This pattern of evolution has been termed epochal evolution. Several studies focused on the GGII.4 genotype and have shown that several epidemic variants can be distinguished within the genotype that succeed one another over time: one variant emerges, replaces the previously circulating variant to become the predominant strain for a few years, and is ultimately replaced with another new variant (7, 13, 21, 33, 34). This phenomenon has been explained by an accumulation of point mutations, essentially in the protruding P2 region of the capsid protein which conduces to genetic drift and an immune evasion. Five lineages have been identified since 1995 that are responsible for the four epidemic peaks observed worldwide in 1995-1996, 2002, 2004, and 2006. The 95/96US cluster observed in the United Kingdom, the United States, Brazil, Canada, Australia, The Netherlands, China, and Germany (25, 43) has been displaced by the variant Farmington Hills, isolated in 2002 in Europe (23) and in the United States (44). Then a new variant termed Hunter (9) became the most common circulating strain worldwide in 2004, displacing the Farmington Hills variant (18). Lately two new variants, 2006a and 2006b, have been described as the global predominant strains (7, 13, 15, 19, 34, 35, 36, 37, 39). Our results show the presence of the 2004 variant Hunter from the very start of our study, in January 2003, as confirmed by the sequencing of the complete ORF2 of two samples collected in that month. This is contrary to the literature that dates the first isolation of this variant to February 2004 in New South Wales (9), 1 year later. The presence of a specific strain in sporadic infantile cases before its global spread has been already pointed out by Medici et al. (24), who proposed that children with sporadic gastroenteritis may act as a reservoir for emerging epidemic NoV strains. In fact our results do not correspond to the general schema of succession of epidemic variants over time or to the worldwide succession of epidemic seasons and global increase of the number of gastroenteritis cases explained by this succession of epidemic variants. Indeed, the systematic sequencing of the polymerase and capsid fragments of all our GGII.4 samples, as well as the sequencing of the complete ORF2 of seven samples, shows that the same variant is found throughout our study, even in the three samples collected in 2006 and 2007, after the global spread of the 2006 variants.
Even if we observed peaks of infection in winter and in summer, we globally observed a decrease of the number of NoV-positive samples, essentially since January 2005. This decrease can be explained in part by the decrease of the number of samplings after May 2004. Nevertheless, a real decrease exists of the ratio between the number of samples tested and the number of samples positive for NoV over time. The reason for this phenomenon is unclear, but the small number of positive samples does not allow us draw conclusions about the circulating strains, especially GGII.4 variants, in 2006 and 2007 in the child population of Monastir and needs more investigation. In addition, it would be interesting to analyze samples from 2002 to see what variant was circulating at that time.
In conclusion, our study shows the predominance of the GGII.4 genotype in sporadic community cases of gastroenteritis in Tunisian children, interestingly with the appearance of the Hunter variant as early as 2003, 1 year before its worldwide emergence. Even if this delay of 1 year between the two events is somewhat surprising, especially considering the importance of the contacts between North Africa and Europe particularly, this reinforces the idea that sporadic gastroenteritis cases may be a reservoir for emerging epidemic NoV strains.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Al-Mashhadani, M. N., O. Nakagomi, W. Dove, H. Ahmed, T. Nakagomi, C. A. Hart, and N. A. Cunliffe. 2008. Norovirus gastroenteritis among children in Iraqi Kurdistan. J. Med. Virol. 80506-509. [DOI] [PubMed] [Google Scholar]
- 2.Ambert-Balay, K., F. Bon, F. Le Guyader, P. Pothier, and E. Kohli. 2005. Characterization of new recombinant noroviruses. J. Clin. Microbiol. 435179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armah, G. E., C. I. Gallimore, F. N. Binka, R. H. Asmah, J. Green, U. Ugoji, F. Anto, D. W. G. Brown, and J. J. Gray. 2006. Characterisation of norovirus strains in rural Ghanaian children with acute diarrhoea. J. Med. Virol. 781480-1485. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti-Ciarlet, A, S. E. Crawford, A. M. Hutson, and M. K. Estes. 2003. The 3′ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein. J. Virol. 7711603-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brachet, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 434659-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buesa, J., B. Collado, P. Lopez-Andujar, R. Abu-Mallouh, J. R. Diaz, A. G. Diaz, J. Prat, S. Guix, T. Llovet, G. Prats, and A. Bosch. 2002. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J. Clin. Microbiol. 402854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buesa, J., R. Montava, R. Abu-Mallouh, M. Fos, J. M. Ribes, R. Bartolome, H. Vanaclocha, N. Torner, and A. Domínguez. 2008. Sequential evolution of genotype GII.4 norovirus variants causing gastroenteritis outbreaks from 2001 to 2006 in eastern Spain. J. Med. Virol. 801288-1295. [DOI] [PubMed] [Google Scholar]
- 8.Bull, R. A., G. S Hansman, L. E. Clancy, M. M. Tanaka, W. D. Rawlinson, and P. A. White. 2005. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 111079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bull, R. A., E. T. V. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castilho, J. G., V. Munford, H. R. Resque, U. Fagundes-Neto, J. Vinjé, and M. L. Rácz. 2006. Genetic diversity of norovirus among children with gastroenteritis in São Paulo State, Brazil. J. Clin. Microbiol. 443947-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dove, W., N. A. Cunliffe, J. S. Gondwe, R. L. Broadhead, M. E. Molyneux, O. Nakagomi, and C. A. Hart. 2005. Detection and characterization of human caliciviruses in hospitalized children with acute gastroenteritis in Blantyre, Malawi. J. Med. Virol. 77522-527. [DOI] [PubMed] [Google Scholar]
- 12.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1861-7. [DOI] [PubMed] [Google Scholar]
- 13.Gallimore. C. I., M. Iturriza-Gomara, J. Xerry, J. Adigwe, and J. J. Gray. 2007. Inter-seasonal diversity of norovirus genotypes: emergence and selection of virus variants. Arch. Virol. 1521295-1303. [DOI] [PubMed] [Google Scholar]
- 14.Hansman, G. S., K. Katayama, N. Maneekarn, S. Peerakome, P. Khamrin, S. Tonusin, S. Okitsu, O. Nishio, N. Takeda, and H. Ushijima. 2004. Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Chiang Mai, Thailand. J. Clin. Microbiol. 421305-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, E. C. M., P. K. C. Cheng, A. W. L. Lau, A. H. Wong, and W. W. L. Lim. 2007. Atypical norovirus epidemic in Hong Kong during summer of 2006 caused by a new genogroup II/4 variant. J. Clin. Microbiol. 452205-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kageyama, T., M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, S. Kojima, R. Takai, T. Oka, N. Takeda, and K. Katayama. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 422988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100107-114. [DOI] [PubMed] [Google Scholar]
- 18.Kroneman, A., H. Vennema, Y. van Duijnhoven, E. Duizer, and M. Koopmans. 23 December 2004, posting date. High number of norovirus outbreaks associated with a GGII.4 variant in the Netherlands and elsewhere: does this herald a worldwide increase? Eurosurveillance Wkly. 8pii=2606. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=2606. [Google Scholar]
- 19.Kroneman, A., H. Vennema, J. Harris, G. Reuter, C. H. von Bonsdorff, K. O. Hedlund, K. Vainio, V. Jackson, P. Pothier, J. Koch, E. Schreier, B. Böttiger, and M. Koopmans. 2006. Increase in norovirus activity reported in Europe. Euro Surveill. 11E061214.1. [DOI] [PubMed] [Google Scholar]
- 20.Lindell, A. T., L. Grillner, L. Svensson, and B. Z. Wirgart. 2005. Molecular epidemiology of norovirus infections in Stockholm, Sweden, during the years 2000 to 2003: association of the GGIIb genetic cluster with infection in children. J. Clin. Microbiol. 431086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindesmith, L. C., E. F., Donaldson, A. D. LoBue, J. L. Cannon, D.-P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5269-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24137-160. [DOI] [PubMed] [Google Scholar]
- 23.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torven, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szucs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363682-688. [DOI] [PubMed] [Google Scholar]
- 24.Medici, M. C., M. Martinelli, L. A. Abelli, F. M. Ruggeri, I. Di Bartolo, M. C. Arcangeletti, F. Pinardi, F. De Conto, G. Izzi, S. Bernasconi, C. Chezzi, and G. Dettori. 2006. Molecular epidemiology of norovirus infections in sporadic cases of viral gastroenteritis among children in northern Italy. J. Med. Virol. 781486-1492. [DOI] [PubMed] [Google Scholar]
- 25.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 1791334-1344. [DOI] [PubMed] [Google Scholar]
- 26.Onishi, N., M. Hosoya, A. Matsumoto, T. Imamura, M. Katayose, Y. Kawasaki, O. Hashimoto, A. Hayashi, H. Ishiko, and H. Suzuki. 2008. Molecular epidemiology of norovirus gastroenteritis in Soma, Japan, 2001-2003. Pediatr. Int. 5065-69. [DOI] [PubMed] [Google Scholar]
- 27.Papaventsis, D. C., W. Dove, N. A. Cunliffe, O. Nakagomi, P. Combe, P. Grosjean, and C. A. Hart. 2007. Norovirus infection in children with acute gastroenteritis, Madagascar, 2004-2005. Emerg. Infect. Dis. 13908-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, M. M., M. A. Widdowson, R. I. Glass, K. Akazawa, J. Vinjé, and U. D. Parashar. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 141224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan, T. G., T. Kuroiwa, K. Kaneshi, Y. Ueda, S. Nakaya, S. Nishimura, A. Yamamoto, K. Sugita, T. Nishimura, F. Yagyu, S. Okitsu, W. E. G. Muller, N. Maneekarn, and H. Ushijima. 2006. Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J. Med. Virol. 78971-978. [DOI] [PubMed] [Google Scholar]
- 30.Ramirez, S., S. De Grazia, G. M. Giammanco, M. Milici, C. Colomba, F. M. Ruggeri, V. Martella, and S. Arista. 2006. Detection of the norovirus variants GGII.4 Hunter and GGIIb/ Hilversum in Italian children with gastroenteritis. J. Med. Virol. 781656-1662. [DOI] [PubMed] [Google Scholar]
- 31.Reuter, G., K. Krisztalovics, H. Vennema, M. Koopmans, and G. Szucs. 2005. Evidence of the etiological predominance of norovirus in gastroenteritis outbreaks—emerging new-variant and recombinant noroviruses in Hungary. J. Med. Virol. 76598-607. [DOI] [PubMed] [Google Scholar]
- 32.Sdiri-Loulizi, K., H. Gharbi-Khélifi, A. de Rougemont, S. Chouchane, N. Sakly, K. Ambert-Balay, M. Hassine, M. N. Guédiche, M. Aouni, and P. Pothier. 2008. Acute infantile gastroenteritis associated with human enteric viruses in Tunisia. J. Clin. Microbiol. 461349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siebenga, J. J., H. Vennema, E. Duizer, and M. P. G. Koopmans. 2007. Gastroenteritis caused by norovirus GGII.4, the Netherlands, 1994-2005. Emerg. Infect. Dis. 13144-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. V. Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 819932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu, E. T. V., T. Nguyen, P. Lee, R. A. Bull, J. Musto, G. Hansman, P. A. White, W. D. Rawlinson, and C. J. McIver. 2007. Norovirus GII.4 strains and outbreaks, Australia. Emerg. Infect. Dis. 131128-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu, E. T. V., R. A. Bull, G. E. Greening, J. Hewitt, J. L. Michael., J. A. Marshall, C. J. McIver, W. D. Rawlinson, and P. A. White. 2008. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin. Infect. Dis. 46413-420. [DOI] [PubMed] [Google Scholar]
- 37.Ueki, Y., M. Shoji, C. Sato, Y. Sato, Y. Okimura, N. Saito, H. Otomo, and H. Onodera. 2007. Norovirus GII/4 variants observed in outbreaks of gastroenteritis in Miyagi prefecture between November and December of 2006. Jpn. J. Infect. Dis. 60240-241. [PubMed] [Google Scholar]
- 38.Vainio, K., and M. Myrmel. 2006. Molecular epidemiology of norovirus outbreaks in Norway during 2000 to 2005 and comparison of four norovirus real-time reverse transcriptase PCR assays. J. Clin. Microbiol. 443695-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verhoef, L., E. Depoortere, I. Boxman, E. Duizer, Y. J. Harris, C. Johnsen, A. Kroneman, S. Le Guyader, L. Maunula, H. Meldal, R. Ratcliff, G. Reuter, E. J. Siebenga, K. Vainio, C. Varela, H. Vennema, and M. Koopmans on behalf of the Food Borne Viruses in Europe Network. 2008. Emergence of new norovirus variants on spring cruise ships and prediction of winter epidemics. Emerg. Infect. Dis. 14238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vinjé, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 174610-615. [DOI] [PubMed] [Google Scholar]
- 41.Vinjé, J., J. Green, D. C. Lewis, C. I. Gallimore, D. W. G. Brown, and M. P. G. Koopmans. 2000. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses.” Arch. Virol. 145223-241. [DOI] [PubMed] [Google Scholar]
- 42.Vinjé, J., A. R. A. Hamidjaja, and M. D. Sobsey. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Med. Virol. 116109-117. [DOI] [PubMed] [Google Scholar]
- 43.White, P. A., G. S. Hansman, A. Li, J. Dable, M. Isaacs, M. Ferson, C. J. McIver, and W. D. Rawlinson. 2002. Norwalk-like virus 95/96-US strain is a major cause of gastroenteritis outbreaks in Australia J. Med. Virol. 68113-118. [DOI] [PubMed] [Google Scholar]
- 44.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 19027-36. [DOI] [PubMed] [Google Scholar]
- 45.Yoon, J. S., S. G. Lee, S. K. Hong, S. A. Lee, W. H. Jheong, S. S. Oh, M. H. Oh, G. P. Ko, C. H. Lee, and S. Y. Paik. 2008. Molecular epidemiology of norovirus infections in children with acute gastroenteritis in South Korea in November 2005 through November 2006. J. Clin. Microbiol. 461474-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]
- 47.Zintz, C., K. Bok, E. Parada, M. Barnes-Eley, T. Berke, M. A. Staat, P. Azimi, X. Jiang, and D. O. Matson. 2005. Prevalence and genetic characterization of caliciviruses among children hospitalized for acute gastroenteritis in the United States. Infect. Gene Evol. 5281-290. [DOI] [PubMed] [Google Scholar]