Abstract
Expression of the retroviral Gag protein leads to formation of virus-like particles in mammalian cells. In vitro and in vivo experiments show that nucleic acid is also required for particle assembly. However, several studies have demonstrated that chimeric proteins in which the nucleocapsid domain of Gag is replaced by a leucine zipper motif can also assemble efficiently in mammalian cells. We have now analyzed assembly by chimeric proteins in which nucleocapsid of human immunodeficiency virus type 1 (HIV-1) Gag is replaced by either a dimerizing or a trimerizing zipper. Both proteins assemble well in human 293T cells; the released particles lack detectable RNA. The proteins can coassemble into particles together with full-length, wild-type Gag. We purified these proteins from bacterial lysates. These recombinant “Gag-Zipper” proteins are oligomeric in solution and do not assemble unless cofactors are added; either nucleic acid or inositol phosphates (IPs) can promote particle assembly. When mixed with one equivalent of IPs (which do not support assembly of wild-type Gag), the “dimerizing” Gag-Zipper protein misassembles into very small particles, while the “trimerizing” protein assembles correctly. However, addition of both IPs and nucleic acid leads to correct assembly of all three proteins; the “dimerizing” Gag-Zipper protein also assembles correctly if inositol hexakisphosphate is supplemented with other polyanions. We suggest that correct assembly requires both oligomeric association at the C terminus of Gag and neutralization of positive charges near its N terminus.
Expression of a single retroviral protein, Gag, in mammalian cells is sufficient for assembly of virus-like particles (VLPs). RNA seems to play an essential role, however, in both the assembly and structure of VLPs. Thus, retrovirus particles always contain RNA; in the absence of genomic RNA, cellular mRNAs replace it in the virus particle (46). RNase treatment of immature murine leukemia virus disrupts the particles (37). Finally, nucleic acid is required for assembly in defined in vitro assembly systems (8, 9).
The contribution of nucleic acid to the assembly and structure of retrovirus particles is not yet understood. As one approach to further understanding the role that nucleic acid binding plays in the assembly process, Zhang et al. (59) replaced the principal nucleic acid-binding domain of the HIV-1 Gag protein, nucleocapsid (NC), with a leucine zipper domain. This chimeric protein was able to assemble efficiently in mammalian cells as evidenced through immunoblotting of released VLPs. This observation was extended by Johnson et al. (28), who used Gag-leucine zipper (dimerizing) chimeras of Rous sarcoma virus and studied the morphologies of the resulting particles. The particles assembled from the chimeric proteins were similar, although not identical, to those formed by wild-type (WT) Gag. The fact that NC could be functionally replaced (with respect to particle assembly) with the dimerizing leucine zipper motif led these investigators to propose that the function of nucleic acid in assembly is to promote dimerization. Additional support for this hypothesis comes from the fact that the minimum length of nucleic acid needed to promote assembly is roughly enough to accommodate two molecules of Gag (30, 31).
Further studies in which the NC domain of HIV-1 Gag has been replaced by leucine zipper motifs have been presented by Accola et al. (1). Interestingly, they found that a Gag-Zipper (Gag-Z) chimera containing a trimeric zipper motif also assembles efficiently. However, these VLPs, as well as those formed by a chimera containing a dimeric zipper motif, were not characterized morphologically.
In the present work, we have extended the analysis of the assembly properties of these HIV-1 Gag-Z chimeras. This study includes the first analysis of recombinant Gag-Z proteins in vitro, as well as detailed characterization of the VLPs formed in mammalian cells. The in vitro assembly results suggest that Gag oligomerization alone is not sufficient to induce particle formation. We raise the possibility here that normal HIV-1 assembly requires neutralization of positive charges in matrix (MA) in addition to nucleic acid-induced oligomerization at the C terminus of the protein.
MATERIALS AND METHODS
Plasmids.
WT HIV-1 Gag protein was expressed in mammalian cells by transfection of the plasmid pCMV55M1-10 (a generous gift from B. Felber, NCI-Frederick), as described previously (47). This plasmid contains silent mutations rendering its expression independent of Rev.
Gag-Z proteins were expressed in mammalian cells from plasmids in which the coding sequences in pCMV55M1-10 3′ of the SphI site in capsid (CA) were replaced with the corresponding sequences from ZWT and ZIL, generous gifts from H. Göttlinger, Dana-Farber Cancer Institute (1). The resulting plasmids encode the Rev-independent Gag up to amino acid 220 (residue 88 of the CA domain), while the remaining, C-terminal sequences are from the Gag-Z chimeras. In turn, the “ZWT” leucine zipper motif (designated Gag-ZLeu in the present work) is from the yeast transcription factor GCN4 (41), while that of ZIle is a modification in which the leucine residues at positions a and d of the zipper heptad repeat are replaced by isoleucines (24, 25, 55); as a free peptide, this sequence forms trimers rather than dimers (35, 42).
Plasmids used for production of Gag-Z proteins in Escherichia coli were constructed by replacing the coding sequences in the Δp6 plasmid (8) 3′ of the SpeI site in CA with the corresponding sequences from ZWT and ZIL. Thus, the residues up to amino acid 241 (residue 109 of the CA domain) in the resulting protein are from Δp6, while those C terminal to this residue are from the Gag-Z clones.
Mutagenesis on all plasmids was performed by QuikChange (Stratagene) as recommended by the manufacturer.
Production of VLPs and immunoblotting.
VLPs were produced by transient transfection of expression constructs into 293T HEK cells using Transit-293 (Mirus) as recommended by the manufacturer. Twenty-four-hour harvests were collected at 48 and 72 h posttransfection. Supernatants were filtered through 0.45-μm filters, and VLPs were isolated by centrifugation through 20% sucrose (wt/wt) in TNE (10 mM Tris-HCl, 0.1 M NaCl, 1 mM EDTA, pH 7.4). The VLP pellets were resuspended in TNE for 1 h on ice in a volume of 10 μl TNE/ml viral supernatant.
Gag proteins in VLPs and cell lysates were detected by immunoblotting. Cells were lysed at 72 h posttransfection. The primary antibody was a goat anti-p24CA HIV (a kind gift of David Ott, AIDS Vaccine Program, SAIC-Frederick). The secondary antibody was a rabbit anti-goat-horseradish peroxidase conjugate (BioChain Institute, Inc.). Both antibodies were used at a dilution of 1:10,000.
Isolation of RNA from VLPs.
RNA was extracted from VLPs exactly as described previously (18, 46).
EM of mammalian cells.
Transfected 293T HEK cells were prepared for thin-section visualization by transmission electron microscopy (EM) as previously described (36, 51).
Gradient purification of VLPs and Ribogreen analysis.
An 11-ml sucrose gradient was prepared using equal volumes of 15% and 60% (wt/wt) sucrose stocks in TNE, which were filtered through 0.45-μm filters. A 100- to 200-μl aliquot of VLPs in TNE was gently placed on top of the gradient. For gradient separation of two sets of VLPs, the VLP samples were mixed prior to loading onto the gradient. The gradient was centrifuged at 40,000 rpm at 4°C for 18 to 24 h (Beckman-Coulter SW41Ti rotor). Fractions of 0.5 ml were collected from the bottom of the centrifuge tube. The densities of the fractions were determined from their refractive indices.
For RNA measurement of gradient fractions, RNA was extracted from the fractions as described elsewhere (18, 46). Extracted RNA was quantitated by the Ribogreen (Invitrogen) assay as recommended by the manufacturer.
Real-time RT-PCR measurements.
Real-time reverse transcription-PCR (RT-PCR) measurements were performed exactly as described previously (46). Samples for the signal recognition particle (SRP), ASB1, PLEKHB2, and PGK1 RNA standard curves (46) and the rRNA standard curve (38) were prepared as previously described. The primers and probe for 18S rRNA are described elsewhere (43). The primers and probe for SRP RNA measurement (Biosource) were as follows: forward primer, 5′-AGGATCGCTTGAGTCCAG; reverse primer, 5′-GTGCGGACACCCGATCG; probe: 5′-6-carboxyfluorescein-CTGGGCTGTAGTGCGCT-6-carboxytetramethylrhodamine. The primers and probe for the mRNAs were supplied as 20× stocks from Applied Biosystems: PGK1 (HS99999906_m1), ASB1 (HS00211548_m1), and PLEKHB2 (HS00215820_m1).
Infectivity measurements.
Infectivity was measured by cotransfecting 293T cells with a Gag- or Gag-Z-expressing plasmid, a pNL4-3-derived vector lacking Vpr and containing the firefly luciferase gene in place of env (a kind gift of Alok Mulky and Vineet KewalRamani, NCI-Frederick), and a vesicular stomatitis virus G-expressing plasmid. Supernatants were collected and used to infect fresh 293T/MCAT (3) cell cultures. Luciferase activity in these cultures was then measured using the luciferase assay system (Promega) as directed by the manufacturer.
Recombinant protein purification.
The HIV-1 Δp6 Gag protein was expressed and purified as previously described (8, 12, 13).
The Gag-Z proteins were expressed and purified from BL21(DE3)pLysS E. coli cells. The culture was allowed to grow at 37°C until the A600 reached approximately 0.8. It was then induced with 0.5 mM isopropyl thiogalactoside (IPTG) (American Bioanalytical) and allowed to shake for another 4 h at 30°C. Cell pellets were frozen at −80°C. Upon thawing, they were resuspended on ice in buffer A [50 mM Tris-HCl, 5 mM β-mercaptoethanol, 1 mM tris(2-carboxyethyl)phosphine, pH 7.4] with 50 mM NaCl, at an approximate ratio of 10 ml/g cells. The resuspended sample was treated with benzonase nuclease (2.5 U/ml) (Novagen), in the presence of 1 mM MgCl2, for 30 min on ice. The NaCl concentration was then increased to a final concentration of 250 mM before disruption of cells by sonication. Upon completion of lysis by sonication, the sample was briefly centrifuged (15 min, 5,000 × g, 4°C) to remove cellular debris. The cleared lysate was then treated with 0.1% polyethyleneimine from a 5% Tris-buffered, pH 7.4 stock for 15 min on ice with stirring. The precipitated debris was removed by centrifugation (15 min, 14,000 × g, 4°C). The desired protein was precipitated from the supernatant fraction by the addition of 0.5 volumes of saturated ammonium sulfate. The lysate-ammonium sulfate mixture was stirred for 1 h on ice, and the protein of interest was then pelleted by centrifugation (30 min, 14,000 × g, 4°C). The protein pellet from this step was resuspended in buffer A with 250 mM NaCl and stored at −80°C to inactivate remaining benzonase. The thawed protein was dialyzed for 6 to 10 h against the same buffer, aliquoted for future use, and stored at −80°C. Examination of Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels indicated that that the Gag-Z proteins were ∼ 50% pure following this procedure. They appear to have negligible nucleic acid contamination, since their A260/A280 ratio was typically ∼0.6; assembly studies were performed with these preparations.
For experiments requiring a higher degree of purity, the protein was further purified on a Superdex-200 10/300 GL (GE Healthcare) column (buffer A with 250 mM NaCl). The resulting chromatogram revealed a slightly asymmetric peak, with a slight trailing off of the protein as elution progressed, possibly indicating a mixture of species or some interaction with the column matrix. Only the apex of the protein peak, i.e., column fractions with absorbance of ≥50% of the peak absorbance, were collected and used. We estimate that these preparations resulted in 90% purity of the Gag-Z proteins. Protein concentrations were calculated using spectrophotometry. The protein sample was diluted in 8 M guanidine-HCl and analyzed at by A280 using extinction coefficients calculated as described previously (20).
In vitro assembly and analysis by EM.
In vitro assembly assays were performed as previously described (8, 12). Supernatant and pellet fractions were separated and further analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) or by EM to visualize particle morphologies. Unless specified otherwise, assembly reaction mixtures contained protein at 1 mg/ml (20 μM), nucleic acid at 0.1 mg/ml, and inositol pentakisphosphate (IP5) or inositol hexakisphosphate (IP6) at 20 μM. For examination by EM, samples were spotted on Formvar-coated grids and stained with 2% (wt/vol) uranyl acetate. Grids were imaged with either a Hitachi H7600 or H7650 electron microscope. Particle sizes were measured using the measurement program of the camera software, AMTv600.
Sedimentation velocity measurements.
Boundary sedimentation velocity analysis was carried out in an Optima XL-A analytical ultracentrifuge (Beckman-Coulter Instruments) as described previously (11). Four hundred microliters of protein solution was centrifuged at 20°C in buffer A with 250 mM NaCl. Absorbance scans were obtained at either 280 nm, 254 nm, or 230 nm, depending on the protein concentration. The sedimentation coefficient was calculated with Sedfit (48).
Cross-linking of proteins.
Cross-linking with dimethyl suberimidate (DMS) (Pierce Chemicals) was performed as previously described (8) with the following changes. Proteins were dialyzed into 50 mM HEPES, pH 7.8. Reactions were allowed to proceed in the presence of 200 μM DMS for 1 h at ambient temperature in the dark. Reactions were terminated by addition of 20 mM glycine (pH 2.0), and samples were analyzed by SDS-PAGE.
Fluorescence anisotropy.
Binding of Δp6 Gag and Gag-Z proteins to nucleic acid was measured by fluorescence anisotropy as previously described (10) with the following changes: 500 nM protein solutions were serially diluted in twofold steps into 10 nM solutions of the fluoresceinated decanucleotide d(TG)5 or d(A)10 in 10 mM HEPES (pH 7.5), 100 μM tris(2-carboxyethyl)phosphine, and 5 mM β-mercaptoethanol with either 25 or 150 mM NaCl. When Δp6 Gag was used, the buffer also contained 1 μM ZnCl2.
RESULTS
Properties of the Gag-Z VLPs.
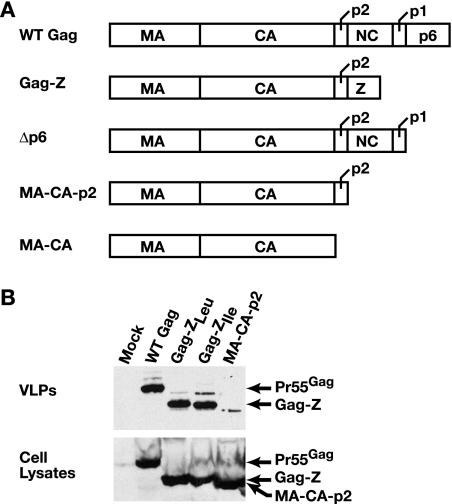
As indicated above, we have characterized chimeric proteins in which the MA, CA, and p2 domains of HIV-1 Gag are fused to either dimerizing (Gag-ZLeu) or trimerizing (Gag-ZIle) zipper motifs, as depicted in Fig. 1A. We tested the ability of the Gag-Z proteins to assemble into VLPs as follows. The plasmids were transfected into 293T cells, and the culture fluid was collected and pelleted through 20% sucrose. The pellets were then analyzed by immunoblotting with anti-p24CA antiserum. As shown in Fig. 1B, both Gag-Z proteins produced VLPs with roughly the same efficiency as WT Gag, in agreement with previous reports (1, 59).
FIG. 1.
(A) Schematic representation of the HIV-1 Gag chimeras utilized in these experiments. (B) Anti-p24CA immunoblot of released VLPs and cell lysates from transfected 293T HEK cells at 72 h posttransfection.
To assess the role of the zipper motifs in the assembly of the Gag-Z proteins, we also transfected a plasmid encoding the truncated protein MA-CA-p2. Figure 1B also shows that this protein was expressed well in the cells, but much lower levels of this protein were released in pelletable form. Taken together, these results confirm that the zipper motifs contribute to the assembly capability of the Gag-Z proteins.
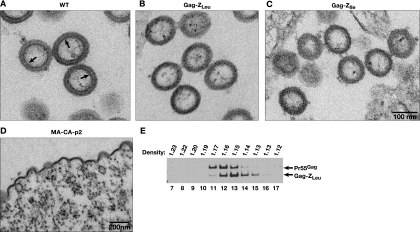
It was also of interest to investigate the morphologies of the Gag-Z VLPs. Figure 2 shows EMs of thin sections of cells transfected with the Gag-Z constructs, in addition to WT Gag and the truncated Gag. Typical WT Gag VLPs are ∼125 nm in diameter, as shown in Fig. 2A. Both Gag-ZLeu (Fig. 2B) and Gag-ZIle (Fig. 2C) chimeras produced spherical particles, ∼125 nm in diameter, which were extremely similar to those produced by WT Gag. The truncated Gag, which fails to effectively release particles, largely accumulates on the plasma membrane of the cell (Fig. 2D), with very little release of VLPs. Upon close inspection of the VLPs, we noted that the WT Gag VLPs have a very distinctive, darkly stained, thin line present on the inner side of the protein ring of the VLP. In contrast, the Gag-Z particles lack this inner stained line (Fig. 2). Thus, while the Gag-Z chimeras assemble into VLPs very similar in overall size and shape to those formed by WT Gag, the structures of the VLPs are not identical.
FIG. 2.
Thin-section EM images of 293T HEK cells at 72 h posttransfection. (A) WT HIV-1 Gag. (B) Gag-ZLeu. (C) Gag-ZIle. (D) MA-CA-p2. Panels A to C are at the same magnification. Arrows in panel A indicate the thin line under the Gag shell in WT VLPs. (E) Anti-p24CA immunoblot analysis of isopycnic sucrose gradient separation of WT Gag and Gag-ZLeu VLPs. Fractions were collected from the bottom of the tube; only fractions 7 to 17 are portrayed here.
The Gag protein of HIV-1, like those of most retroviruses, is modified by N-terminal myristylation (Myr+) (49, 52). This modification is crucial for targeting the protein to the plasma membrane and in turn for release of assembled particles (6, 22, 45). We tested the ability of Gag-Z proteins lacking myristate at their N termini to assemble in mammalian cells. Myristylation was blocked by replacing the glycine codon just 3′ of the initiator methionine codon with an alanine codon. When the resulting G2A mutant Gag-Z proteins were expressed in 293T cells, no VLPs were released; however, examination of thin sections by EM showed that VLPs were abundant in the cytoplasm of these cells. Similar VLPs were also observed in cells expressing full-length G2A Gag (data not shown).
We also characterized the WT and Gag-Z VLPs with respect to buoyant densities. WT retrovirus particles have a density of 1.14 to 1.18 g/ml (53). WT and Gag-ZLeu VLPs were pelleted, and the pellets were resuspended, mixed, and overlaid on a 15 to 60% sucrose gradient. After centrifugation, fractions were collected and analyzed by immunoblotting with anti-p24CA antiserum. As shown in Fig. 2E, the Gag-ZLeu VLPs (detected as a ∼45-kDa band) overlap in density with the WT Gag (∼55-kDa) VLPs, but the peak for the chimeras tends to be at a somewhat lower density. Specifically, the peak fraction of WT Gag VLPs is at 1.16 g/ml, while that of Gag-ZLeu VLPs typically ranged between 1.14 and 1.15 g/ml. The same difference in density has also been observed with Gag-ZIle (data not shown).
RNA content of Gag-Z VLPs.
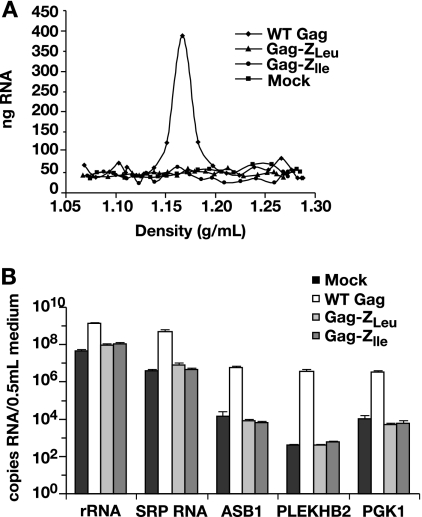
As the Gag-Z proteins lack NC, the primary nucleic acid-binding domain of Gag, it was of considerable interest to determine whether VLPs assembled from these proteins contain RNA. We purified VLPs for these experiments by isopycnic banding in a sucrose gradient. VLPs from WT Gag, Gag-ZLeu, and Gag-ZIle were equilibrated on 15 to 60% sucrose gradients. A parallel sample from a “mock” transfection with a control vector, BlueScript, was used as a negative control. The gradient was fractionated from the bottom into approximately 20 samples. RNA was extracted from each and quantitated using the Ribogreen assay. VLPs were localized in the gradients by immunoblotting as in Fig. 2E. As seen in Fig. 3A, RNA in the WT Gag gradient is in a sharp peak at 1.16 g/ml, as expected; this profile is superimposable with that of Pr55Gag (data not shown). In contrast, while the Gag-Z VLPs were readily detected at ∼1.15 g/ml by immunoblotting (not shown), there was no RNA peak in the Gag-Z gradients; indeed, the RNA profiles were not significantly different from that of the mock-transfected sample. From this assay, we estimate that Gag-Z VLPs contain no more than 10% total RNA compared to WT RNA encapsidation levels.
FIG. 3.
(A) RNA quantitation by Ribogreen measurements of the sucrose gradient fractionation of WT, Gag-ZLeu, and Gag-ZIle VLPs, as well as a mock transfection. (B) Real-time RT-PCR quantitation of rRNA, SRP RNA, ASB1 mRNA, PLEKHB2 mRNA, and PGK1 mRNA in WT, Gag-ZLeu, and Gag-ZIle VLPs, as well as a “VLP” preparation from a mock transfection. Error bars indicate standard deviations.
We further analyzed the Gag-Z VLPs by assaying for specific RNA species using real-time RT-PCR, which is potentially far more sensitive than the Ribogreen assay. We measured rRNA, the most abundant RNA in the cell; SRP RNA, which is known to be encapsidated in WT VLPs (4, 14, 39, 40, 46); and three mRNA species. Two of these, i.e., ASB1 and PLEKHB2, are selectively packaged in retrovirus particles, while the third, i.e., PGK1, is not (46).
As shown in Fig. 3B, the amount of rRNA detected in the Gag-Z chimeras was ∼10-fold less than that found in WT VLPs and not significantly different from that in the “VLP” preparation from the mock-transfected cells. Similarly, SRP RNA was ∼100-fold less in the Gag-Z VLPs than in the WT VLPs, and the three mRNA species were ≥1,000-fold less in the Gag-Z VLPs. In each case, RNA levels for the Gag-Z VLPs were very near those of background levels. Although the Gag-Z proteins might be encapsidating an RNA species we have not tested, the data show that the Gag-Z VLPs contain far less RNA than WT VLPs and are consistent with the possibility that they assemble without incorporating any RNA.
Interaction of Gag-Z proteins with WT Gag.
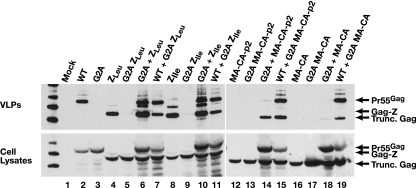
It seemed possible that the Gag-Z proteins, which appear to assemble without RNA, might fail to interact with WT Gag, which requires RNA for assembly. We tested the ability of the Gag-Z proteins to coassemble with WT Gag by expressing a Gag-Z protein together with WT Gag; in each case, one of the two proteins was a G2A mutant. Equal amounts of the two plasmids were cotransfected into 293T cells. VLPs were then harvested and analyzed by immunoblotting for the presence of the G2A protein. As can be seen in Fig. 4, all of the proteins were present at substantial levels in the cell lysates. No detectable VLPs were released when G2A proteins were expressed alone, but with each combination, e.g., Myr+ WT Gag and G2A Gag-Z, or the reciprocal pair, i.e., G2A WT Gag together with Myr+ Gag-Z, approximately equal amounts of the two proteins were present in the VLPs. Thus, the Gag-Z proteins are able to coassemble efficiently with the WT Gag despite the absence of the NC domain.
FIG. 4.
Anti-p24CA immunoblot analysis of released VLPs and cell lysates resulting from cotransfection of myristylated (Myr+) and G2A unmyristylated WT Gag, Gag-Z proteins, and truncated Gag proteins in 293T HEK cells.
We also tested two truncated Gag proteins, i.e., MA-CA-p2 and MA-CA, for their ability to coassemble with WT Gag. Figure 4 shows that neither of these proteins assembles into substantial levels of VLPs when expressed alone in 293T cells, nor do myristylated (“Myr+”) truncated proteins efficiently rescue G2A WT Gag into VLPs (lanes 14 and 18). In contrast, Gag efficiently rescued G2A MA-CA-p2 (lane 15) but not G2A MA-CA (lane 19). Thus, the zipper moieties are essential for the ability of the Gag-Z proteins to rescue G2A WT Gag, while the truncated protein containing p2 can evidently interact with WT Gag.
The effects of incorporation of Gag-Z proteins upon the specific infectivity of WT virions were also determined. We observed a mild dominant-negative effect of the Gag-Z proteins (data not shown), further indicating that they can coassemble with WT Gag.
Properties of recombinant Gag-Z proteins.
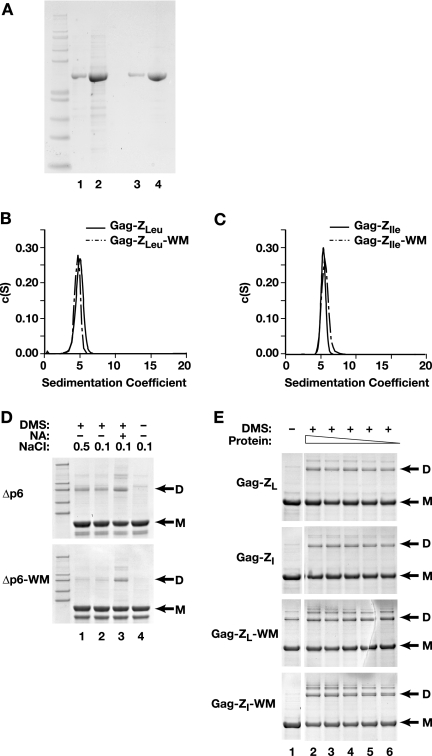
Analysis of VLP assembly using recombinant Gag proteins in a defined in vitro system has been a valuable experimental approach to retrovirus assembly. We and others have previously characterized the properties of recombinant HIV-1 Gag protein (7, 8, 23). This protein, the “WT Gag” control for recombinant protein experiments, lacks the p6 domain and is designated Δp6. (This truncation has been shown to have no significant effect on immature particle morphology [27, 56].) In addition, none of the recombinant proteins studied here has the myristate modification found on Gag made in mammalian cells. We expressed the two Gag-Z proteins in E. coli and purified them as described in Materials and Methods. Interestingly, they were soluble in the bacterial lysates; in other words, they did not assemble spontaneously following expression in bacteria. Typical preparations are shown in a Coomassie blue-stained SDS-polyacrylamide gel in Fig. 5A.
FIG. 5.
(A) Coomassie blue-stained SDS-PAGE of recombinant Gag-ZLeu protein, purified from E. coli. Lane 1 (1 μg) and lane 2 (10 μg) show the (NH4)2SO4-precipitated protein used for in vitro assembly reactions. Lane 3 (1 μg) and lane 4 (10 μg) show the fast protein liquid chromatography-purified fraction used for analytical ultracentrifugation and fluorescence anisotropy. Similar results were obtained with Gag-ZIle (not shown). Protein molecular weight marker, Mark 12 (Invitrogen). (B) Sedimentation velocity results for the recombinant Gag-ZLeu and Gag-ZLeu-WM proteins, at 5.4 μM. (C) Sedimentation velocity results for the recombinant Gag-ZIle and Gag-ZIle-WM proteins, at 5.4 μM. (D) Coomassie blue-stained SDS-PAGE of the DMS cross-linking reactions of Δp6 Gag and Δp6 Gag-WM proteins. Protein molecular weight marker, Mark 12 (Invitrogen). Δp6 Gag is ∼50 kDa. Lane 1, DMS cross-linked reaction in 0.5 M NaCl; lane 2, DMS cross-linked reaction in 0.1 M NaCl; lane 3, DMS cross-linked reaction in the presence of nucleic acid (0.1 mg/ml yeast tRNA) in 0.1 M NaCl; lane 4, no DMS. All protein concentrations were 20 μM. (E) Coomassie blue-stained SDS-PAGE of the DMS cross-linking reactions of Gag-ZLeu, Gag-ZIle, Gag-ZLeu-WM, and Gag-ZIle-WM. Lane 1 is the control reaction, non-cross-linked sample. Lanes 2 to 6 are DMS cross-linked reactions in 0.1 M NaCl, in the absence of nucleic acid. Protein reaction concentrations: lane 2, 43.5 μM; lane 3, 21.7 μM; lane 4, 10.8 μM; lane 5, 5.4 μM; lane 6, 2.7 μM.
The zipper motifs present in these Gag chimeras were shown to exist as dimers (ZLeu) and trimers (ZIle) by X-ray crystallography of the free zipper peptides (24, 41). However, it was possible that the Gag moiety might somehow influence the oligomerization of the Gag-Z proteins. We therefore attempted to characterize the state of the two proteins in solution. As one experimental approach, we characterized the Gag-ZLeu and Gag-ZIle chimeras by sedimentation velocity analysis. In repeated tests of both Gag-Z proteins, using protein concentrations ranging from 1.7 μM to 43.5 μM, the S values of both proteins varied from ∼3.0 to ∼6.0. In general, they tended to increase with increasing protein concentration. There was slight variability, however, in S values between different protein preparations. Figure 5B and C show representative analyses at a protein concentration of 5.4 μM, where both proteins sediment with S values near 5.0. We also noted that in almost every experiment, the Gag-ZIle protein exhibited a slightly higher S value than the Gag-ZLeu protein (as in Fig. 5B and C, in which the Gag-ZIle profile has a peak at 5.4S and the Gag-ZLeu at 5.0S). Taken together, these observations suggest that the proteins are similar in their oligomeric status but that there is a tendency for formation of some larger forms in addition to the major oligomeric species.
We have reported that Δp6 Gag is in monomer-dimer equilibrium in solution, with a Kd of ∼10−5 M (13). This interaction is due to the dimer interface in its CA domain. In contrast, the “WM” mutant of Δp6 Gag, in which two residues at the dimer interface, i.e., Trp 316 and Met 317, are replaced with alanines, is almost entirely monomeric in solution (11). We therefore tested WM mutants of the Gag-Z proteins to determine whether the dimer interface is contributing significantly to oligomerization of these proteins. As shown in Fig. 5B and C, the sedimentation profiles of the two WM Gag-Z proteins are nearly superimposable upon those of the Gag-Z proteins themselves. Thus, the oligomerization of the Gag-Z proteins is evidently mediated by the zipper domains and is independent of the dimer interface in CA.
The S values obtained for the Gag-Z proteins are far higher than that of the monomeric WM Δp6 Gag, which sediments at 2.6S (11). As anticipated, therefore, they appear to be oligomeric in solution. As an independent test of this conclusion, we also analyzed them by DMS cross-linking. Controls included Δp6 Gag and the WM mutant of Δp6. As shown in Fig. 5D, cross-linking generated dimers of Δp6 in either 0.1 or 0.5 M NaCl, but almost no cross-linked dimers were observed with WM Δp6. (As a positive control on the latter negative result, we also tested the ability of WM Δp6 to be cross-linked in the presence of nucleic acid. These conditions lead to the close juxtaposition of the mutant protein molecules in assembled VLPs. As expected, cross-linked dimers were observed in this case [lane 3].) These results show that Δp6 can be cross-linked in solution because of the activity of the dimer interface, i.e., Trp 316 and Met 317.
Both Gag-Z proteins were also cross-linked by DMS (Fig. 5E); the efficiency of cross-linking did not vary as the protein concentration was decreased from 43.5 to 2.7 μM. The oligomerization detected in these proteins did not depend on the dimer interface in CA, as WM mutants of the Gag-Z proteins were cross-linked as efficiently as the Gag-Z proteins themselves (Fig. 5E). These results strongly support the idea that the Gag-Z proteins oligomerize via their zipper motifs. The similarity of the results with Gag-ZLeu and Gag-ZIle, as well as the preponderance of the dimeric cross-linking product, suggests that both the Gag-Z proteins are largely dimeric in solution.
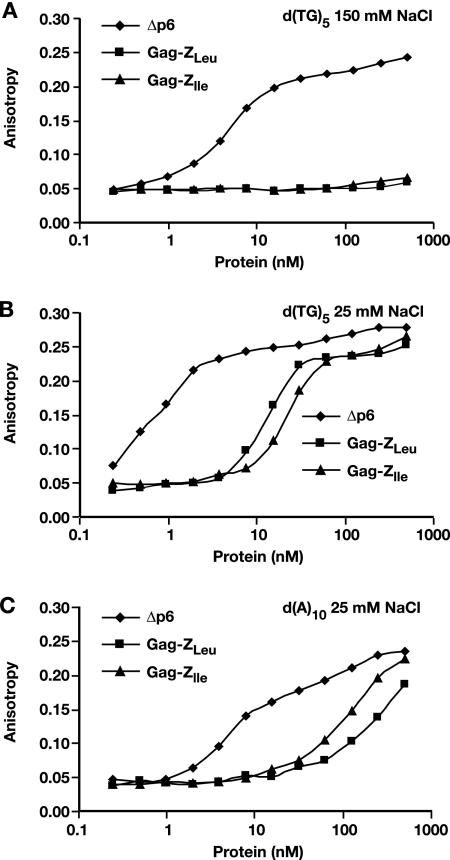
It was of considerable interest to determine whether the recombinant Gag-Z proteins could bind nucleic acids, despite the lack of an NC domain. We tested their ability to bind the single-stranded oligodeoxynucleotides d(TG)5 and d(A)10 using fluorescence anisotropy measurements. At 0.15 M NaCl, no binding of either Gag-Z protein to d(TG)5 was detected, although binding by Δp6 was evident (Fig. 6A).
FIG. 6.
Fluorescence anisotropy measurements of oligodeoxynucleotide binding to recombinant Δp6 Gag and Gag-Z proteins. (A) Ten nanomolar d(TG)5 at 150 mM NaCl. (B) Ten nanomolar d(TG)5 at 25 mM NaCl. (C) Ten nanomolar d(A)10 at 25 mM NaCl.
We also performed similar assays at lower salt concentrations. We found that the Gag-Z proteins could bind d(TG)5 at 25 mM NaCl (Fig. 6B); weaker binding was also observed at 50 and 100 mM NaCl (data not shown). In each case, the affinity of the Gag-Z proteins for d(TG)5 was considerably lower than that of Δp6, since the increase in anisotropy was seen only at Gag-Z concentrations more than 10-fold higher than with the Δp6 control.
Finally, we measured the ability of the proteins to bind d(A)10 in 25 mM NaCl. While the affinity of Δp6 for this oligodeoxynucleotide is significantly lower than its affinity for d(TG)5, the binding of the Gag-Z proteins to d(A)10 was far weaker, with no significant interaction observed at protein concentrations of <30 to 60 nM (Fig. 6C). Taken together, the data show that the Gag-Z proteins can each bind nucleic acids, albeit with a significantly lower affinity than that of Δp6 Gag.
Assembly properties of the recombinant Gag-Z proteins.
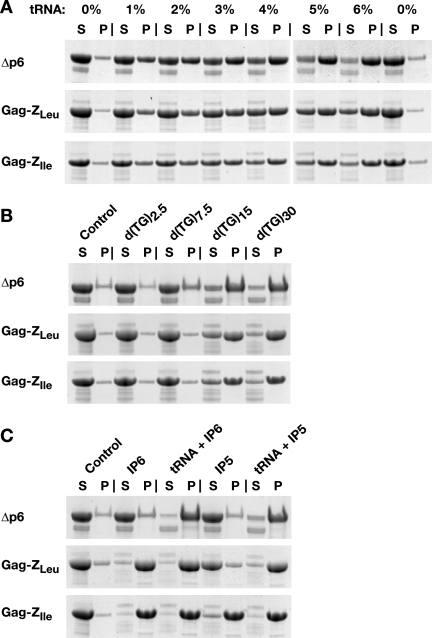
Previous studies have shown that addition of nucleic acid induces Δp6 to assemble into VLPs (8). We tested the ability of nucleic acids to promote the assembly of the Gag-Z proteins. Figure 7A shows the results of a titration in which increasing amounts of tRNA were added to Δp6 and the Gag-Z proteins, and the efficiency of assembly was evaluated by a pelleting assay. It can be seen that the proteins become pelletable in the presence of the RNA. However, low levels of RNA render a smaller proportion of the Gag-Z proteins pelletable than Δp6; for example, the majority of Δp6 is assembled in only 3 to 4% tRNA (i.e., 30 to 40 μg tRNA to 1 mg protein), while ≥∼6% tRNA is required for efficient assembly of the Gag-Z proteins.
FIG. 7.
Coomassie blue-stained SDS-PAGE pelleting assay of in vitro assembly reactions. (A) In vitro assembly of recombinant Δp6 Gag and Gag-Z proteins with various amounts of yeast tRNA. (B) In vitro assembly of recombinant Δp6 Gag and Gag-Z proteins with various lengths of d(TG)n oligodeoxynucleotides. (C) In vitro assembly of recombinant Δp6 Gag and Gag-Z proteins with inositol phosphates with or without yeast tRNA. S, supernatant fraction. P, pellet fraction.
We also tested the ability of d(TG)n oligodeoxynucleotides of different lengths to support the assembly of the Gag-Z proteins (8). As shown in Fig. 7B, Δp6 Gag pelleted efficiently in the presence of either d(TG)30 or d(TG)15. Limited pelleting (∼25%) was seen with d(TG)7.5, and negligible pelleting was detected with the shorter oligodeoxynucleotide d(TG)2.5. In contrast, the Gag-Z proteins assembled efficiently only in the presence of the longest oligodeoxynucleotide, d(TG)30. There was still significant, although not complete, pelleting seen with d(TG)15, but only negligible amounts of pelleting were seen with the shorter oligodeoxynucleotides. Thus, the Gag-Z proteins require longer d(TG)n oligonucleotides than Δp6 for efficient pelleting.
We also found that d(A)n oligodeoxynucleotides, which are far less effective than d(TG)n oligodeoxynucleotides at supporting Δp6 assembly (8, 15), did not promote detectable levels of pelleting of the Gag-Z proteins under our assembly conditions (data not shown).
IP5 and IP6 modulate the assembly of Δp6 in the presence of nucleic acids (7). Specifically, VLPs formed by Δp6 in nucleic acid alone are far smaller than authentic virus particles; supplementation of these reactions with IP5 “corrects” the radius of curvature of the particles, so that VLPs formed in both nucleic acid and IP5 are the correct size. The effects of IP6 are similar, though not identical, to those of IP5 in these experiments. We also tested the effects of these compounds on assembly of the Gag-Z proteins, both in the presence and the absence of tRNA. Remarkably, either IP5 or IP6 alone supported assembly of the Gag-Z proteins, while Δp6 remained soluble under these conditions (Fig. 7C). A larger fraction of the Gag-Z proteins assembled in IP6 than in IP5, and somewhat more Gag-ZIle than Gag-ZLeu assembled with the inositol phosphates. All three proteins were quantitatively assembled in RNA plus IP5 or RNA plus IP6.
As both RNA and inositol phosphates are polyanions, we tested the ability of other polyanions to induce assembly of the Gag-Z proteins. We found that the two proteins remained almost entirely soluble in the presence of a 1:1 molar ratio of inositol hexasulfate (IS6) or even a 10-fold molar excess of IS6. In contrast, they pelleted upon addition of 10% or 50% heparin (i.e., with a heparin/protein mass ratio of 1:10 or 1:2) (data not shown).
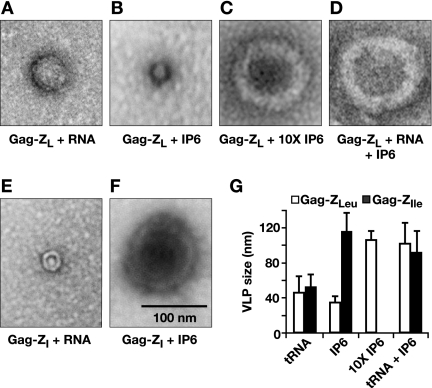
Finally, the VLPs assembled from the Gag-Z proteins in the presence of these cofactors were examined by negative staining and EM. We found that Gag-ZLeu, like Δp6, formed small VLPs (∼45 nm in diameter) in RNA alone (Fig. 8A) or 1 equivalent of IP6 alone (Fig. 8B) but formed full-size VLPs in 10 equivalents of IP6 (Fig. 8C) or 1 equivalent of IP6 plus either 10% RNA (Fig. 8D), 10% heparin, or 9 equivalents of IS6 (data not shown). In contrast, Gag-ZIle formed small VLPs in RNA alone (Fig. 8E) but, unlike either of the other proteins, full-size VLPs in 1 equivalent of IP6 alone (Fig. 8F). While the Gag-Z proteins pelleted in 10% or 50% heparin, the pellets contained only aggregates, not recognizable VLPs. The sizes of the VLPs formed with the two cofactors are tabulated in Fig. 8G.
FIG. 8.
EMs of in vitro-assembled products from Gag-Z proteins. (A) Gag-ZLeu plus tRNA. (B) Gag-ZLeu plus 1 equivalent IP6. (C) Gag-ZLeu plus 10 equivalents IP6. (D) Gag-ZLeu plus tRNA plus 1 equivalent IP6. (E) Gag-ZIle plus tRNA. (F) Gag-ZIle plus 1 equivalent IP6. All six images are at the same magnification. (G) Summary of in vitro-assembled Gag-Z VLP sizes. Twenty to 50 particles were measured from each set of reactions. Error bars indicate standard deviations.
DISCUSSION
Nucleic acid is required for retroviral assembly (8, 9, 37). As one approach to understanding the role of nucleic acid in assembly, Zhang et al. (59) replaced the NC domain (the principal nucleic acid-binding domain) of HIV-1 Gag with a leucine zipper motif. Remarkably, they found that the resulting chimeric protein was still capable of efficient assembly into VLPs in mammalian cells. The present study constitutes a detailed analysis of the assembly properties of these chimeric proteins.
Briefly, we found that both of the chimeric proteins assemble in human cells with roughly the same efficiency as WT Gag. The zipper moieties are essential for their assembly competence, since a truncated Gag containing neither NC nor a zipper motif could not assemble efficiently (Fig. 1B and 2). The VLPs are very similar in morphology to those formed by WT Gag. Their buoyant density is slightly lower than that of the WT controls (Fig. 2). They appear to lack RNA (Fig. 3). The Gag-Z proteins can coassemble into VLPs together with WT Gag (Fig. 4).
We also characterized recombinant Gag-Z proteins purified from E. coli. Both of the proteins are evidently oligomeric in solution over a considerable concentration range (Fig. 5). They can bind nucleic acids, despite the absence of the NC domain (Fig. 6). Significantly, they do not assemble spontaneously but (like WT Gag) require cofactors for VLP formation. Either nucleic acid or inositol phosphates can serve as assembly cofactors. The “trimerizing” Gag-Z protein, Gag-ZIle, forms full-size VLPs upon addition of one equivalent of IP6, while Gag-ZLeu requires IP6 plus additional polyanions for full-size VLP assembly (Fig. 7 and 8).
Assembly of Gag-Z proteins in mammalian cells.
The striking morphological resemblance of Gag-Z VLPs to WT Gag VLPs shows that the overall morphology and radius of curvature are determined by the MA-CA-p2 regions of Gag, in agreement with prior findings in other systems (2, 5). However, one deviation from the morphology of WT VLPs was the absence of a darkly stained ring on the interior of the shell of the VLPs. Since we could not detect RNA in the Gag-Z VLPs, it seems likely that this ring is RNA bound to the NC domain of WT Gag.
Data both from Ribogreen analysis of sucrose-gradient fractions and from assays for specific RNA species by the far more sensitive real-time RT-PCR technique showed in all cases that the RNA content of the Gag-Z VLPs was very near background levels; the results are consistent with the idea that these proteins assemble into VLPs without incorporating RNA. This conclusion might be expected, as these proteins lack the NC domain, and is consistent with a prior report by Zennou et al. (58).
The Gag-Z VLPs have a slightly lower buoyant density, i.e., 1.14 to 1.15 g/ml, than that of WT VLPs (1.16 g/ml) (Fig. 2E). It has previously been suggested (49) that a reduction in buoyant density in some mutant VLPs (“I domain” mutants) implies that the mutant Gag proteins are not as tightly packed as those in WT VLPs. However, the packing of the Gag-Z proteins may actually be identical to that of WT Gag. Under our experimental conditions, the buoyant density of a retrovirus particle simply reflects its mass per unit volume. The Gag-Z VLPs are roughly the same size and shape as WT VLPs and thus are roughly the same volume. Their total mass derives principally from their lipid membrane and their Gag-Z proteins (54). In turn, the mass of protein is the mass of a monomer times the number of monomers. As each Gag-Z monomer is approximately 45 kDa, rather than the 55 kDa of a WT Gag molecule, the reduction in buoyant density in Gag-Z VLPs could be due solely to the lower molecular weight of each Gag-Z monomer, and Gag-Z VLPs could contain, on average, the same number of monomers as WT VLPs. Thus, the spacing between monomers may be the same as in WT VLPs and is presumably determined solely by the MA-CA-p2 portion of Gag, which is common to the Gag-Z and WT proteins (2, 5).
The ability of the Gag-Z proteins to coassemble with WT Gag proteins shows again that the protein-protein interactions involved in assembly are all within the MA-CA-p2 portion of Gag (2). It also argues strongly against the idea that the role of nucleic acid in assembly is to act as a “scaffold” to which all of the coassembling proteins bind (37).
Assembly of Gag-Z proteins in a defined system in vitro.
Perhaps the most remarkable and unexpected observation in this study is the fact that purified recombinant Gag-Z proteins require cofactors for assembly in vitro. The efficient assembly of retroviral Gag-Z proteins in eukaryotic cells, despite the absence of the NC domain, has prompted the hypothesis that nucleic acids promote assembly of WT Gags simply by promoting oligomerization (28, 30, 31). However, our results indicate that the oligomerization mediated by the zipper motifs is not, in fact, sufficient for assembly. (It might be suggested that the absence of myristate from the recombinant proteins affects their assembly properties, so that the in vitro results do not accurately represent the behavior of the myristylated proteins in cells. However, this is unlikely, since unmyristylated G2A mutant Gag-Z proteins assemble within the cytoplasm of human cells [data not shown].)
What cofactors might be used by the Gag-Z proteins for assembly in eukaryotic cells? The apparent absence of RNA in the particles suggests that some other cofactor, perhaps an inositol phosphate-related molecule or another polyanion, is used. (We cannot exclude the possibility that the particles contain DNA.)
We do not know what residues in the MA-CA-p2 portion of the Gag-Z proteins contact nucleic acids, but MA has a positively charged face (26, 32) and has been reported to bind nucleic acids (29, 44). Thus, we imagine that the Gag-Z proteins bind nucleic acids via their MA domains. In any case, the fact that the Gag-Z proteins can bind nucleic acid (Fig. 6) implies that regions of Gag other than NC could conceivably participate in the packaging of RNA during normal assembly. Analysis of NC-RNA interactions may thus be inadequate as an approach to the study of genomic RNA packaging.
Measurements of both binding (Fig. 6) and assembly (Fig. 7) showed that the Gag-Z proteins have a higher affinity for oligodeoxynucleotides with the sequence d(TG)n than for those with d(A)n. A similar preference was previously observed with HIV-1 NC (16, 17) and Gag (15) and indeed is seen with lysine20 as well (A. Stephen and R. Fisher, unpublished results). It seems possible that the propensity of adenosine residues to stack upon each other and/or the resulting rigidity of d(A)n interferes with the binding of proteins to this oligonucleotide (21, 33).
We attempted to characterize the state of the Gag-Z proteins in solution using sedimentation velocity analysis and chemical cross-linking. Despite some variability in the sedimentation velocity data, the results all show that both proteins are oligomeric over a wide concentration range. The results with the two proteins are not identical, but they are so similar that it seems very unlikely that the Gag-ZIle protein forms an oligomer that is 50% more massive than that of Gag-ZLeu, as expected from the properties of the free zipper moieties (24, 25, 41, 42). We therefore believe that both proteins are predominantly dimeric in free solution. Presumably, other domains of Gag interfere with the addition of a third monomer to a Gag-ZIle dimer, essentially by steric hindrance.
We have previously reported (8) that Δp6 assembles into very small VLPs (only 25 to 30 nm in diameter) in nucleic acid alone and into full-size VLPs in nucleic acid plus IP5 (a compound closely related to IP6) (7). As Gag is ∼25 nm long in authentic immature VLPs (19, 56, 57), Δp6 is almost certainly folded over in the VLPs formed with nucleic acid alone in vitro, as it is in solution (11). Presumably, basic residues in both the MA and NC domains are interacting with the nucleic acid in these VLPs. Addition of IP5 seems to somehow promote the extension of Δp6 into a rod-shaped protein in the VLPs (7). Perhaps in the presence of both nucleic acid and IP6, the NC domain of Δp6 remains bound to nucleic acid while IP6 binds the MA domain; this decoupling of the two domains allows the protein to extend into a rod (13).
In the absence of nucleic acid, IP6 induces trimerization of Δp6 in solution, and basic residues in both MA and NC contribute to the binding of IP6 to Δp6 in 0.5 M NaCl (13). In contrast, IP6 does not bind to, or cause trimerization of, Δp6 which lacks most of the MA domain (“Δ16-99”) under these conditions. Because of the extremely high charge density of IP6, we speculate that it neutralizes repulsive intermolecular interactions between positively charged residues in the MA domain and that this neutralization permits trimeric MA-MA interactions in solution. Trimers of MA have also been observed when the protein is N-terminally myristylated (50) and in crystals of MA (26). Bacterially expressed, histidine-tagged MA is also evidently trimeric at physiological ionic strength (34). A functional linkage between this trimerization of Δp6 in solution and its assembly into full-size VLPs in nucleic acid plus IP5 is suggested by the fact that both require an intact dimer interface in the CA domain (11, 13).
The VLPs assembled in IP6 alone (at 1 mol per mol of protein) are small in the case of Gag-ZLeu but full size in Gag-ZIle. This is the only major difference that we observed between the two Gag-Z proteins, and it is particularly surprising since both proteins are apparently almost totally dimeric in free solution. Perhaps the difference between the two Gag-Z proteins becomes apparent only once IP6 neutralizes charges in MA, thus relieving the steric hindrance that interferes with trimerization of Gag-ZIle. However, Gag-ZLeu does form full-size VLPs if the IP6 is supplemented with additional polyanion; the latter requirement appears to be quite nonspecific, as it can be fulfilled by RNA, IS6, or heparin as well as IP6.
The ability of the Gag-Z proteins to assemble in IP6 alone (Fig. 7) is a striking difference from the behavior of Δp6. One possible explanation is that assembly requires both oligomerization at the C-terminal end of the protein and elimination of intermolecular repulsion at the N-terminal end; in turn, the repulsion at the N terminus can be neutralized by an inositol phosphate or abolished by deletion (as in Δ16-99), while the C-terminal oligomerization can be induced either by juxtaposition on nucleic acid, as in Δp6, or by association through a zipper motif. With respect to assembly of full-size VLPs, the data also suggest that dimeric association at the C terminus leads to a greater requirement for charge neutralization than does trimeric association. The mechanisms behind these rather complex patterns are not yet clear.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
We thank Barbara Felber, Henrich Göttlinger, Vineet KewalRamani, Alok Mulky, Delphine Muriaux, David Ott, and Samuel J. Rulli, Jr., for reagents. We also thank Demetria Harvin and Anne Kamata for technical assistance.
Footnotes
Published ahead of print on 10 December 2008.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 745395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ako-Adjei, D., M. C. Johnson, and V. M. Vogt. 2005. The retroviral capsid domain dictates virion size, morphology, and coassembly of Gag into virus-like particles. J. Virol. 7913463-13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57659-666. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, J. M., W. E. Levinson, D. Sullivan, L. Fanshier, N. Quintrell, and J. Jackson. 1970. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 42927-937. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, J. A., M. C. Johnson, M. N. Simon, S. D. Fuller, and V. M. Vogt. 2006. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J. Mol. Biol. 355157-168. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., R. J. Fisher, E. M. Towler, S. Fox, H. J. Issaq, T. Wolfe, L. R. Phillips, and A. Rein. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 9810875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 732270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 696487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruceanu, M., M. A. Urbaneja, C. V. Hixson, D. G. Johnson, S. A. Datta, M. J. Fivash, A. G. Stephen, R. J. Fisher, R. J. Gorelick, J. R. Casas-Finet, A. Rein, I. Rouzina, and M. C. Williams. 2006. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 34593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta, S. A., J. E. Curtis, W. Ratcliff, P. K. Clark, R. M. Crist, J. Lebowitz, S. Krueger, and A. Rein. 2007. Conformation of the HIV-1 Gag protein in solution. J. Mol. Biol. 365812-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta, S. A., and A. Rein. 2009. Preparation of recombinant HIV-1 Gag protein and assembly of virus-like particles in vitro. Methods Mol. Biol. 485197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta, S. A., Z. Zhao, P. K. Clark, S. Tarasov, J. N. Alexandratos, S. J. Campbell, M. Kvaratskhelia, J. Lebowitz, and A. Rein. 2007. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J. Mol. Biol. 365799-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erikson, E., R. L. Erikson, B. Henry, and N. R. Pace. 1973. Comparison of oligonucleotides produced by RNase T1 digestion of 7 S RNA from avian and murine oncornaviruses and from uninfected cells. Virology 5340-46. [DOI] [PubMed] [Google Scholar]
- 15.Feng, Y. X., T. Li, S. Campbell, and A. Rein. 2002. Reversible binding of recombinant human immunodeficiency virus type 1 Gag protein to nucleic acids in virus-like particle assembly in vitro. J. Virol. 7611757-11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher, R. J., M. J. Fivash, A. G. Stephen, N. A. Hagan, S. R. Shenoy, M. V. Medaglia, L. R. Smith, K. M. Worthy, J. T. Simpson, R. Shoemaker, K. L. McNitt, D. G. Johnson, C. V. Hixson, R. J. Gorelick, D. Fabris, L. E. Henderson, and A. Rein. 2006. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 34472-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher, R. J., A. Rein, M. Fivash, M. A. Urbaneja, J. R. Casas-Finet, M. Medaglia, and L. E. Henderson. 1998. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 721902-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 675443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller, S. D., T. Wilk, B. E. Gowen, H. G. Kräusslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7729-738. [DOI] [PubMed] [Google Scholar]
- 20.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182319-326. [DOI] [PubMed] [Google Scholar]
- 21.Goddard, N. L., G. Bonnet, O. Krichevsky, and A. Libchaber. 2000. Sequence dependent rigidity of single stranded DNA. Phys. Rev. Lett. 852400-2403. [DOI] [PubMed] [Google Scholar]
- 22.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 865781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grättinger, B. Müller, S. Fuller, and H. G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harbury, P. B., P. S. Kim, and T. Alber. 1994. Crystal structure of an isoleucine-zipper trimer. Nature 37180-83. [DOI] [PubMed] [Google Scholar]
- 25.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 2621401-1407. [DOI] [PubMed] [Google Scholar]
- 26.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 933099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 752985-2997. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 7611177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lochrie, M. A., S. Waugh, D. G. Pratt, Jr., J. Clever, T. G. Parslow, and B. Polisky. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 252902-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 7852-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 765452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massiah, M. A., M. R. Starich, C. Paschall, M. F. Summers, A. M. Christensen, and W. I. Sundquist. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244198-223. [DOI] [PubMed] [Google Scholar]
- 33.Mills, J. B., E. Vacano, and P. J. Hagerman. 1999. Flexibility of single-stranded DNA: use of gapped duplex helices to determine the persistence lengths of poly(dT) and poly(dA). J. Mol. Biol. 285245-257. [DOI] [PubMed] [Google Scholar]
- 34.Morikawa, Y., W. H. Zhang, D. J. Hockley, M. V. Nermut, and I. M. Jones. 1998. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J. Virol. 727659-7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller, K. M., K. M. Arndt, and T. Alber. 2000. Protein fusions to coiled-coil domains. Methods Enzymol. 328261-282. [DOI] [PubMed] [Google Scholar]
- 36.Muriaux, D., S. Costes, K. Nagashima, J. Mirro, E. Cho, S. Lockett, and A. Rein. 2004. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 7812378-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 985246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muriaux, D., J. Mirro, K. Nagashima, D. Harvin, and A. Rein. 2002. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 7611405-11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onafuwa-Nuga, A. A., S. R. King, and A. Telesnitsky. 2005. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J. Virol. 7913528-13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onafuwa-Nuga, A. A., A. Telesnitsky, and S. R. King. 2006. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA 12542-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254539-544. [DOI] [PubMed] [Google Scholar]
- 42.O'Shea, E. K., R. Rutkowski, and P. S. Kim. 1989. Evidence that the leucine zipper is a coiled coil. Science 243538-542. [DOI] [PubMed] [Google Scholar]
- 43.Oshima, M., D. Muriaux, J. Mirro, K. Nagashima, K. Dryden, M. Yeager, and A. Rein. 2004. Effects of blocking individual maturation cleavages in murine leukemia virus Gag. J. Virol. 781411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purohit, P., S. Dupont, M. Stevenson, and M. R. Green. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7576-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 837246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rulli, S. J., Jr., C. S. Hibbert, J. Mirro, T. Pederson, S. Biswal, and A. Rein. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 816623-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 714892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 781606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 50.Tang, C., E. Loeliger, P. Luncsford, I. Kinde, D. Beckett, and M. F. Summers. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 101517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tobin, G. J., K. Nagashima, and M. A. Gonda. 1996. Immunologic and ultrastructural characterization of HIV pseudovirions containing Gag and Env precursor proteins engineered in insect cells. Methods 10208-218. [DOI] [PubMed] [Google Scholar]
- 52.Veronese, F. D., T. D. Copeland, S. Oroszlan, R. C. Gallo, and M. G. Sarngadharan. 1988. Biochemical and immunological analysis of human immunodeficiency virus gag gene products p17 and p24. J. Virol. 62795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27-69. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 54.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 737050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weissenhorn, W., L. J. Calder, A. Dessen, T. Laue, J. J. Skehel, and D. C. Wiley. 1997. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc. Natl. Acad. Sci. USA 946065-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Krausslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright, E. R., J. B. Schooler, H. J. Ding, C. Kieffer, C. Fillmore, W. I. Sundquist, and G. J. Jensen. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 262218-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zennou, V., D. Perez-Caballero, H. Göttlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 7812058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 721782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]