Abstract
Noncoding RNAs are a feature of many herpesvirus genomes. They include microRNAs, whose function is the subject of intense investigation, in addition to longer RNA molecules such as the Epstein-Barr virus-encoded RNAs and herpesvirus saimiri U RNAs, which have been known for some time but whose function is still not well defined. Murine gammaherpesvirus 68 (MHV-68) encodes eight viral tRNA-like molecules (vtRNAs) of unknown function. Investigating the kinetics of expression of the vtRNAs, we observed that they were present directly after infection with the virus. This strongly suggested that vtRNAs were part of the virion structure, which was confirmed by their detection within various purified, RNase-treated preparations. Although both viral and cellular mRNAs were also detected within the MHV-68 virion, the major RNA species present were small RNAs of around 70 nucleotides in length. Interestingly, incorporation of viral mRNA was not related to the relative abundance in infected cells, as M11 mRNA, which is present at low abundance, was found in virions. MHV-76, which lacks the genes encoding the vtRNAs, also incorporated small RNA molecules within the virion, suggesting a requirement for these molecules for virion maturation. In productively infected cells the vtRNAs localized predominantly within the cytoplasm, although they also exhibited a globular pattern of nuclear staining. Their presence in the cytoplasm is consistent with interaction with virion components prior to maturation of virus particles. The significance of these findings for virion architecture and function is discussed.
Noncoding RNA molecules have been recognized as a feature of the genomes of members of the herpesvirus family for many years (1, 17, 28). They have been identified in numerous viruses including the human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) and are commonly expressed during latent infection. The noncoding RNAs are expressed at high levels, suggesting that they have important roles in virus pathogenesis, but despite considerable interest, their function(s) is not well defined. The most intensely studied herpesvirus noncoding RNAs are the EBV-encoded RNAs (EBERs). These are RNA polymerase (RNA pol) III transcripts present in both lytic and latent infections which appear to have multiple functions including resistance to apoptosis and control of the immune response as well as a potential role in tumorigenesis (1, 7, 19, 23, 34). They have been detected in both the nucleus and the cytoplasm of infected cells (25). Herpesvirus saimiri also encodes a family of small noncoding RNAs, the small nuclear Sm binding RNAs herpesvirus saimiri U RNAs (HSUR) 1 to 7, which are expressed in transformed cells (14, 17). In contrast to the EBERs, these are transcribed from RNA pol II promoters and accumulate in the nucleus. A role for these RNAs in mRNA stability has been proposed (18), and there is evidence that two of these RNAs, HSUR 1 and 2, regulate transcription of genes involved in T-cell activation (8). The polyadenylated nuclear RNA (PAN RNA) encoded by KSHV is also an RNA pol II transcript present in the nucleus but is expressed only during lytic infection (29, 35). PAN RNA is the most abundant viral RNA species in the nucleus of productively infected cells. Its function is unknown, but a role in splicing has been suggested (35).
Murine gammaherpesvirus 68 (MHV-68 or γHV68; also called MuHV-4) is widely used as a mouse model for the study of replication and pathogenesis of the gammaherpesviruses. Like EBV, MHV-68 encodes a set of RNA pol III-driven noncoding RNAs. These eight transcripts, which encode tRNA-like molecules capable of forming a cloverleaf secondary structure, are located in the left end of the genome, a region known to be involved in pathogenesis (3). The viral tRNA-like molecules (vtRNAs) each contain RNA pol III promoter elements and are transcribed monocistronically. Interestingly, the primary transcripts are processed to produce both the vtRNAs and a set of microRNA molecules (20). At least four of the vtRNAs are not aminoacylated by cellular aminoacyl-tRNA synthetases, and they are therefore not thought to function as classical tRNAs during protein synthesis. However, they are recognized and processed as tRNAs to some extent by the host cell, with the addition of 3′ CCA termini (3). The vtRNAs are expressed to high levels during both lytic and latent infection, and it is therefore likely that they play a role during both infectious events, although no function has been demonstrated so far.
Herpesvirus lytic replication involves the temporally regulated expression of three sets of genes, with the immediate-early genes expressed first, then the early genes, and finally, following viral DNA replication, the late genes. Expression of the vtRNAs has been previously classified as immediate-early (11), and yet their exact timing of expression has not been investigated. The initial objective of this study was therefore to characterize the expression pattern of the vtRNAs during lytic infection. However, a preliminary investigation suggested that the vtRNAs were present within the viral stock used and hence either contaminated the stock or formed part of the virus particle. Consequently, we investigated whether the vtRNAs were packaged within the virus particle and hence were an integral component of the infectious virion.
MATERIALS AND METHODS
Cell lines.
The murine epithelial cell line C127 (ATCC CRL1616) was maintained in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin (Merck BDH), and 100 U/ml streptomycin (Merck BDH) (DME10). Baby hamster kidney fibroblast cells (BHK-21) were maintained in Glasgow's modified Eagle's medium, supplemented with 10% (vol/vol) tryptose phosphate broth, 10% (vol/vol) newborn calf serum, 2 mM l-glutamine (VWR, United Kingdom), 100 U/ml penicillin, and 100 U/ml streptomycin. Immortalized murine embryonic fibroblasts derived from type I interferon (IFN) receptor-deficient mouse embryos by stable transfection with the simian virus 40 T antigen (αβ SV1) were maintained in DME10.
Preparation of purified virus stocks.
Murid herpesvirus 4 strain MHV-68 clone g2.4 (30) and MHV-76 (15) cell-associated virus stocks were prepared by infection of BHK-21 cells at a multiplicity of infection (MOI) of 0.001 PFU/cell as previously described (31). Supernatant virus was prepared by infection of αβ SV1 at an MOI of 0.001 PFU/cell. Following complete cytopathic effect, the cell suspension was centrifuged at 2,000 × g for 20 min. The resulting supernatant was centrifuged at 20,000 × g for 2 h at 4°C in order to pellet the virus.
Virus stocks were purified by layering onto a 20% sucrose cushion and spun at 141,000 × g for 1 h 20 min at 4°C. The pellet was resuspended in 1 ml RNase One buffer (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9) for treatment with RNase One (New England Biolabs) or phosphate-buffered saline (PBS) for further purification. For purification through a discontinuous sucrose gradient, the viral stock was applied onto 55%, 30%, and 10% sucrose layered in descending order and banded by ultracentrifugation at 141,000 × g for 18 h at 4°C, following which a viral band was clearly visible. The band was removed, sterile PBS was added to a total volume of 20 ml, and the purified virus was concentrated by ultracentrifugation at 141,000 × g for 1 h 20 min and resuspended in 1 ml RNase One buffer. Purified virus stocks were treated with 700 units RNase One for 5 h at 37°C.
RT-PCR.
RNA was extracted using RNAwiz (Ambion) according to the manufacturer's instructions, and the RNA was resuspended in 30 μl RNase-free water. Contaminating DNA was removed using DNA free (Ambion) according to the manufacturer's instructions. For the removal of DNA from purified virions, two DNase treatments were carried out, with an incubation at 95°C for 3 min in between. Random priming reverse transcription was carried out using Superscript II (Invitrogen) according to the manufacturer's instructions. PCR was carried out using the previously described primers to the vtRNAs, M3, M11, DNA polymerase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-actin (10, 11, 24). PCR mixtures contained between 1.5 and 5 mM MgCl2; 100 μM each of dATP, dCTP, dGTP, and dTTP; 50 pmol of each primer; PCR buffer (20 mM Tris [pH 8.4], 50 mM KCl); and 5 U Taq DNA polymerase (Invitrogen). Reverse transcriptase PCR (RT-PCR) products were separated by agarose gel electrophoresis.
Total RNA labeling.
RNA isolated from virions was radiolabeled with 32P-cytidine-3′,5′-bisphosphate (pCp) using T4 RNA ligase (New England Biolabs). RNA was incubated with T4 reaction buffer (50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 10 mM dithiothreitol, pH 7.8), 10% (vol/vol) dimethyl sulfoxide, 20 U SUPERase-In RNase inhibitor (Ambion), 250 μCi [32P]pCp, and 20 units T4 RNA ligase for 30 min at 37°C. Unincorporated nucleotides were removed with Micro-Bio-Spin chromatography columns. The samples were fractionated on either a 1% (wt/vol) agarose gel containing 6.7% (vol/vol) formaldehyde in 1× morpholinepropanesulfonic acid (MOPS) buffer or a Novex Tris-buffered EDTA (TBE)-urea-10% polyacrylamide gel (Invitrogen). All gels were fixed in 10% acetic acid-20% methanol for 20 min, and agarose gels were dried for 2 h at 80°C. For detection of the radiolabeled RNA, the gels were exposed to Hyperfilm ECL (GE Healthcare).
Northern blot analysis.
For detection of vtRNA1, RNA was run on a Novex TBE-urea-10% polyacrylamide gel (Invitrogen) and electrophoretically transferred to a Zetaprobe membrane (Bio-Rad) at 200 mA for 1 h in 0.25× TBE, using a semidry blotter (Amcos), and the RNA was UV cross-linked to the membrane. Labeled vtRNA1 probe was produced by incorporation of [32P]CTP using the mirVana microRNA probe construction kit (Ambion, United Kingdom) using a synthetic oligonucleotide, 5′-GCCAGAGTAGCTCAATTGGTCCTGTCT-3′. Hybridization was carried out in Ultrahyb (Ambion) according to the manufacturer's directions.
In situ hybridization.
In situ hybridization was carried out using digoxigenin-labeled antisense probes derived from pEH1.4 (a gift from S. Efstathiou) (3), using a digoxigenin RNA labeling kit (Roche). Cytospins were prepared from C127 cells infected with MHV-68 or MHV-76 at an MOI of 5 PFU/cell, for 24 h. Cells were permeabilized in 0.5% Triton X-100 in PBS and rinsed in PBS and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Labeled probe (200 ng/ml) was diluted in hybridization buffer [50% (vol/vol) formamide, 5× salts (0.05 M EDTA, 0.05 M PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 0.6 M NaCl, pH 6.8), 0.1× sodium dodecyl sulfate, 5× Denhardt's solution, 0.25 mg/ml salmon sperm DNA, 0.25 mg/ml yeast tRNA, 20 U/ml heparin, and 5 mg/ml dextran sulfate] and heated to 95°C for 2 min. Dithiothreitol was added to 10 mM. Hybridization was carried out at 55°C overnight. Unbound probe was removed by washing with 2× SSC and 1× SSC at 37°C and 0.2× SSC at 55°C. Indirect fluorescence detection of the digoxigenin-labeled probe was carried out using a primary antidigoxigenin antibody (Roche), a secondary biotinylated antibody (Vector Labs), and streptavidin-Alexa Fluor 488 (Invitrogen). The slides were mounted in Vectashield mounting medium for fluorescence with propidium iodide (Vector Labs).
RESULTS
Incorporation of the vtRNAs within MHV-68 cell lysate stocks.
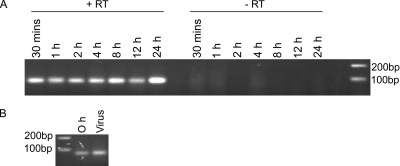
To determine the exact timing of expression of the vtRNAs, C127 cells were infected with virus stocks prepared from lysates of MHV-68-infected BHK cells. RT-PCR carried out on the extracted RNA indicated the presence of vtRNA1 from 30 min to 24 h postinfection (hpi) (Fig. 1A). The presence of the vtRNA as early as 30 min following addition of the virions was surprising. Although it was possible that viral gene expression could have taken place by this time, it was perhaps more likely that the vtRNAs were present within the virus preparation. In order to investigate whether this was the case, RNA was extracted from both the cell lysate MHV-68 stock and infected cells at 0 hpi and was found to contain vtRNA1 by RT-PCR (Fig. 1B). Given that the virus stock used for the infection was produced from a crude cellular lysate of infected BHK-21 cells, it was possible that the vtRNAs could have been present in the stock. However, as human cytomegalovirus (HCMV), herpes simplex virus type 1 (HSV-1), and KSHV virions have all been found to incorporate RNA (2, 4, 26), it was possible that the vtRNAs were packaged within the MHV-68 virion.
FIG. 1.
(A) Expression of vtRNA1 in C127 cells infected with MHV-68. C127 cells were infected at an MOI of 5 PFU/cell, and at various times, the RNA was harvested and subjected to RT-PCR for vtRNA1. Experiments were carried out in the presence (+RT) or absence (−RT) of reverse transcriptase. (B) RNA was isolated from the virus stock and from infected cells at 0 hpi and subjected to RT-PCR for vtRNA1.
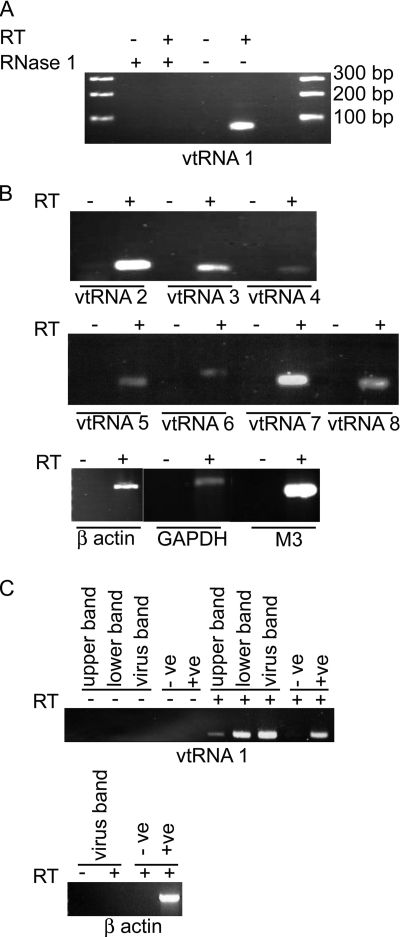
The BHK lysate-derived virions were purified by ultracentrifugation on a 20% sucrose cushion and treated with RNase One to remove any remaining contaminating RNA from the outside of the virus particles. The virions were then disrupted, and the RNA was extracted. RT-PCR demonstrated the presence of vtRNA1 within the virus preparation (Fig. 2A). Following treatment of the extracted RNA with RNase One, no PCR product was visible, demonstrating the ability of the enzyme to digest vtRNA1. In addition, PCR amplification was not a result of contaminating genomic DNA, as no PCR product was present when the reverse transcriptase was absent from the reaction. PCRs were also carried out on the cDNA for the remaining vtRNAs, which were all found to be present (Fig. 2B). In addition to the vtRNAs, both viral (M3) and cellular (GAPDH and β-actin) mRNAs could be found within the sucrose cushion-purified virus stock.
FIG. 2.
vtRNAs are present in purified MHV-68 virions. (A and B) Cell-associated MHV-68 virions were purified through a sucrose cushion, and RNA was extracted and subjected to RT-PCR. (A) RNA was subjected to RT-PCR for vtRNA1 or treated with RNase One followed by RT-PCR, showing that the vtRNA1 product was derived from RNA. (B) RT-PCR of virion RNA showing the presence of vtRNA2 to -8 and mRNAs for β-actin, GAPDH, and M3 in virion preparations. (C) RT-PCR on RNA extracted from MHV-68 grown in BHK-21 cells and purified through a sucrose gradient. RT-PCR was carried out for vtRNA1 and β-actin on virus taken from the upper band, lower band, and virus band. Experiments were carried out in the presence (+) and absence (−) of reverse transcriptase. Positive (+ve) and negative (−ve) controls were included in the reactions.
The cell lysate virus stock was further purified through a 5 to 55% sucrose gradient, and the obvious, opaque virus band (designated the virus band) was harvested. The band immediately above the virus band (the upper band) and the lower band, which included the pellet, were also collected, and all three were concentrated by ultracentrifugation. Following RNase treatment, the virions were disrupted and the RNA was extracted. RT-PCR demonstrated the presence of vtRNA1 in all three preparations (Fig. 2C). However, no viral or cellular mRNA transcripts, such as M3 or β-actin, could be detected in any of the bands by RT-PCR. Although the sucrose gradient-purified virus stock appeared to be cleaner than that purified through a sucrose cushion, as judged by the removal of the mRNA molecules, electron micrographs of the virus preparations revealed that they still contained cellular debris (data not shown). In addition, a 2-log decrease in virus titer following ultracentrifugation through the sucrose gradient resulted in a lower RNA yield, and hence, it is not clear whether the mRNAs could not be detected due to their levels falling below the limit of detection or as a result of further purification of the virus stock.
vtRNAs are present within a highly purified supernatant virus stock.
To determine if the vtRNAs were present within virions released into the cellular supernatant, MHV-68 was grown within type I IFN receptor-deficient murine embryonic fibroblasts, which produce high levels of extracellular virus (J. Stewart, personal communication). This allowed for a much easier purification of the viral stock, as contaminating cytoplasmic material was absent. The preparation was purified by ultracentrifugation through a 20% sucrose cushion, resulting in a high-titer (>1010 PFU/ml) virus stock. Transmission electron microscopy of the resulting virus stock indicated that it contained high levels of enveloped virions and relatively little contaminating debris (data not shown). RT-PCR demonstrated the presence of vtRNA1 within the RNase One-treated supernatant virus stock (Table 1).
TABLE 1.
RNA species present in purified MHV-68 and MHV-76 virions and in a mock preparation
| RNA species | Presence in virion preparation
|
||
|---|---|---|---|
| MHV-68 | MHV-76 | Mock | |
| vtRNA1 | + | − | − |
| vtRNA2 | + | − | − |
| vtRNA3 | + | − | − |
| vtRNA4 | + | − | − |
| vtRNA5 | + | − | − |
| vtRNA6 | + | − | − |
| vtRNA7 | − | − | − |
| M3 | + | − | − |
| ORF65 | + | + | − |
| M11 | + | + | − |
| DNA polymerase | + | + | − |
| β-Actin | + | + | − |
| GAPDH | + | − | − |
MHV-76 is a deletion mutant of MHV-68 which lacks four genes found within the left end of the genome (M1 to M4) along with the eight vtRNAs and nine microRNAs. MHV-76 produces yields of virus similar to and exhibits growth kinetics identical to those of MHV-68 in vitro but is attenuated in vivo (15). The incorporation of RNA molecules within MHV-76 virions was analyzed alongside that in MHV-68 virions in order to investigate RNA packaging in the absence of the vtRNAs. In addition, a mock preparation was produced following the same protocol. To determine the specific RNA species present within MHV-68 and MHV-76 viral particles, RT-PCR was carried out on the RNA extracted from the extracellular virions. PCRs were carried out on the cDNA using primers specific for all eight vtRNAs and cellular (GAPDH and β-actin) and viral mRNA molecules. M3 and ORF65 mRNAs are present at high levels at the late times of infection when the virus particles are maturing. M11 and DNA polymerase (ORF9), although expressed, are present at much lower levels (11). The results of the PCR are summarized in Table 1. All the vtRNAs were found to be present within the MHV-68 viral preparation, except for vtRNA7, which, despite numerous assays, was never detected. As predicted, none of the vtRNAs were present within either an MHV-76 or a mock-infected preparation. The viral mRNA M3 could be detected within MHV-68, but once again not in MHV-76, which is as expected given that it lacks the M3 gene. Additional viral mRNAs could be detected within both MHV-68 and MHV-76, including ORF65, and surprisingly, given their apparent low level of expression, M11 and DNA polymerase mRNAs were also found to be packaged. The detection of cellular mRNAs within the viral stocks appeared to vary, with both GAPDH and β-actin being present within the MHV-68 preparation. However, GAPDH mRNA could not be detected by RT-PCR within the MHV-76 preparation. No cellular RNA molecules could be detected by RT-PCR within the mock-infected preparation. Hence, it appears that the presence of RNA molecules within the virus preparations was due to their protection within the virion and not a result of contamination from cellular RNA.
Small RNA molecules are the major RNA species packaged within the MHV-68 virion.
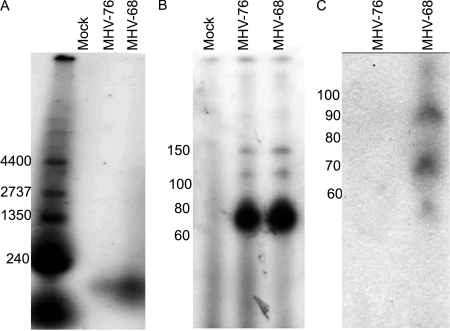
Given that both MHV-68 and MHV-76 package an assortment of RNA species, the nature of the RNA present was determined by total RNA labeling using radiolabeled ribonucleotides. Despite the presence of mRNA molecules within the MHV-68 virion, as determined by RT-PCR, the only RNAs detected using T4 labeling of RNA extracted from supernatant virus were around 100 nucleotides in length (Fig. 3A). No other RNA species could be detected using this method, indicating that both MHV-76 and MHV-68 preferentially package small RNA molecules. The absence of any RNA species in the mock-infected preparation indicates protection of the RNA within the virions and not contamination from cellular RNA molecules.
FIG. 3.
Analysis of the RNA species present within purified virus stocks. MHV-68 and MHV-76 were grown in mouse embryonic fibroblast cells lacking the type I IFN receptor and purified from cell-free supernatants by centrifugation though a sucrose cushion. Virions were treated with RNAse One prior to extraction of RNA. RNA was labeled with [32P]CTP and fractionated on a 3% agarose gel (A), a 10% polyacrylamide-urea gel together with a mock preparation (B), and a Northern blot of purified virion RNAs probed with [32P]CTP-labeled RNA probe specific for the first 20 nucleotides of vtRNA1 (C). Numbers at left of each panel are molecular sizes in nucleotides.
To determine the exact size of the RNA species present, the experiment was repeated but this time the labeled RNA was run on a 10% polyacrylamide-urea gel. The major RNA species extracted from both MHV-68 and MHV-76 viral preparations were approximately 70 nucleotides in length (Fig. 3B), indicating that in the absence of the vtRNAs (i.e., in MHV-76) the virus is able to package RNA molecules of the same size. Once again, no labeled RNA could be detected within the mock-infected preparation.
To investigate whether the small polynucleotides present within MHV-68 represent the vtRNAs, Northern blot analysis was carried out. RNA was extracted from both MHV-76 and MHV-68. Hybridization was carried out using an in vitro-transcribed [32P]CTP-labeled probe specific for the first 20 nucleotides of vtRNA1. Two RNA species of approximately 70 and 90 nucleotides in length were detected within MHV-68 virions (Fig. 3C). No hybridization to MHV-76 RNA was detected. The 70-nucleotide fragment corresponds to fully processed vtRNA. It is possible that the 90-nucleotide fragment represents RNA that has been semiprocessed from the longer primary transcript.
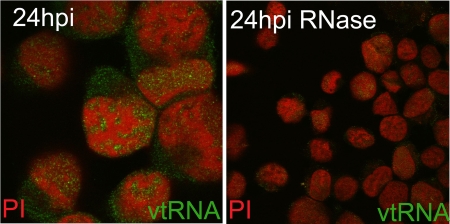
The vtRNAs localize mainly to the cytoplasm during lytic infection.
To investigate the subcellular localization of the vtRNAs in greater detail, in situ hybridization was carried out on C127 cells infected with MHV-68 for 24 h (Fig. 4). High levels of vtRNAs could be detected within the cytoplasm, where they exhibited a diffuse pattern of staining. In addition the vtRNAs could be found to localize to specific areas within the nucleus. The exclusion of host chromatin, based on the propidium iodide staining (27), from areas of vtRNA staining suggests their localization to viral replication compartments where herpesvirus DNA replication, transcription, and DNA encapsidation take place (5, 9, 13, 21, 22). However, the lack of a specific antibody to viral proteins found within replication compartments compounded the difficulty of the assignment of these structures. Treatment of the slides with RNase resulted in loss of the signal, indicating specific binding to the vtRNAs and not viral DNA.
FIG. 4.
In situ hybridization for vtRNA1 to -4. C127 cells were infected at an MOI of 5 PFU/cell for 24 h and probed with a digoxigenin-labeled RNA probe specific for vtRNA1 to -4. Detection of the digoxigenin-labeled probe was carried out using biotinylated antidigoxigenin and streptavidin-Alexa Fluor 488. Sections were counterstained with propidium iodide (PI) and imaged using a Leica TCS-NT confocal microscope. Treatment with RNase was also carried out prior to hybridization.
DISCUSSION
In this study, we demonstrate that the vtRNAs are packaged within the MHV-68 virion particle, as determined by their presence within different viral preparations produced from both cell-associated and extracellular virions. Extensive treatment of the virions with a nuclease to digest RNA exterior to the virus particles prior to RNA isolation makes it unlikely that the results are due to contaminating RNA. It was possible that homogenization of infected cells resulted in the production of membranous vesicles which may also contain vtRNAs; however, the presence of the vtRNAs within extracellular virions which did not contain homogenized cellular material argues against this. We also demonstrate that, within infected cells, the vtRNAs localize to both the nucleus and the cytoplasm, although at least at late times in infection they were found at higher levels within the cytoplasm.
Selective packaging of vtRNAs.
Although mRNA molecules were detected within the MHV-68 particle, the vtRNAs were present with a greater abundance, as they were the only RNA species detected by RT-PCR analysis on lower-titer sucrose gradient-purified viral stocks. In addition, direct labeling of the total RNA present within the virion indicated that the major encapsidated RNA species are small RNA molecules of approximately 70 nucleotides in length.
Cellular and viral mRNAs have previously been found to be packaged within the HCMV and HSV virions (4, 12, 26). In addition a recent study by Bechtel et al. demonstrated that along with 10 viral mRNA molecules, the noncoding PAN RNA is encapsidated by KSHV (2). However, here we demonstrated that, at least in the case of MHV-68, small RNAs are selectively packaged within the virion. Interestingly, a deletion mutant of MHV-68 which lacks the vtRNAs (MHV-76) also appears to encapsidate small RNA molecules of the same size. Given that the most abundant RNA species within cells of approximately 70 nucleotides in length are tRNAs, it is likely that the encapsidated RNA species within MHV-76 are cellular tRNAs. It is worth noting that we were unable to detect vtRNA7 within purified virions. vtRNA7 is the only vtRNA that contains a predicted intron and is thought to fold into only a presplicing tRNA but not fully mature tRNA (3), further supporting the preferential packaging of mature tRNAs within the virion. All previous studies of RNA packaging by herpesviruses were based on the detection of specific transcripts by RT-PCR, Northern blotting, and microarray analysis but not the total RNA present, and hence, it is not clear whether the high level of encapsidation of small RNA is unique to MHV-68.
Mechanism of RNA packaging.
A matter of controversy is the mechanism by which RNA is incorporated within herpesvirus particles. In the case of HCMV, it has been demonstrated that viral and cellular RNAs are packaged in proportion to the relative quantities present within infected cells at the time of virion assembly (32). However, viral mRNA packaging by HSV-1 and KSHV does not correspond to mRNA expression levels within infected cells. A superficial analysis of the RNA species present indicates that the very highly expressed vtRNAs are the most abundant viral RNA within the virion. However, the M11 transcript is one of the least abundantly expressed genes within infected cells (11), and yet this was also present within the virion. Therefore, this study suggests that for MHV-68, the viral RNA is not packaged within the virion in proportion to the expression level within the cell. An alternative theory is that RNA is selectively packaged, perhaps through binding to viral proteins, in a way analogous to that of genomic RNA packaging by RNA viruses. The seven vtRNAs that are packaged are 75% homologous at the sequence level as well as shared predicted structure. It is possible that they are recognized via sequence as well as structural elements. Virion proteins that are capable of binding packaged RNA have been identified in both HSV-1 and HCMV (26, 32).
Functions of packaged RNA.
A key issue arising from this study is the possible function of encapsidated RNA and in particular the vtRNAs. It has been shown elsewhere that a number of virion mRNAs delivered into the cell are translated (2). However, given that the vtRNAs do not encode protein, they are not delivered into the host cell to allow immediate translation. It is possible that, whatever role the vtRNAs play during infection, it is beneficial for the virus to deliver the vtRNAs immediately upon infection of the host cell. This does not explain the apparent encapsidation of small RNA molecules by MHV-76, although it is possible that in the absence of the vtRNAs the virus still maintains the mechanism responsible for vtRNA packaging and cellular tRNAs are incorporated in their place, effectively by mistake.
A further hypothesis is that the packaged RNA has a structural role, acting either as a scaffold during virion assembly or as a form of molecular packaging maintaining the integrity of the virion. tRNAs of both viral and cellular origin have been proposed to act as nucleating agents during the assembly of the plant virus brome mosaic virus (6). In retroviruses, RNA is important for virion assembly (16, 33) and has also been found to be an integral part of the structure of retrovirus particles (16). If RNA, and in particular low-molecular-weight RNA, is important in virion assembly and structure, this would explain the incorporation of cellular tRNAs within the MHV-76 virion. It is of relevance that MHV-76 is not attenuated in exponentially growing tissue culture cells but is severely attenuated within the lymphoid system in vivo, where cellular tRNAs levels may be more limited (15).
In conclusion, we have demonstrated the encapsidation of small RNA molecules within the virion of MHV-68. The presence of these molecules argues for a specific interaction with virion components and a function in early events in virus infection. Further work is under way to determine whether the vtRNAs interact with specific viral proteins and to identify their role in virus pathogenesis.
Acknowledgments
This work was funded by the Wellcome Trust. Anna R. Cliffe was funded by a Wellcome Trust Studentship.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Arrand, J. R., and L. Rymo. 1982. Characterization of the major Epstein-Barr virus-specific RNA in Burkitt lymphoma-derived cells. J. Virol. 41376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechtel, J., A. Grundhoff, and D. Ganem. 2005. RNAs in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 7910138-10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowden, R. J., J. P. Simas, A. J. Davis, and S. Efstathiou. 1997. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J. Gen. Virol. 781675-1687. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 2882373-2376. [DOI] [PubMed] [Google Scholar]
- 5.Bush, M., D. R. Yager, M. Gao, K. Weisshart, A. I. Marcy, D. M. Coen, and D. M. Knipe. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 651082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, J. K., B. S. Lee, S. N. Shim, M. Li, and J. U. Jung. 2000. Identification of the novel K15 gene at the rightmost end of the Kaposi's sarcoma-associated herpesvirus genome. J. Virol. 74436-446. [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke, P. A., M. Schwemmle, J. Schickinger, K. Hilse, and M. J. Clemens. 1991. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 19243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, H. L., J. R. Lytle, H. E. Mischo, M. J. Li, J. J. Rossi, D. P. Silva, R. C. Desrosiers, and J. A. Steitz. 2005. Small nuclear RNAs encoded by Herpesvirus saimiri upregulate the expression of genes linked to T cell activation in virally transformed T cells. Curr. Biol. 15974-979. [DOI] [PubMed] [Google Scholar]
- 9.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55857-868. [DOI] [PubMed] [Google Scholar]
- 10.Dutia, B. M., D. J. Roy, B. Ebrahimi, B. Gangadharan, S. Efstathiou, J. P. Stewart, and A. A. Nash. 2004. Identification of a region of the virus genome involved in murine gammaherpesvirus 68-induced splenic pathology. J. Gen. Virol. 851393-1400. [DOI] [PubMed] [Google Scholar]
- 11.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 8499-109. [DOI] [PubMed] [Google Scholar]
- 12.Greijer, A. E., C. A. Dekkers, and J. M. Middeldorp. 2000. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J. Virol. 749078-9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, S. I., S. C. Murthy, J. J. Trimble, R. C. Desrosiers, and J. A. Steitz. 1988. Four novel U RNAs are encoded by a herpesvirus. Cell 54599-607. [DOI] [PubMed] [Google Scholar]
- 15.Macrae, A. I., B. M. Dutia, S. Milligan, D. G. Brownstein, D. J. Allen, J. Mistrikova, A. J. Davison, A. A. Nash, and J. P. Stewart. 2001. Analysis of a novel strain of murine gammaherpesvirus reveals a genomic locus important for acute pathogenesis. J. Virol. 755315-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 985246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murthy, S., J. Kamine, and R. C. Desrosiers. 1986. Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J. 51625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myer, V. E., S. I. Lee, and J. A. Steitz. 1992. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl. Acad. Sci. USA 891296-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nanbo, A., and K. Takada. 2002. The role of Epstein-Barr virus-encoded small RNAs (EBERs) in oncogenesis. Rev. Med. Virol. 12321-326. [DOI] [PubMed] [Google Scholar]
- 20.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2269-276. [DOI] [PubMed] [Google Scholar]
- 21.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 711124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 672163-2177. [DOI] [PubMed] [Google Scholar]
- 23.Rosa, M. D., E. Gottlieb, M. R. Lerner, and J. A. Steitz. 1981. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol. 1785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy, D. J., B. C. Ebrahimi, B. M. Dutia, A. A. Nash, and J. P. Stewart. 2000. Murine gammaherpesvirus M11 gene product inhibits apoptosis and is expressed during virus persistence. Arch. Virol. 1452411-2420. [DOI] [PubMed] [Google Scholar]
- 25.Schwemmle, M., M. J. Clemens, K. Hilse, K. Pfeifer, H. Troster, W. E. Muller, and M. Bachmann. 1992. Localization of Epstein-Barr virus-encoded RNAs EBER-1 and EBER-2 in interphase and mitotic Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 8910292-10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 758105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 785591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spivack, J. G., and N. W. Fraser. 1988. Expression of herpes simplex virus type 1 latency-associated transcripts in the trigeminal ganglia of mice during acute infection and reactivation of latent infection. J. Virol. 621479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, R., S. F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 9311883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunil-Chandra, N. P. 1991. Studies on the pathogenesis of a murine gammaherpesvirus (MHV-68). Ph.D. dissertation. University of Cambridge, Cambridge, United Kingdom.
- 31.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 732347-2356. [DOI] [PubMed] [Google Scholar]
- 32.Terhune, S. S., J. Schroer, and T. Shenk. 2004. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J. Virol. 7810390-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, S. W., and A. Aldovini. 2002. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 7611853-11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, Y., S. Maruo, M. Yajima, T. Kanda, and K. Takada. 2007. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J. Virol. 8111236-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong, W., and D. Ganem. 1997. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J. Virol. 711207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]