Abstract
The papillomavirus E2 proteins regulate viral replication, gene transcription, and genome maintenance by interacting with other viral and host proteins. From a yeast two-hybrid screen, we identified the cellular protein Tax1BP1 as a novel binding partner of human papillomavirus type 18 (HPV18) E2. Tax1BP1 also interacts with the HPV16 and bovine papillomavirus type 1 (BPV1) E2 proteins, with the C-terminal region of Tax1BP1 interacting with the N-terminal transactivation domain of BPV1 E2. Tax1BP1 complexes with p300 and acts synergistically as a coactivator with p300 to enhance E2-dependent transcription. Using chromatin immunoprecipitation assays, we show that Tax1BP1 and E2 localize to the long control region on the BPV1 genome. Tax1BP1 was recently reported to bind ubiquitin and to function as an essential component of an A20 ubiquitin-editing complex. We demonstrate that Tax1BP1 plays a role in the regulation of the steady-state level of E2 by preventing its proteasomal degradation. These studies provide new insights into the regulation of E2 functions.
The papillomavirus E2 protein is a key regulator of viral DNA replication, gene expression, and genome maintenance. The E2 proteins are structurally and functionally conserved across different papillomaviruses and are composed of an N-terminal transactivation domain (TAD) and a C-terminal dimerization and DNA binding domain (DBD) separated by a less conserved proline-rich hinge region (reviewed in reference 32). The N-terminal TAD is essential for E2 functions and interacts with numerous viral and cellular proteins, including the viral E1 protein, TFIIB, GPS2/AMF-1, MKlp2, CHlR1, Brd4, Brahma, NAP-1, p300/CBP, and p/CAF (20, 23, 24, 29, 33, 36, 38, 44, 45). The E2 DBD binds to the E2-responsive elements, specific palindromic sequences (ACCN6GGT) located mainly in the long control region (LCR) of the viral genome (1). Upon binding to the E2-responsive element, E2 activates gene transcription from viral early and late promoters. Besides the 410-amino-acid (aa) transcriptional activator E2, the open reading frame of the bovine papillomavirus type 1 (BPV1) E2 also encodes a transcriptional repressor, E2R (aa 162 to 410), which is expressed from the C-terminal part of the E2 open reading frame and represses E2-dependent transcription due to a lack of functional TAD (21).
Tax1-binding protein 1 (Tax1BP1) (also named TXBP151 and T6BP) was originally identified as a binding partner of the human T-cell leukemia virus type 1 Tax oncoprotein (6, 28). The N-terminal region (aa 1 to 150) of Tax1BP1 contains a SKIP carboxyl homology (SKICH) domain. The central region (aa 150 to 600) is predicted to form three coiled-coil domains and is involved in self-dimerization. The C-terminal region (aa 601 to 789) encodes two zinc fingers. Tax1BP1 is highly conserved across the species, with human Tax1BP1 sharing 79% and 81% identity with rat and mouse orthologs, respectively. It is a nuclear protein and is abundantly expressed in most human tissues and established cell lines (4, 6).
Tax1BP1 is a negative regulator of NF-κB signaling. It interacts with TRAF6 in an interleukin 1-dependent manner and reduces the polyubiquitination of RIP1 and TRAF6 signaling intermediates by recruiting the ubiquitin-editing enzyme A20 (16, 28, 39). The second zinc finger at the C terminus of Tax1BP1 contains a ubiquitin-binding domain. Mutations in this region abolished ubiquitin binding and TRAF6 association. Moreover, through its two zinc finger domains and the associated “PPXY” motifs, Tax1BP1 recruits the E3 ligase Itch to form a functional A20 ubiquitin-editing complex, which is essential for the termination of NF-κB signaling (40). Tax1BP1 knockout mice exhibit elevated NF-κB activation in response to tumor necrosis factor alpha and interleukin 1 stimulation. In addition, Tax1BP1 also serves as a transcriptional coactivator of glucocorticoid receptor by stimulating glucocorticoid response element-dependent reporter through its N-terminal activation domain (4).
To better understand the regulation of E2 functions, we performed a yeast two-hybrid screen of a HeLa cDNA library using human papillomavirus type 18 (HPV18) E2 as bait and identified Tax1BP1 as a novel E2-interacting protein. Here, we show that Tax1BP1 interacts with both HPV and BPV E2 proteins. Tax1BP1 functions synergistically as a coactivator with p300 in E2-dependent transcription. In addition, Tax1BP1 interaction significantly extends the half-life of E2 proteins by preventing their proteasomal degradation.
MATERIALS AND METHODS
Plasmids and antibodies.
The full-length Tax1BP1 human cDNA construct was purchased from ATCC (catalog no. 10436569; human 6055189). FLAG-tagged full-length Tax1BP1 (aa 1 to 789), TXBP-Nter (aa 1 to 420), and TXBP-Cter (aa 421 to 789) were cloned in frame into the p3XFLAG-CMV-7.1 vector (Sigma) by PCR amplification using specific primers containing NotI and BamHI restriction sites. The FLAG-tagged Tax1BP1 ubiquitin-binding domain mutants (UBZ1, UBZ2, and UBZ*) were generated by PCR (16). The BPV1 and HPV16 E2 expression plasmids have been previously described (3, 36). The hemagglutinin (HA)-tagged HPV18 E2 expression plasmid was constructed by PCR amplification of the HPV18 E2 coding sequence from pSGE2-18 using specific primers containing BamHI and EcoRI restriction sites and inserting it in frame into a pcDNA-HA expression vector (43).
The following antibodies were used: mouse anti-FLAG (M2; Sigma), rabbit anti-FLAG (F7425; Sigma), anti-c-Myc (9E10; Santa Cruz), anti-HA (12CA5 [34]), anti-EE (13), B201 (a mouse monoclonal antibody whose epitope maps between aa 160 and 220 of BPV1 E2), B202 (a mouse monoclonal antibody whose epitope maps between aa 285 and 325 of BPV1 E2), II-1 (rabbit anti-BPV1 E2 antiserum), TVG261 (mouse anti-HPV16 E2 monoclonal antibody), and anti-β-actin (Sigma). Goat anti-mouse- and goat anti-rabbit-horseradish peroxidase were purchased from Jackson ImmunoResearch. The rabbit polyclonal TXBP151-C antibody was kindly provided by Kuan-Teh Jeang (6).
Yeast two-hybrid assay.
The initial yeast two-hybrid screen was performed in Saccharomyces cerevisiae strain DBY1 transformed with a HeLa cDNA library fused to the herpesvirus VP16 activation domain, along with an E2-dependent lacZ reporter, pBSY72, in which the URA3 gene was replaced by the HIS4 gene (pBSY72-H4) (3). pRS314G-18E2 was constructed by inserting HPV18 E2 into a yeast centromere vector, pRS314 (41). Yeast transformants were selected on minimal medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
To confirm Tax1BP1 interaction with BPV1 E2, pYEplac112G, encoding galactose-inducible BPV1 E2, and pGADT7, encoding Tax1BP1, were transformed into DBY1 cells containing pBSY72. Transformants were selected on galactose-X-Gal medium.
Cell culture and transient transfections.
Human C33A cervical carcinoma cells and mouse mammary tumor fibroblast (C127)-derived A3 cells (generously provided by M. Botchan [25]), which stably maintain BPV1 genomes, were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) with 10% fetal calf serum. The cells were washed and transfected in serum-free DMEM using Lipofectamine 2000 (Invitrogen) and refed with DMEM with 10% fetal calf serum.
Coimmunoprecipitation assays and Western blot analysis.
Cells were lysed on ice for 20 min in lysis buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 20 mM NaF, 50 mM KH2PO4, 1% Triton X-100, 10% glycerol, 2 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor cocktail [Roche]). The lysates were cleared by centrifugation and mixed with equal volumes of binding buffer (50 mM Tris-HCl, pH 8.0, 100 mM KCl, 0.1 mM EDTA, 0.2% NP-40, 0.1% bovine serum albumin, 2.5% glycerol, 2 mM DTT, 1 mM PMSF, and protease inhibitor cocktail). Protein A/G-Sepharose beads and the respective antibodies were added, incubated overnight at 4°C, and washed with wash buffer (100 mM Tris-HCl, pH 8.0, 100 to 300 mM NaCl, 0.5% NP-40, 2 mM DTT, and 1 mM PMSF). Coimmunoprecipitated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride membranes (Millipore), and detected by chemiluminescence (Pierce).
Chromatin immunoprecipitation (ChIP) assays.
Forty-eight hours posttransfection, cells were fixed with 1% formaldehyde at room temperature for 10 min. The cells were rinsed with phosphate-buffered saline and lysed on ice for 10 min in lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris-HCl, pH 8.0, and protease inhibitor cocktail). The sonicated cell lysates were cleared by centrifugation, diluted 10-fold in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.0), and incubated overnight at 4°C with the respective antibodies and protein A/G-Sepharose beads (preblocked with 1% bovine serum albumin and sheared salmon sperm DNA in Tris-EDTA buffer [GE Healthcare]). The beads were washed sequentially in low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0), and Tris-EDTA buffer. Protein-DNA complexes were eluted from the beads at room temperature for 10 min in elution buffer (1% SDS, 0.1 M NaHCO3) and then reverse cross-linked overnight at 65°C. DNA was recovered using a PCR purification kit (Qiagen) and was subjected to PCR amplification of the BPV1 LCR using the following primers: BPV1LCRS (5′-AAAGTTTCCATTGCGTCTGG-3′) and BPV1LCRAS (5′-GCTTTTTATAGTTAGCTGGCTATTTT-3′).
Proteasome inhibition assays.
Approximately 5 × 105 C33A cells were transfected with 100 ng of expression constructs for wild-type BPV1 E2, the truncation mutant E2TAD or E2R, and 1 μg of FLAG-tagged full-length Tax1BP1 or 500 ng of TXBP-Nter and TXBP-Cter. Forty hours posttransfection, the proteasome inhibitor MG132 dissolved in dimethyl sulfoxide (DMSO) was added to the cells for 6 to 8 h at a final concentration of 10 μM. The cells were lysed in lysis buffer (2% SDS, 50 mM Tris-HCl, pH 6.8). The protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce).
Cycloheximide chase assays.
Approximately 5 × 105 C33A cells were transfected with 100 ng of pCG-E2b and 1 μg of FLAG-tagged full-length Tax1BP1 or 500 ng of TXBP-Nter and TXBP-Cter. Forty hours posttransfection, the cells were treated with cycloheximide at a final concentration of 50 μg/ml and lysed immediately and after 0.5, 1, 2, 4, and 8 h in lysis buffer (2% SDS, 50 mM Tris-HCl, pH 6.8). The protein concentration was determined by a bicinchoninic acid assay.
Reporter assays.
Approximately 5 × 105 C33A cells were transfected using Lipofectamine 2000 with 100 ng of the E2-dependent reporter plasmid pGL2-4BS-Luc, which contains four E2 binding sites fused to the simian virus 40 minimal promoter. Other expression vectors included 10 ng of pCG-E2b, 500 ng of the p300 expression construct pCMV-p300NHA, 200 ng of the adenovirus E1A 12S protein expression construct pCMV-E1A12S (36), and 1 μg of FLAG-tagged full-length or truncated Tax1BP1 constructs. Luciferase activities were measured at 24 h posttransfection with the luciferase assay system (Promega).
RNA extraction and RT-PCR.
Total RNA was extracted using TRIzol reagent (Invitrogen) and treated with RQ1 RNase-free DNase (Promega) to remove transfected plasmid DNA. After phenol-chloroform extraction, 250 ng of RNA was subjected to reverse transcription (RT)-PCR using ImProm-II reverse transcriptase (Promega). BPV1 E2 cDNA was amplified by PCR in 30 cycles using the following primers: BS16&17sense (5′-GGTGGTAGAGGTGGAGTTTGATG-3′) and BS16&17anti (5′-AGTAGTAGAGCCCAGTTCCGTCAG-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was amplified by PCR in 25 cycles using the following primers: G3S (5′-ACCACAGTCCATGCCATCAC-3′) and G3A (5′-TCCACCACCCTGTTGCTGTA-3′).
In vivo ubiquitination assays.
C33A cells were transfected with expression constructs for BPV1 E2 and FLAG-tagged full-length Tax1BP1 and HA-tagged ubiquitin. Forty hours posttransfection, the proteasome inhibitor MG132 dissolved in DMSO was added to the cells for 6 to 8 h at a final concentration of 10 μM. The cells were lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl) with 0.5% SDS. The SDS was then diluted to 0.1% with lysis buffer. The lysates were sonicated using a Sonic Dismembrator (Fisher Scientific), boiled for 2 min, and cleared by centrifugation. Protein A/G-Sepharose beads and rabbit anti-E2 (II-1) antiserum or rabbit immunoglobulin G (IgG) were added, incubated overnight at 4°C, and washed with lysis buffer. The coimmunoprecipitated proteins were resolved by SDS-PAGE and detected by Western blotting.
siRNA knockdown.
Tax1BP1-small interfering RNA 1 (siRNA-1) and Tax1BP1-siRNA-2 sequences have been described by Iha et al. (16). Silencer Negative Control no. 1 siRNA was purchased from Applied Biosystems (AM4635). Approximately 3 × 105 C33A cells were transfected using Lipofectamine 2000 with Tax1BP1-siRNA-1 and -2 (75 nM each) or negative control siRNA (150 nM) for 24 h, followed by a second siRNA transfection, along with 200 ng of pCG-E2b for 48 h prior to RT-PCR and Western blot analyses as described above. Tax1BP1 cDNA was amplified by PCR in 25 cycles using the forward primer 5′-CAAGGTTGGATGGAGTACTGCTCGTGATTATTAC-3′ and the reverse primer 5′-GTAAGACCCTTTTGTTCTGC-3′.
RESULTS
Papillomavirus E2 interacts with Tax1BP1.
TXBP151 is the synonymic name of Tax1BP1, also known as TRAF6-binding protein (T6BP). TXBP151 (GenBank accession no. U33821) and T6BP (GenBank accession no. AF268075) are 747-aa proteins that differ by only two residues (aa 233 and 234) (6, 28). A more recent cDNA sequence (GenBank accession no. BC050358) encodes a larger 789-aa Tax1BP1. Sequence alignment showed that 42 aa between aa 603 and 646 of Tax1BP1 are absent in TXBP151, which might represent a splicing variant. The full-length Tax1BP1 used throughout this study refers to the 789-aa protein, and TXBP151 refers to the 747-aa protein.
To identify novel cellular proteins that interact with HPV18 E2, a yeast two-hybrid screen of a HeLa cDNA library was performed using full-length HPV18 E2 as bait. The cDNA insert from one candidate was found to encode the C-terminal 222 aa of TXBP151. Since the BPV1 E2 protein shares a high degree of functional and structural homology with HPV E2 proteins, this interaction was analyzed in the yeast two-hybrid assay using full-length Tax1BP1 and various BPV1 E2 mutants (Table 1). The transactivation-defective E2 mutant F87L was chosen to replace the wild-type full-length E2 as bait to avoid the general transcriptional-activation property of the wild-type E2. The C-terminal aa 541 to 886 of MKlp2, which bind BPV1 E2, were used as a positive control (46). BPV1 E2 F87L interacted with full-length Tax1BP1, but the deletion mutants E2 55-410 and E2 162-410 did not.
TABLE 1.
Yeast two-hybrid assay results using full-length Tax1BP1 and various BPV1 E2 mutants
| pYEplac112G | pGADT7 insert | X-Gal staina |
|---|---|---|
| E2 F87L | Empty vector | − |
| E2 F87L | MKlp2 541-886 | ++ |
| E2 F87L | Tax1BP1 FLb | + |
| E2 55-410 | Tax1BP1 FL | − |
| E2 162-410 | Tax1BP1 FL | − |
−, negative; +, positive; ++, strongly positive.
FL, full length.
To confirm this interaction in mammalian cells, we first examined the interaction of HPV E2 with Tax1BP1 in coimmunoprecipitation assays. Human C33A cervical carcinoma cells were transfected with expression vectors for full-length HPV16 E2 or HA-HPV18 E2 and FLAG-Tax1BP1. HPV16 and HPV18 E2 proteins were immunoprecipitated from the FLAG-Tax1BP1-transfected extract only using anti-FLAG M2 antibody, but not the anti-EE control antibody. Neither HPV16 nor HPV18 E2 was immunoprecipitated from the extracts that did not include FLAG-Tax1BP1 (Fig. 1A and B). We also tested complexes of HPV16 E2 with endogenous Tax1BP1. HPV16 E2 was expressed in C33A cells and lysates reacted with an anti-Tax1BP1 antibody (TXBP151-C), followed by immunoblotting for HPV16 E2. This experiment revealed that HPV16 E2 coprecipitated with endogenous Tax1BP1 (Fig. 1C).
FIG. 1.
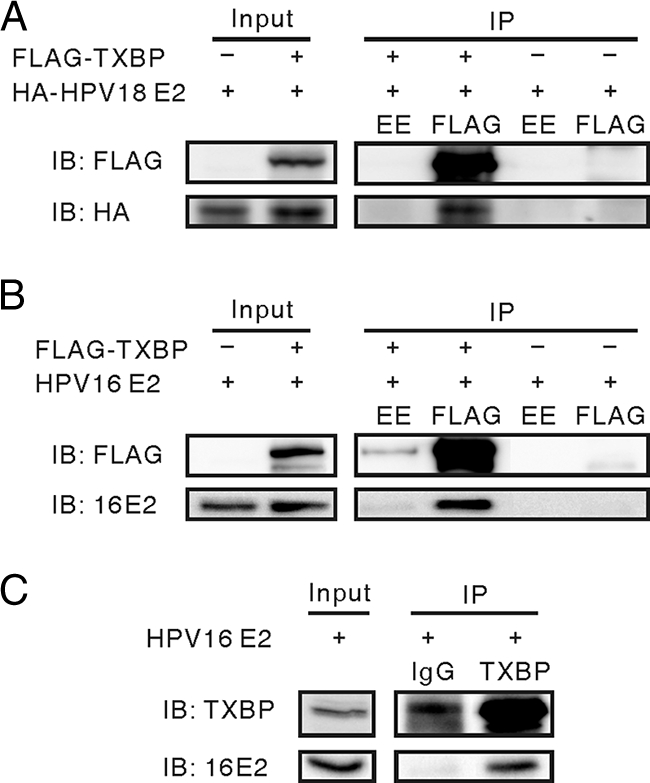
Tax1BP1 interacts with HPV16 and HPV18 E2 proteins. (A and B) C33A cells were transfected with expression vectors for FLAG-Tax1BP1 (TXBP) or control vector, along with either HA-tagged HPV18 E2 (A) or HPV16 E2 (B) expression plasmids. Cell extracts were immunoprecipitated (IP) with anti-FLAG (M2) or anti-EE control antibodies and immunoblotted (IB) with antibodies against HA (12CA5) (A) or HPV16 E2 (TVG261) (B). (C) Coimmunoprecipitation of endogenous Tax1BP1 with HPV16 E2. C33A cell extracts expressing HPV16 E2 were immunoprecipitated with an anti-Tax1BP1 antibody (TXBP151-C) or rabbit IgG and immunoblotted for HPV16 E2 (TVG261).
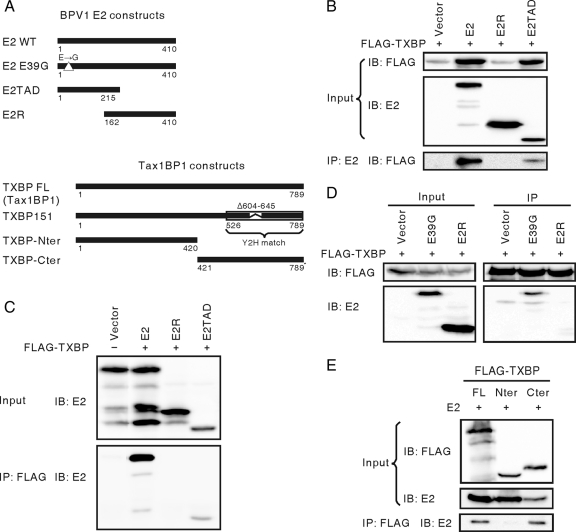
Next, we examined the interaction of BPV1 E2 with Tax1BP1. C33A cells were transfected with expression vectors for FLAG-Tax1BP1, full-length BPV1 E2, or the truncation mutant E2TAD (aa 1 to 215) or E2R (aa 162 to 410), which encode the BPV1 E2 TAD and DBD, respectively (Fig. 2A). FLAG-Tax1BP1 coprecipitated with full-length E2 and E2TAD, but not with E2R (Fig. 2B). Reciprocal coimmunoprecipitations were also performed. Consistently, both full-length E2 and E2TAD coprecipitated with FLAG-Tax1BP1 (Fig. 2C). These data demonstrated that Tax1BP1 interacts with BPV1 E2 and that this association requires the N-terminal TAD of BPV1 E2.
FIG. 2.
Tax1BP1 interacts with BPV1 E2. (A) Schematic diagrams of BPV1 E2 and Tax1BP1 constructs. Y2H, matched region in the initial yeast two-hybrid screen; WT, wild type. (B and C) C33A cells were transfected with expression vectors for FLAG-Tax1BP1 and the specified BPV1 E2 constructs. Reciprocal coimmunoprecipitations were performed by either immunoprecipitating (IP) BPV1 E2 (B201) and immunoblotting (IB) for FLAG-Tax1BP1 (M2) (B) or immunoprecipitating FLAG-TXBP (M2) and immunoblotting for E2 (II-1) (C). (D) C33A cells were transfected with expression vectors for FLAG-Tax1BP1 and E2 E39G, E2R, or control vector. Cell extracts were immunoprecipitated with anti-FLAG M2 antibody and immunoblotted for E2 (II-1). (E) C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-tagged full-length Tax1BP1 (TXBP FL) or the truncation mutant TXBP-Nter or TXBP-Cter. Cell extracts were immunoprecipitated with anti-FLAG M2 antibody and immunoblotted for E2 (II-1).
When coexpressed with transactivation-competent E2, the expression level of Tax1BP1 is markedly increased due to the general transcriptional activation of BPV1 E2 (Fig. 2B, top, Tax1BP1 input) (15). The lack of coimmunoprecipitation of E2R with Tax1BP1 could be due to the low expression level of Tax1BP1. In addition, since the E2 TAD interacts with Tax1BP1, it is possible that the transactivation function of E2 might be required for Tax1BP1 association. To address these issues, Tax1BP1 was transfected into C33A cells with the transactivation-defective E2 mutant E39G or E2R. When similar levels of Tax1BP1 were coexpressed with E2 E39G, E2R, or control vector, only E2 E39G coprecipitated with Tax1BP1 (Fig. 2D). E2R failed to coprecipitate with Tax1BP1, even though the level of E2R protein was markedly greater than that of E2 E39G. This result supports the conclusion that E2 interaction with Tax1BP1 requires its N-terminal TAD and that the overlapping region between E2TAD and E2R (aa 162 to 215) is not sufficient for Tax1BP1 association. Furthermore, the transactivation property of E2 is not required for Tax1BP1 association.
The initial yeast two-hybrid screen identified the C-terminal 222 aa of TXBP151 as the region that interacts with HPV18 E2, so this region was likely to interact with BPV E2. Tax1BP1 is predicted to fold into coiled-coil structures in its central portion. The N-terminal 1 to 420 residues encompass the first two coiled-coil structures, which contain an activation domain and a self-association domain. The C-terminal 421 to 789 residues encompass the third coiled-coil structure and two zinc finger domains (4, 28). Based on this structural prediction, N-terminal (TXBP-Nter) and C-terminal (TXBP-Cter) truncation protein expression constructs were made (Fig. 2A). C33A cells were transfected with full-length BPV1 E2 and FLAG-tagged full-length Tax1BP1, TXBP-Nter, or TXBP-Cter. As expected, E2 coimmunoprecipitated with full-length Tax1BP1 and TXBP-Cter, but not with TXBP-Nter (Fig. 2E).
Tax1BP1 synergizes with p300 to enhance E2-dependent transcription.
Cellular proteins, such as Gps2/AMF-1, Brd4, TFIIB, NAP-1, Brahma, CBP/p300, and p/CAF, were reported to interact with E2 and to enhance E2-dependent transcription. To determine the biological function of Tax1BP1 in E2-dependent transcription, C33A cells were transfected with an E2-dependent luciferase reporter and vectors expressing BPV1 E2, HA-p300, or FLAG-Tax1BP1. The E2 reporter exhibited minimal basal activity, which was unaffected by the overexpression of p300 or Tax1BP1. E2 strongly activated the reporter 20-fold. Coexpression of p300 increased E2 activity by 20%. Coexpression of Tax1BP1 increased E2 activity by 80%. When coexpressed with p300, Tax1BP1 synergistically increased E2 activity 1.7-fold (Fig. 3A).
FIG. 3.
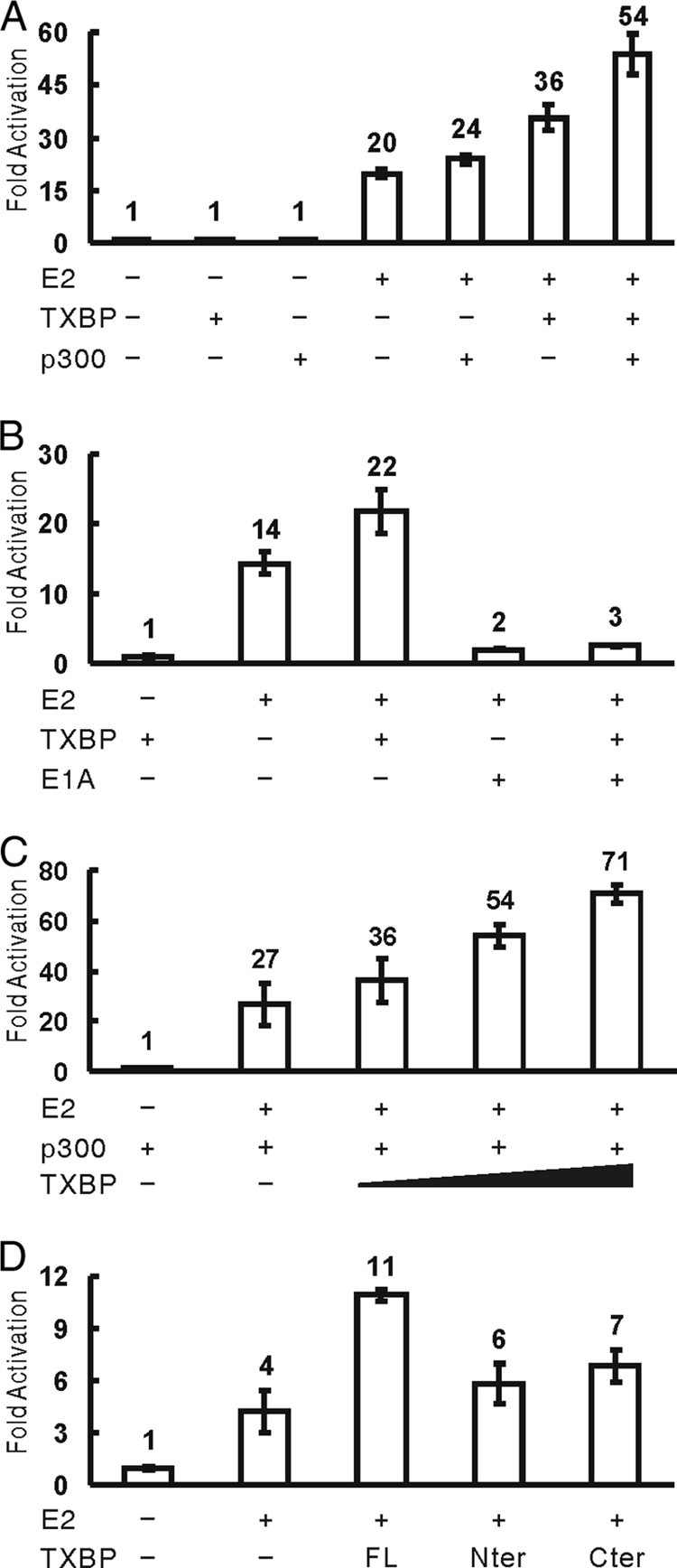
Tax1BP1 and p300 synergistically enhance E2-dependent transcription. The E2-dependent luciferase reporter was cotransfected into C33A cells with various expression plasmids. Luciferase activities were measured 24 h posttransfection and normalized against reporter alone. Each sample was analyzed in triplicate (the error bars indicate standard deviations). (A) Tax1BP1 and p300 synergistically enhance E2-dependent transcription. Expression vectors for BPV1 E2, HA-p300, and FLAG-Tax1BP1 alone or in combination were transfected as indicated. (B) The coactivator function of Tax1BP1 is p300 dependent. Expression vectors for BPV1 E2, FLAG-Tax1BP1, and adenovirus E1A 12S protein were transfected as indicated. (C) Tax1BP1 enhances E2 transcriptional activity in a dose-dependent manner. Expression vectors for BPV1 E2, HA-p300, and increasing amounts of FLAG-Tax1BP1 (125 ng, 500 ng, and 1 μg) were transfected. (D) Full-length Tax1BP1 is required for its coactivator function. Expression vectors for BPV1 E2 and FLAG-tagged full-length Tax1BP1 or the truncation constructs TXBP-Nter and TXBP-Cter were transfected.
E2-dependent transcription requires p300 and can be inhibited by the adenovirus E1A 12S protein (19, 33, 36). In agreement with previous studies, E1A inhibited the transcriptional activity of E2 regardless of Tax1BP1 expression (Fig. 3B). An E1A deletion mutant [E1A Del (2-36)], which is defective in p300/CBP binding, failed to inhibit E2 transactivation (data not shown). Tax1BP1 enhanced p300-dependent E2 transcriptional activity in a dose-dependent manner (Fig. 3C). Neither TXBP-Nter nor TXBP-Cter enhanced E2 activity significantly (Fig. 3D). These results suggest that Tax1BP1 cooperates with p300 in E2-dependent transcription.
Based on these observations, we sought to test whether Tax1BP1 and p300 form complexes in vivo. C33A cells were transfected with expression vectors for FLAG-Tax1BP1 and HA-tagged p300 or a Renilla luciferase (HA-RLuc) control. p300 coprecipitated Tax1BP1, but HA-RLuc did not (Fig. 4). This demonstrates that Tax1BP1 and p300 form complexes independent of BPV1 E2.
FIG. 4.
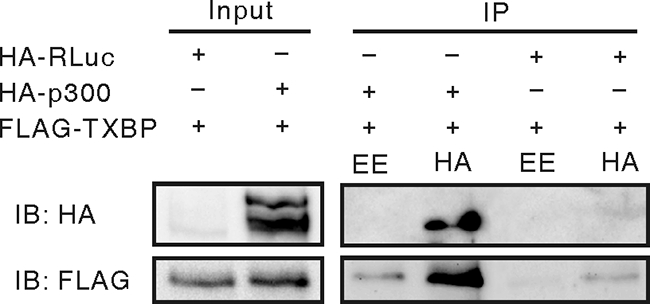
Tax1BP1 interacts with p300 in vivo. C33A cells were transfected with expression vectors for FLAG-Tax1BP1 and HA-p300 or HA-luciferase control (HA-RLuc). Cell extracts were immunoprecipitated (IP) with anti-HA (12CA5) or anti-EE control antibodies and immunoblotted (IB) for FLAG-Tax1BP1 (M2).
Tax1BP1 and E2 are present at the BPV1 LCR.
E2 regulates viral gene expression by binding to E2 binding sites in the viral LCR and forming complexes with components of the basic transcriptional machinery and transcription coactivators. Given that Tax1BP1 interacts with BPV1 E2 and enhances E2-dependent transcription, it is possible that Tax1BP1 is present, along with E2, at the LCR. C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-Tax1BP1, as well as a BPV1 genome-containing plasmid. ChIP assays were performed using mouse anti-E2 (B201 and B202), anti-FLAG (M2), or control antibodies, followed by PCR amplification of the BPV1 LCR. Antibodies against E2 and FLAG-Tax1BP1 precipitated the viral LCR, but the control antibodies did not (Fig. 5A). Similar results were obtained using a set of rabbit antibodies against E2 (II-1) and FLAG-Tax1BP1 (F7425 [Sigma] and TXBP151-C). To test whether endogenous Tax1BP1 is present at the BPV1 LCR, ChIP assays were performed on the A3 cell line harboring episomal BPV1 genomes. Antibodies against BPV1 E2 and endogenous Tax1BP1 precipitated the viral LCR (Fig. 5B). These results are consistent with participation of Tax1BP1 in E2-dependent transcription.
FIG. 5.

E2 and Tax1BP1 are present at the BPV1 LCR. (A) A BPV1 genome-containing plasmid was cotransfected into C33A cells with expression vectors for BPV1 E2 and FLAG-Tax1BP1. Cell extracts were immunoprecipitated (IP) with mouse or rabbit antibodies (Ab) against FLAG, E2, or Tax1BP1 or control antibodies. The LCR of the BPV1 genome was amplified by PCR. (B) BPV1 genome-containing A3 cell extracts were immunoprecipitated with anti-E2 (II-1) or anti-Tax1BP1 (TXBP151-C) antibodies or rabbit IgG. The LCR of the BPV1 genome was amplified by PCR.
Tax1BP1 stabilizes E2 by preventing its proteasome-mediated degradation.
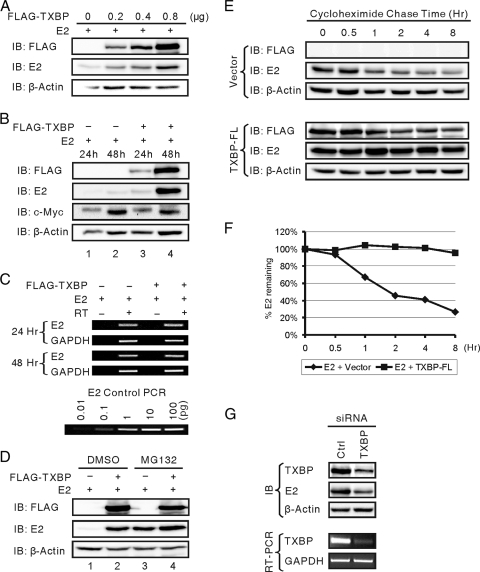
HPV18 E2 and BPV1 E2 are degraded by the ubiquitin-proteasome pathway (2, 37). We noticed in our experiments that transient expression of Tax1BP1 increased the BPV1 E2 protein level in a dose-dependent manner (Fig. 6A). Two possible explanations could account for this observation. First, Tax1BP1 may increase E2 transcription due to its coactivator function. Secondly, Tax1BP could increase E2 protein stability. To examine the first possibility, E2 transcripts from FLAG-Tax1BP1- and BPV1 E2-transfected cells were amplified by semiquantitative RT-PCR at 24 and 48 h posttransfection. The expression levels of transiently expressed proteins were monitored by Western blotting. Compared to control vector-transfected cells, Tax1BP1 slightly increased the E2 protein level after 24 h (Fig. 6B, lane 1 versus lane 3). By 48 h posttransfection, the E2 level in Tax1BP1-transfected cells was significantly higher than that in control vector-transfected cells (Fig. 6B, lane 2 versus lane 4). However Tax1BP1 did not alter the level of E2 transcripts at either time point, suggesting that the accumulation of steady-state E2 protein in the presence of overexpressed Tax1BP1 is not likely a transcriptional effect on cytomegalovirus promoter-driven E2 expression, but rather, is caused by increased E2 stability (Fig. 6C).
FIG. 6.
Tax1BP1 stabilizes E2 by preventing its proteasomal degradation. (A) Tax1BP1 increases the level of BPV1 E2 protein in a dose-dependent manner. C33A cells were transfected with expression vectors for BPV1 E2 and increasing amounts of FLAG-Tax1BP1. Cell extracts were resolved on SDS-PAGE and immunoblotted (IB) for FLAG-Tax1BP1 (M2), E2 (B201), or β-actin. (B and C) C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-Tax1BP1 or control vector. (B) At 24 and 48 h posttransfection, cell extracts were resolved on SDS-PAGE and immunoblotted for FLAG-Tax1BP1 (M2), E2 (B201), c-Myc (9E10), or β-actin. (C) At 24 and 48 h posttransfection, total RNA was extracted using TRIzol reagent. RNase-free DNase digestion of the isolated RNA was performed to remove the contamination of E2 plasmid DNA. E2 and GAPDH mRNAs were amplified by semiquantitative RT-PCR. PCR using increasing amounts of E2 plasmid DNA as templates were also performed as a control. (D) Tax1BP1 prevents proteasomal degradation of BPV1 E2. C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-Tax1BP1 or control vector. At 40 h posttransfection, the cells were treated with 10 μM MG132 or DMSO (mock) for 8 h. Cell extracts were resolved on SDS-PAGE and immunoblotted for FLAG-Tax1BP1 (M2), E2 (B201), or β-actin. (E) Tax1BP1 extends the half-life of BPV1 E2. C33A cells were transfected as described for panel D. At 40 h posttransfection, the cells were treated with cycloheximide (50 μg/ml) for up to 8 h. Cell extracts were resolved on SDS-PAGE and immunoblotted for FLAG-Tax1BP1 (M2), E2 (B201), or β-actin. (F) Graphical presentation of data presented in panel E. The level of E2 was quantitated and normalized using β-actin. (G) siRNA knockdown of endogenous Tax1BP1 reduces BPV1 E2 stability. C33A cells were transfected with control or Tax1BP1 siRNA and expression vectors for BPV1 E2. Total RNA was extracted. Tax1BP1 and GAPDH mRNAs were amplified by RT-PCR. Duplicated cell extracts were resolved on SDS-PAGE and immunoblotted for endogenous Tax1BP1 (TXBP151-C), E2 (B201), or β-actin.
Recent studies have shown that Tax1BP1 is an essential subunit of the A20 ubiquitin-editing enzyme complex (40). Tax1BP1 reduces polyubiquitination of RIP1 and TRAF6 by recruiting the ubiquitin-editing enzyme A20 to these molecules (6, 16, 39). To examine whether Tax1BP1 nonspecifically stabilizes any ubiquitinated protein, the same cell lysates were probed for c-Myc, which is degraded by the ubiquitin-proteasome pathway (9). Overexpression of Tax1BP1 did not change the levels of endogenous c-Myc at 24 and 48 h posttransfection (Fig. 6B). Based on these findings, we hypothesized that Tax1BP1 specifically prevents E2 degradation by the ubiquitin-proteasome pathway. To test this hypothesis, C33A cells were transfected with expression vectors for BPV1 E2 or FLAG-Tax1BP1 or with control vector for 40 h and then treated with the proteasome inhibitor MG132 or DMSO (mock treatment) for 8 h. The levels of E2 and Tax1BP1 were examined by Western blotting. MG132 treatment greatly increased the E2 protein level in vector-transfected cells, which was similar to that in Tax1BP1-transfected cells (Fig. 6D, lane 1 versus lane 3 and lane 3 versus lane 4). MG132 treatment did not significantly further increase the E2 protein level in Tax1BP1-transfected cells (Fig. 6D, lane 2 versus lane 4). These results indicate similar rates of E2 protein synthesis in vector- and Tax1BP1-transfected cells. They also suggest that the elevated E2 protein level caused by Tax1BP1 overexpression is primarily due to the inhibition of E2 proteasomal degradation by Tax1BP1 (Fig. 6D, lane 2 versus lane 3).
To confirm that the increase in the E2 protein level was indeed caused by changes to E2 stability, we performed cycloheximide chase experiments using C33A cells transfected with BPV1 E2 in the presence and absence of overexpressed Tax1BP1. Forty hours posttransfection, cells were treated with cycloheximide and lysed at different time points up to 8 h. In control vector-transfected cell extracts, E2 protein degraded with a half-life of 1.5 h, while overexpression of Tax1BP1 greatly extended its half-life to more than 8 h (Fig. 6E and F). To determine the role of endogenous Tax1BP1 in E2 stabilization, siRNA knockdown of endogenous Tax1BP1 was performed in C33A cells. siRNA effectively reduced the level of endogenous Tax1BP1 as determined by the level of Tax1BP1 transcript and protein (Fig. 6G). The level of E2 protein was dramatically reduced as a result of siRNA knockdown, which is consistent with the overexpression data and demonstrated the role of Tax1BP1 in E2 stabilization.
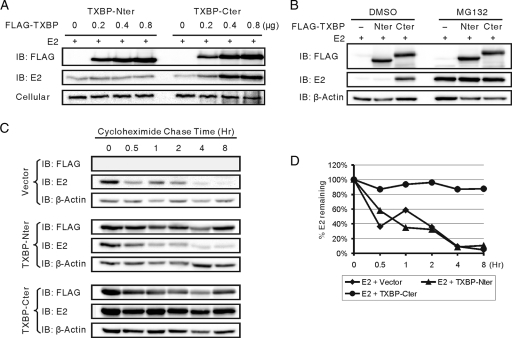
To determine the region on Tax1BP1 that is responsible for E2 stabilization, increasing amounts of the Tax1BP1 truncation mutants TXBP-Nter and TXBP-Cter and BPV1 E2 were transfected into C33A cells. The level of E2 protein was monitored by Western blotting. TXBP-Cter increased the E2 protein level in a dose-dependent manner, but TXBP-Nter did not (Fig. 7A). MG132 treatment of vector-, TXBP-Nter-, and TXBP-Cter-transfected cells caused E2 to accumulate to similar levels (Fig. 7B). Cycloheximide chase experiments revealed that the half-lives of E2 in vector- and TXBP-Nter-transfected cells were approximately 1 h (similar to that shown in Fig. 6E) but were significantly extended in TXBP-Cter-transfected cells (Fig. 7C and D).
FIG. 7.
The C-terminal region of Tax1BP1 stabilizes E2. (A) TXBP-Cter increases the level of BPV1 E2 protein in a dose-dependent manner. C33A cells were transfected with expression vectors for BPV1 E2 and increasing amounts of the FLAG-tagged Tax1BP1 truncation construct TXBP-Nter or TXBP-Cter. Cell extracts were resolved on SDS-PAGE and immunoblotted (IB) for FLAG-Tax1BP1 (M2), E2 (B201), or β-actin. (B) TXBP-Cter prevents proteasomal degradation of BPV1 E2 protein. C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-tagged TXBP-Nter, TXBP-Cter, or control vector. At 40 h posttransfection, the cells were treated with 10 μM MG132 or DMSO (mock) for 6 h and analyzed by Western blotting as described for Fig. 6D. (C) TXBP-Cter extends the half-life of BPV1 E2 protein. C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-tagged TXBP-Nter, TXBP-Cter, or control vector. At 40 h posttransfection, cycloheximide chase experiments were performed and analyzed as described for Fig. 6E. (D) Graphical presentation of data presented in panel C. The levels of E2 were quantitated and normalized using β-actin.
HPV18 E2 is degraded by the ubiquitin-proteasome pathway through its TAD (2). The TAD of HPV18 E2 is more efficiently ubiquitinated than the hinge/DBD. Since BPV1 E2TAD, but not E2R, interacts with Tax1BP1, we predicted that only E2TAD would be stabilized by Tax1BP1. As expected, both full-length Tax1BP1 and TXBP-Cter stabilized E2TAD, but not E2R (Fig. 8A and B). Similarly, full-length Tax1BP1 also stabilized HPV16 E2 (data not shown).
FIG. 8.
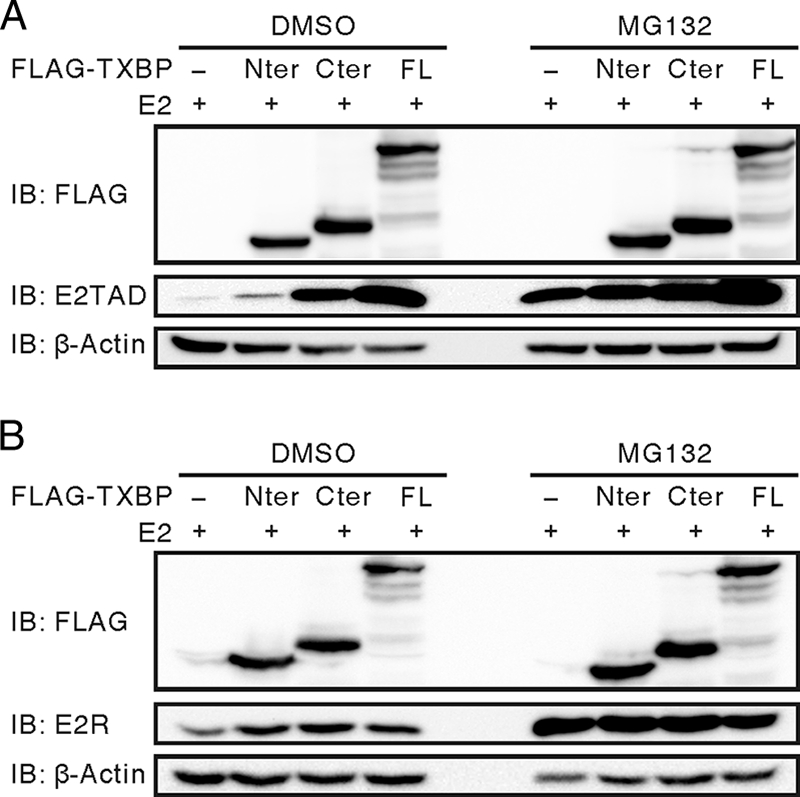
Tax1BP1 stabilizes the N-terminal TAD of BPV1 E2. C33A cells were transfected with expression vectors for E2TAD (A) or E2R (B) and FLAG-tagged full-length Tax1BP1, the truncation constructs TXBP-Nter and TXBP-Cter, or control vector. At 40 h posttransfection, the cells were treated with 10 μM MG132 or DMSO (mock) for 6 h and analyzed by Western blotting (IB) as described for Fig. 6D.
E2 stabilization by Tax1BP1 is independent of Tax1BP1 ubiquitin binding.
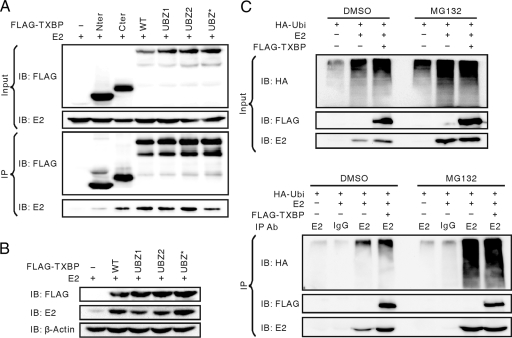
The C terminus of Tax1BP1 was recently shown to harbor a ubiquitin-binding domain in the second zinc finger (the UBZ2 domain) (16). The UBZ2 domain Tax1BP1 mutants were defective in ubiquitin binding and failed to coimmunoprecipitate TRAF6, implying that Tax1BP1 might interact with ubiquitinated TRAF6. To examine whether Tax1BP1 interacts with ubiquitinated E2, Tax1BP1 UBZ domain mutants were tested for E2 interaction and stabilization. The two conserved Phe residues were mutated to Ala in each of the two C-terminal zinc fingers (UBZ1, F737A; UBZ2, F764A). Two cysteine residues in the UBZ2 domain were also mutated (UBZ*, C757/760A). Both Tax1BP1 UBZ2 and UBZ* were defective in ubiquitin binding, whereas UBZ1 was not. BPV1 E2 coprecipitated with all UBZ domain mutants, as well as wild-type Tax1BP1 and TXBP-Cter, but not with TXBP-Nter (Fig. 9A). These UBZ mutants also stabilized E2 significantly (Fig. 9B). Therefore, stabilization and binding of E2 by Tax1BP1 does not require binding to ubiquitin. To further determine whether Tax1BP1 affects the ubiquitination status of E2, an in vivo ubiquitination assay was performed in Tax1BP1- and HA-ubiquitin-coexpressed cell lysates. Surprisingly, Tax1BP1 increased not only the level of nonubiquitinated E2, but also the level of polyubiquitinated E2 species (Fig. 9C). When the proteasomal function was inhibited by MG132, nonubiquitinated and polyubiquitinated E2 species accumulated to similar levels regardless of Tax1BP1 overexpression.
FIG. 9.
E2 stabilization by Tax1BP1 is independent of Tax1BP1 ubiquitin binding. (A) Ubiquitin-binding-defective Tax1BP1 mutants interact with BPV1 E2. C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-tagged full-length wild-type Tax1BP1 (WT); the ubiquitin-binding-defective mutants UBZ1, UBZ2, and UBZ*; or the truncation mutant TXBP-Nter or TXBP-Cter. To ensure equal levels of E2 inputs, the cells were treated with 10 μM MG132 for 6 h. Cell extracts were immunoprecipitated with anti-FLAG M2 antibody and immunoblotted (IB) for E2 (II-1). (B) Ubiquitin-binding-defective Tax1BP1 mutants increase the level of BPV1 E2 protein. C33A cells were transfected with expression vectors for BPV1 E2 and FLAG-tagged wild-type Tax1BP1 (WT), UBZ1, UBZ2, UBZ*, or control vector. Cell extracts were resolved on SDS-PAGE and immunoblotted for FLAG-Tax1BP1 (M2), E2 (B201), or β-actin. (C) in vivo ubiquitination assay. C33A cells were transfected with expression constructs for BPV1 E2, FLAG-tagged full-length Tax1BP1, and HA-tagged ubiquitin. Forty hours posttransfection, the proteasome inhibitor MG132 dissolved in DMSO was added to the cells for 6 h at a final concentration of 10 μM. Cell extracts were immunoprecipitated with anti-E2 antibody (II-1) or rabbit IgG and immunoblotted for HA-ubiquitin (12CA5), FLAG-Tax1BP1 (M2), or E2 (B201).
DISCUSSION
The identification and characterization of papillomavirus E2-interacting proteins has provided valuable information for our understanding of E2 functions. Here, we reported that Tax1BP1 interacts with BPV and HPV E2 proteins in yeast and mammalian cells, complexes with p300, and acts synergistically with p300 to enhance E2-dependent transcription. In addition, Tax1BP1 stabilizes E2 by preventing its proteasomal degradation. Since HPV and BPV E2 proteins have relatively short half-lives, interaction with Tax1BP1 may illustrate another mechanism to regulate their activities.
The E2 proteins are relatively well conserved in both the N-terminal TAD and the C-terminal DBD among different papillomaviruses (8, 31). While we used HPV18 E2 as bait in a yeast two-hybrid screen, we also showed that Tax1BP1 interacts with HPV16 and BPV1 E2 proteins. The interaction is mediated through the N-terminal TAD of BPV1 E2 and the C-terminal region of Tax1BP1. Even though these proteins efficiently coimmunoprecipitated in vivo, we were unable to detect direct binding between the glutathione S-transferase fusion E2 and in vitro-translated Tax1BP1, possibly due to a lack of required posttranslational modifications of the E2 protein produced in bacteria or other factors that facilitate their association in vivo.
The papillomavirus E2 proteins are degraded by the ubiquitin-proteasome pathway. We demonstrated that full-length Tax1BP1 and TXBP-Cter stabilize both full-length BPV1 E2 and E2TAD by preventing their degradation. Tax1BP1 did not increase the E2R protein level, nor did Tax1BP1 stabilize c-Myc, which has a short half-life and is also degraded through the ubiquitin-proteasome pathway. These results suggest that Tax1BP1 does not stabilize E2 as a result of interfering with the proteasomal degradation of any ubiquitinated protein but rather plays a specific role in regulating E2 stability. We also observed that E2R is expressed at a much higher level than full-length E2 or E2TAD, which is consistent with a previous report that N-terminally truncated HPV18 E2 is more stable than full-length E2 and the N-terminal TAD of HPV18 E2 is highly ubiquitinated compared to the hinge/DBD (2). Altogether, these findings suggest that the ubiquitination of the N-terminal TAD of BPV1 E2 targets the protein for proteasomal degradation.
Tax1BP1 is known to serve as an assembly platform for protein deubiquitination. As reported, Tax1BP1 binds ubiquitinated TRAF6 and recruits A20 to reduce TRAF6 ubiquitination (16). However, our data do not support the hypothesis that Tax1BP1 might utilize a similar mechanism to stabilize E2 by preventing its ubiquitination. First, both ubiquitin-binding-defective Tax1BP1 mutants (UBZ2 and UBZ*) coimmunoprecipitate and stabilize BPV1 E2, indicating that the association of Tax1BP1 with E2 is not mediated by ubiquitin (Fig. 9). Secondly, Tax1BP1 expression did not cause a decrease in polyubiquitinated E2 accompanied by an increase in nonubiquitinated E2 in the in vivo ubiquitination assay. However, since proteasome functions are regulated by opposing deubiquitinating activities and ubiquitin ligase activities in a dynamic balance (5), we cannot rule out the possibility that the increased nonubiquitinated E2 species favor the ubiquitin ligase activities until equilibrium is reached.
The histone acetyltransferases CBP/p300 and p/CAF are multifunctional regulators in the cells of higher eukaryotes. As transcription coactivators, they promote gene transcription by bridging between DNA-bound transcription factors and the RNA polymerase II holoenzymes and by acetylating histones and other transcription factors. CBP/p300 and p/CAF bind papillomavirus E2 and activate E2-dependent transcription (19, 23, 24, 33). We found that Tax1BP1 interacts with p300 and that the coactivator function of Tax1BP1 is p300 dependent. Therefore, Tax1BP1 may be present in complexes with E2 and p300 and enhance E2-dependent transcription.
ChIP assays showed that both Tax1BP1 and E2 precipitated the BPV1 LCR. There are 12 E2 binding sites in the LCR (27). We were unable to effectively sonicate viral minichromosomes into smaller LCR-containing DNA fragments to distinguish whether Tax1BP1 preferentially associated with E2 at a specific site. We also cannot rule out the possibility that Tax1BP1 may be tethered by E2 to E2 binding sites outside the LCR. Since most of the high-affinity E2 binding sites are located within the LCR, it is reasonable to expect that most Tax1BP1-E2 complexes bind to these high-affinity sites. This model is also consistent with the coactivator function of Tax1BP1 in E2-dependent transcription.
In addition, CBP/p300 and p/CAF may exert their coactivator functions indirectly by regulating the turnover of transcription factors. p300 and p/CAF were shown to stabilize Smad7, SREBP, and E2F by acetylation (7, 10, 30, 42). p300 also stabilizes MDM2 and ATF4 independently of its acetylase activity, possibly by sequestration of MDM2 and ATF4 into nuclear-body-like structures, thereby protecting these molecules from proteasomal degradation (22, 47). The pleiotropic functions of these histone acetyltransferases are illustrated by the effects of p300 on p53 stability. p53 was the first nonhistone protein reported to be acetylated by CBP/p300 (14, 18). Acetylation of p53 inhibits its ubiquitination by Mdm2 (26). Acetylated p53 is found in promyelocytic leukemia bodies with CBP in Ras-overexpressing cells (35). In contrast, p300 participates in MDM2-mediated p53 degradation (12). Recent data also show that p300 possesses E4 ubiquitin ligase activity, which promotes the polyubiquitination of p53 for proteasomal degradation (11). Since the same lysine residues can be acetylated or ubiquitinated, what controls the switch between acetylation and ubiquitination by p300 remains undetermined (17). Interestingly Tax1BP1 also localizes in the intranuclear speckles that partially overlap with promyelocytic leukemia bodies (4). By analogy, Tax1BP1 may bridge p300 and E2 and sequester E2 into a nuclear compartment that is not accessible to proteasomal degradation. Studies are under way to determine whether Tax1BP1 regulates the subcellular localization of E2 and the potential involvement of p300 in Tax1BP1-mediated E2 stabilization.
The stabilization of E2 by Tax1BP1 raised the question of whether the observed coactivator function of Tax1BP1 could simply be due to an increased E2 level. Though this is possible, the elevated E2 cannot be the only contributing factor because TXBP-Cter also stabilizes E2 but did not enhance E2-dependent transcription (Fig. 3D). These results distinguish the function of Tax1BP1 in transcriptional coactivation from that of E2 stabilization. It is possible that the initial complexes of Tax1BP1 with E2 and p300 at the viral LCR transiently stimulate E2 transcriptional activity. E2 is then either modified or targeted to a different subcellular compartment, where it is rendered transcriptionally inactive. Further studies are needed to elucidate the functional link between these two properties of Tax1BP1.
Acknowledgments
We thank Kuan-Teh Jeang for providing the rabbit TXBP151-C antibody and Michael Botchan for providing the A3 cell line. We greatly appreciate the many useful discussions with members of our laboratory.
This work was supported by NIH grant R01 CA58376.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Androphy, E. J., D. R. Lowy, and J. T. Schiller. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature 32570-73. [DOI] [PubMed] [Google Scholar]
- 2.Bellanger, S., C. Demeret, S. Goyat, and F. Thierry. 2001. Stability of the human papillomavirus type 18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J. Virol. 757244-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiding, D. E., M. J. Grossel, and E. J. Androphy. 1996. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology 22134-43. [DOI] [PubMed] [Google Scholar]
- 4.Chin, K. T., A. C. Chun, Y. P. Ching, K. T. Jeang, and D. Y. Jin. 2007. Human T-cell leukemia virus oncoprotein tax represses nuclear receptor-dependent transcription by targeting coactivator TAX1BP1. Cancer Res. 671072-1081. [DOI] [PubMed] [Google Scholar]
- 5.Crosas, B., J. Hanna, D. S. Kirkpatrick, D. P. Zhang, Y. Tone, N. A. Hathaway, C. Buecker, D. S. Leggett, M. Schmidt, R. W. King, S. P. Gygi, and D. Finley. 2006. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 1271401-1413. [DOI] [PubMed] [Google Scholar]
- 6.De Valck, D., D. Y. Jin, K. Heyninck, M. Van de Craen, R. Contreras, W. Fiers, K. T. Jeang, and R. Beyaert. 1999. The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene 184182-4190. [DOI] [PubMed] [Google Scholar]
- 7.Giandomenico, V., M. Simonsson, E. Gronroos, and J. Ericsson. 2003. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell. Biol. 232587-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giri, I., and M. Yaniv. 1988. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 72823-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory, M. A., and S. R. Hann. 2000. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol. Cell. Biol. 202423-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gronroos, E., U. Hellman, C. H. Heldin, and J. Ericsson. 2002. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10483-493. [DOI] [PubMed] [Google Scholar]
- 11.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300342-344. [DOI] [PubMed] [Google Scholar]
- 12.Grossman, S. R., M. Perez, A. L. Kung, M. Joseph, C. Mansur, Z. X. Xiao, S. Kumar, P. M. Howley, and D. M. Livingston. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell 2405-415. [DOI] [PubMed] [Google Scholar]
- 13.Grussenmeyer, T., K. H. Scheidtmann, M. A. Hutchinson, W. Eckhart, and G. Walter. 1985. Complexes of polyoma virus medium T antigen and cellular proteins. Proc. Natl. Acad. Sci. USA 827952-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90595-606. [DOI] [PubMed] [Google Scholar]
- 15.Haugen, T. H., L. P. Turek, F. M. Mercurio, T. P. Cripe, B. J. Olson, R. D. Anderson, D. Seidl, M. Karin, and J. Schiller. 1988. Sequence-specific and general transcriptional activation by the bovine papillomavirus-1 E2 trans-activator require an N-terminal amphipathic helix-containing E2 domain. EMBO J. 74245-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iha, H., J. M. Peloponese, L. Verstrepen, G. Zapart, F. Ikeda, C. D. Smith, M. F. Starost, V. Yedavalli, K. Heyninck, I. Dikic, R. Beyaert, and K. T. Jeang. 2008. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 27629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 216236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 201331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruppel, U., A. Muller-Schiffmann, S. E. Baldus, S. Smola-Hess, and G. Steger. 2008. E2 and the co-activator p300 can cooperate in activation of the human papillomavirus type 16 early promoter. Virology 377151-159. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, R. A., S. R. Naidu, X. Wang, A. N. Imbalzano, and E. J. Androphy. 2007. Interaction of papillomavirus E2 protein with the Brm chromatin remodeling complex leads to enhanced transcriptional activation. J. Virol. 812213-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 5069-78. [DOI] [PubMed] [Google Scholar]
- 22.Lassot, I., E. Estrabaud, S. Emiliani, M. Benkirane, R. Benarous, and F. Margottin-Goguet. 2005. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J. Biol. Chem. 28041537-41545. [DOI] [PubMed] [Google Scholar]
- 23.Lee, D., S. G. Hwang, J. Kim, and J. Choe. 2002. Functional interaction between p/CAF and human papillomavirus E2 protein. J. Biol. Chem. 2776483-6489. [DOI] [PubMed] [Google Scholar]
- 24.Lee, D., B. Lee, J. Kim, D. W. Kim, and J. Choe. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 2757045-7051. [DOI] [PubMed] [Google Scholar]
- 25.Lehman, C. W., D. S. King, and M. R. Botchan. 1997. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J. Virol. 713652-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., J. Luo, C. L. Brooks, and W. Gu. 2002. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 27750607-50611. [DOI] [PubMed] [Google Scholar]
- 27.Li, R., J. Knight, G. Bream, A. Stenlund, and M. Botchan. 1989. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding sites in the BPV-1 genome. Genes Dev. 3510-526. [DOI] [PubMed] [Google Scholar]
- 28.Ling, L., and D. V. Goeddel. 2000. T6BP, a TRAF6-interacting protein involved in IL-1 signaling. Proc. Natl. Acad. Sci. USA 979567-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lusky, M., and E. Fontane. 1991. Formation of the complex of bovine papillomavirus E1 and E2 proteins is modulated by E2 phosphorylation and depends upon sequences within the carboxyl terminus of E1. Proc. Natl. Acad. Sci. USA 886363-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride, A. A., J. C. Byrne, and P. M. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride, A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 26618411-18414. [PubMed] [Google Scholar]
- 33.Muller, A., A. Ritzkowsky, and G. Steger. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 7611042-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niman, H. L., R. A. Houghten, L. E. Walker, R. A. Reisfeld, I. A. Wilson, J. M. Hogle, and R. A. Lerner. 1983. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc. Natl. Acad. Sci. USA 804949-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406207-210. [DOI] [PubMed] [Google Scholar]
- 36.Peng, Y. C., D. E. Breiding, F. Sverdrup, J. Richard, and E. J. Androphy. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 745872-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penrose, K. J., and A. A. McBride. 2000. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J. Virol. 746031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rehtanz, M., H. M. Schmidt, U. Warthorst, and G. Steger. 2004. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol. Cell. Biol. 242153-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shembade, N., N. S. Harhaj, D. J. Liebl, and E. W. Harhaj. 2007. Essential role for TAX1BP1 in the termination of TNF-α-, IL-1- and LPS-mediated NF-κB and JNK signaling. EMBO J. 263910-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shembade, N., N. S. Harhaj, K. Parvatiyar, N. G. Copeland, N. A. Jenkins, L. E. Matesic, and E. W. Harhaj. 2008. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat. Immunol. 9254-262. [DOI] [PubMed] [Google Scholar]
- 41.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonsson, M., C. H. Heldin, J. Ericsson, and E. Gronroos. 2005. The balance between acetylation and deacetylation controls Smad7 stability. J. Biol. Chem. 28021797-21803. [DOI] [PubMed] [Google Scholar]
- 43.Sverdrup, F., and S. A. Khan. 1994. Replication of human papillomavirus (HPV) DNAs supported by the HPV type 18 E1 and E2 proteins. J. Virol. 68505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao, J. M., D. E. Breiding, and E. J. Androphy. 1998. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 721013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]
- 46.Yu, T., Y. C. Peng, and E. J. Androphy. 2007. Mitotic kinesin-like protein 2 binds and colocalizes with papillomavirus E2 during mitosis. J. Virol. 811736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng, S. X., Y. Jin, D. T. Kuninger, P. Rotwein, and H. Lu. 2003. The acetylase activity of p300 is dispensable for MDM2 stabilization. J. Biol. Chem. 2787453-7458. [DOI] [PubMed] [Google Scholar]