Abstract
Studies of the hepatitis C virus (HCV) life cycle have been aided by development of in vitro systems that enable replication of viral RNA and production of infectious virus. However, the functions of the individual proteins, especially those engaged in RNA replication, remain poorly understood. It is considered that NS4B, one of the replicase components, creates sites for genome synthesis, which appear as punctate foci at the endoplasmic reticulum (ER) membrane. In this study, a panel of mutations in NS4B was generated to gain deeper insight into its functions. Our analysis identified five mutants that were incapable of supporting RNA replication, three of which had defects in production of foci at the ER membrane. These mutants also influenced posttranslational modification and intracellular mobility of another replicase protein, NS5A, suggesting that such characteristics are linked to focus formation by NS4B. From previous studies, NS4B could not be trans-complemented in replication assays. Using the mutants that blocked RNA synthesis, defective NS4B expressed from two mutants could be rescued in trans-complementation replication assays by wild-type protein produced by a functional HCV replicon. Moreover, active replication could be reconstituted by combining replicons that were defective in NS4B and NS5A. The ability to restore replication from inactive replicons has implications for our understanding of the mechanisms that direct viral RNA synthesis. Finally, one of the NS4B mutations increased the yield of infectious virus by five- to sixfold. Hence, NS4B not only functions in RNA replication but also contributes to the processes engaged in virus assembly and release.
Recent estimates predict that the prevalence of hepatitis C virus (HCV) infection is approximately 2.2% worldwide, equivalent to about 130 million persons (22). The virus typically establishes a chronic infection that frequently leads to serious liver disease (1), and current models indicate that both morbidity and mortality as a consequence of HCV infection will continue to rise for about the next 20 years (10, 11, 29).
HCV is the only assigned species of the Hepacivirus genus within the family Flaviviridae. The virus can be classified into six genetic groups or clades (numbered 1 to 6) and then further separated into subtypes (e.g., 1a, 1b, 2a, 2b, etc.) (53, 55). HCV has a single-stranded, positive-sense RNA genome that is approximately 9.6 kb in length (reviewed in reference 46). Genomic RNA carries a single open reading frame flanked by 5′ and 3′ nontranslated regions, which are important for both replication and translation (19, 20, 34, 47, 56). Viral RNA is translated by the host ribosomal machinery, and the resultant polyprotein is co- and posttranslationally cleaved to generate the mature viral proteins. The structural proteins (core, E1, and E2) and a small hydrophobic polypeptide called p7 are produced by the cellular proteases signal peptidase and signal peptide peptidase (28, 45, 54). Two virus-encoded proteases, the NS2-3 autoprotease and the NS3 serine protease (5, 13, 26), are responsible for maturation of the nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). With the exception of NS2, the NS proteins are necessary for genome replication (8, 40) and form replication complexes (RCs), which are located at the endoplasmic reticulum (ER) membrane (14, 24, 52, 57, 59). The functions of all viral constituents of RCs have not been characterized in detail. It is known that NS5B is the RNA-dependent RNA polymerase (6), while NS3 possesses helicase and nucleoside triphosphatase activities in addition to acting as a protease (32, 58). However, the precise roles of the other proteins remain to be firmly established.
Expression of NS4B, one of the replicase proteins, generates rearrangements at the ER membrane that have been termed the “membranous web” (14, 24) and “membrane-associated foci” (MAFs) (25). Detection of viral RNA at such foci suggests that NS4B is involved in creating the sites where genome synthesis occurs (18, 24, 59). It is predicted that NS4B has an amphipathic α-helix within its N-terminal region, which is followed by four transmembrane domains (TMDs) in the central portion of the protein (17, 42). As a result, the majority of NS4B is likely to be tightly anchored to membranes, and experimental evidence indicates that it has characteristics consistent with an integral membrane protein (27). It is thought that after membrane association, NS4B rearranges membranes into a network, thereby generating foci which act as a “scaffold” to facilitate RNA replication. The mechanisms engaged in formation of foci are not known but include the notion that the NS4B N terminus can translocate into the ER lumen, resulting in rearrangement of cellular membranes (41, 42). Alternatively, palmitoylation, a lipid modification, might facilitate polymerization of NS4B, in turn promoting formation of RCs on the ER membrane (68).
Apart from inducing membranous changes required for replication, NS4B may perform other tasks in HCV RNA synthesis. For example, studies of cell culture adaptive mutations in subgenomic replicons (SGRs) have identified amino acid changes that can stimulate RNA production (39), suggesting that NS4B may exert a regulatory role in determining replication efficiency. In support of a regulatory function, replacement of NS4B sequences in an SGR from strain H77 (a genotype 1a strain) with those from strain Con-1 (a genotype 1b strain) gave higher levels of replication than for a wild-type (wt) strain H77 SGR (7). The corresponding replacement of strain Con-1 NS4B sequences with those from strain H77 reduced the replication efficiency of a Con-1 SGR (7). Moreover, interactions of NS4B with the RC can affect the behavior of other replicase proteins. For example, NS4B is needed for hyperphosphorylation of NS5A (35, 48) and restricts its intracellular movement (30).
To try to gain greater insight into the functional organization of the components that constitute RCs, trans-complementation assays using defective and helper SGRs have been established (2, 64). Such studies reveal that the only protein capable of trans-complementation is NS5A, while active replication cannot be restored for replicons harboring deleterious mutations in NS3, NS4B, and NS5B. These data led to the conclusion that functional NS5A may be able to exchange between RCs (2), whereas, by inference, such exchange would not be possible for other HCV replicase proteins. In transient-replication assays, complementation by NS5A also relied on its expression as part of a polyprotein (minimally NS3-NS5A), and production of the protein alone failed to restore replication for an inactive SGR (2). However, in a separate study, stable expression of wt NS5A was capable of complementing a defective replicon (64). Thus, different assay systems can give dissimilar results for complementation by NS5A.
In this study, we have created a series of mutations in the NS4B gene of HCV strain JFH1 (31) to explore the function of the protein in the HCV life cycle. We focused our attention on the C-terminal portion of NS4B, downstream from the predicted TMD regions, since it is relatively well conserved and is predicted to lie on the cytosolic side of the ER membrane (15, 42). Our analysis examines the impact of mutations on replication efficiency and the intracellular characteristics of the mutants compared to the behavior of the wt protein. In addition, we have utilized this series of mutants to reassess trans-complementation of NS4B in replication assays. Finally, we also analyze the impact of mutations which do not affect replication on the production of infectious virus to determine whether NS4B plays a role in virus assembly and release.
MATERIALS AND METHODS
Construction of plasmids and mutations in the NS4B and NS5A genes.
The construction of plasmids pLuc-JFH1GFP and pLuc-JFH1GND has been described previously (30). pCMV-JFH1Poly was created by first amplifying the NS3-NS5B coding region in pLuc-JFH1GFP by PCR. For cloning purposes, EcoRI and KpnI sites were introduced at the 5′ and 3′ ends of the NS3 and NS5B genes, respectively, and the amplified product was inserted into plasmid pCMV10. Plasmid pCMV-NS5A-GFP was generated by amplification of a fragment (which incorporated XmaI sites at the 5′ and 3′ termini) encoding the NS5A-green fluorescent protein (GFP) fusion protein from pLuc-JFH1GFP and insertion of the fragment into pCMV10. To create pCMV-JFH1Poly-Δ4B, two fragments were amplified from pCMV-JFH1Poly. Fragment 1 consisted of the sequences encoding NS3 and NS4A, with an EcoRI site introduced at the 5′ end of NS3 and an FspI site at the 3′ end of the NS4A coding region. Fragment 2 contained NS5A up to the AgeI site, which marks the insertion site for the GFP gene, and an FspI site was introduced at the 5′ end of the NS5A coding region. Both fragments were ligated into a pCMV-JFH1Poly backbone cut with EcoRI and AgeI (at the insertion site for GFP). The creation of an FspI site at the junction between the NS4A and NS5A coding regions in pCMV-JFH1Poly-Δ4B altered the nucleotide sequence from TGCGCC to TGCGCA (coordinates 5483 to 5488 in the JFH1 genome), generating a Cys-Ala cleavage site between the two proteins. This alteration in the nucleotide sequence substitutes alanine for serine at the P1′ position in the NS4B-NS5A cleavage site. However, alanine is present at the junction between NS4A and NS4B and, along with serine, is highly favored at the P1′ position (33). Mutations in NS4B were generated using the QuikChange mutagenesis kit (Stratagene) and introduced into plasmid pGEM-NS4BCT. pGEM-NS4BCT was generated by introducing a BamHI fragment (site coordinates 6007 and 6273 on the JFH1 genomic sequence), encoding the C-terminal 87 amino acids of NS4B, into the pGEM-T-Easy vector (Promega). Each mutated NS4B fragment was then reintroduced into both the pLuc-JFH1GFP replicon and pCMV-JFH1Poly. To introduce the NS4B mutants into the full-length JFH1 and J6-JFH1 genomes, the C-terminal region of each mutant NS4B was excised from the appropriate pLuc-JFH1GFP SGR using BamHI and ligated into pJFH1 cut with the same enzyme. J6-JFH1 is a chimeric cDNA in which strain JFH1 sequences from core to the loop region between TMDs 1 and 2 in NS2 (amino acids 1 to 864 in the JFH1 polyprotein sequence) are replaced with those from strain HC-J6 (51, 67). Plasmid pLuc-JFH1S232I was generated by site-directed mutagenesis of pLuc-JFH1 (62) using the QuikChange mutagenesis kit (Stratagene). Nucleotide sequences were determined to verify the authenticity of mutants and the orientation of insertions.
Maintenance of tissue culture cells.
Huh-7 cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum as described previously (60). The Huh-7-derived cell line termed 2/1 (62), which supports replication of subgenomic RNA from strain JFH1, was maintained in DMEM containing 10% fetal calf serum and 100 μg/ml G418.
Antibodies.
The NS5A antibody used for Western blot analysis was kindly provided by Steve Griffin and Mark Harris (44). Mouse monoclonal antibody J2, which is specific for double-stranded RNA (dsRNA), was purchased from Scicons (Hungary). Rabbit antiserum R1063 was raised against a pool of three synthetic peptides composed of NS4B-specific sequences (NRLIAFASRGNHV, THYVPESDAAAR, and LLRRLHQWISSEC) (S. Gretton, G. Hope, and J. McLauchlan, unpublished).
Transfection of plasmid DNA.
All transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were seeded at approximately 8 × 105 in 35-mm dishes and allowed to adhere before addition of transfection reagents. Cells were examined 18 to 20 h after transfection.
In vitro transcription and electroporation of RNA.
Constructs containing subgenomic (JFH1) and full-length (JFH1 and J6-JFH1) sequences were linearized with XbaI and treated with mung bean nuclease as described previously (62, 65). RNA was transcribed in vitro from linearized constructs using the T7 RiboMAX Express large-scale RNA production system (Promega) and introduced into Huh-7 cells by electroporation as described previously (45).
Infection of cells with HCV.
Huh-7 cells were electroporated with either wt or mutant JFH1 and J6-JFH1 RNAs and maintained at 37°C. Supernatant containing virus was removed from cells at 24, 48, and 72 h, and the 50% tissue culture infectious dose (TCID50) was obtained in Huh-7 cells using the limiting-dilution assay (37). Infected cells were detected using a polyclonal anti-NS5A antiserum.
Determination of luciferase activity in transient-replication assays.
wt and mutant Luc-JFH1GFP RNAs were electroporated into Huh-7 cells and seeded into 2-cm2 wells in 24-well plates. Cell extracts were prepared at 4, 24, 48, and 72 h after electroporation. Luciferase activities were determined using the luciferase assay system (Promega) and a Biotrace M3 luminometer (Biotrace Ltd.). Data points were obtained in duplicate, and experiments were repeated at least three times; representative data are given for each experiment.
Indirect immunofluorescence.
Huh-7 cells were fixed in methanol for 20 min at −20°C. After being washed with phosphate-buffered saline (PBS), cells were incubated with primary antibody (diluted in PBS) for 2 h at room temperature. Cells were washed in PBS and then incubated with secondary antibody conjugated to the appropriate fluorophore for 1 h at room temperature. After incubation with secondary antibody, cells were washed with PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI) at a final concentration of 1 μg/ml for 2 min to stain nuclei. Following further washes with PBS, cells were rinsed with H2O before being mounted onto slides with Citifluor (Citifluor Ltd.). Images were captured using a Zeiss LSM510 META inverted confocal microscope and associated software. For the data on NS5A distribution shown in Fig. 2C, at least 250 randomly selected cells were scored into two categories according to the distribution of GFP fluorescence.
FIG. 2.
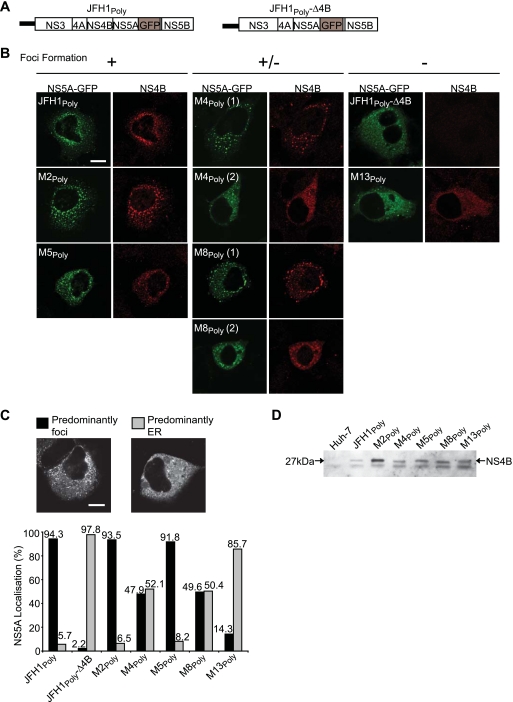
The NS4B C terminus contains determinants for focus formation. (A) Schematic representations of the HCV polyproteins encoded by pCMV-JFH1Poly and pCMV-JFH1Poly-Δ4B, both of which express NS5A-GFP. (B) Huh-7 cells were transfected with the indicated pCMV-JFH1Poly plasmids. At 20 h after transfection, cells were fixed and probed with NS4B antiserum R1063 by indirect immunofluorescence. Subcellular localization patterns for NS4B (red) and NS5A-GFP (green) are shown. Localization patterns for both proteins were separated into three phenotypic categories: proteins predominantly in foci (+), proteins either in foci or with a distribution consistent with an ER-like pattern (+/−), and proteins with a distribution consistent with an ER-like pattern (−). (C and D) Huh-7 cells were transfected with the pCMV-JFH1Poly plasmids and either fixed (C) or used to prepare extracts (D) at 20 h posttransfection. In panel C, distribution patterns for NS5A-GFP were assessed as either predominantly in foci (left image) or at the ER membrane (right image); the graph shows the percentage of NS5A-GFP in both patterns for each construct. In panel D, cell extracts were probed with NS4B antiserum R1063 by Western blot analysis. The species corresponding to NS4B is indicated by an arrow. Scale bars in panels B and C represent 10 μm.
Fluorescence recovery after photobleaching analysis.
Huh-7 cells were seeded onto 35-mm glass-bottom microwell dishes (MatTek Cultureware) and transfected with plasmids expressing GFP-tagged proteins. Fluorescence recovery after photobleaching (FRAP) analysis and image recording were conducted with a Zeiss LSM510 META inverted confocal microscope at 37°C in a humidified chamber with an atmosphere of 5% CO2. Images were recorded with a Plan-Apochromat ×63 lens (numerical aperture, 1.4). Prior to photobleaching, the cell medium was replaced with prewarmed (to 37°C) DMEM lacking phenol red and supplemented with 2% fetal calf serum. For photobleaching, selected regions of cells (38 μm2 in area) were bleached at 100% laser power (488-nm laser line) for approximately 1 s. Before and after photobleaching, images were taken at 1-s intervals using 2% laser power. The intensity of fluorescence for each cell was expressed as a percentage of the prebleach level and plotted against time. The data shown in Fig. 3 represent average values generated from 10 cells.
FIG. 3.
The NS4B C terminus influences mobility and hyperphosphorylation of NS5A. Huh-7 cells were transfected with the pCMV-JFH1Poly plasmid series as well as constructs expressing DNase X and NS5A-GFP. At 24 h posttransfection, cells were either examined by FRAP analysis (A and C) or extracts were prepared for Western blot analysis (B and D). (A and C) Fluorescence recovery curves for NS5A-GFP expressed by the indicated constructs. The time at which regions of interest were photobleached is indicated by an arrow. Numbers in parentheses correspond to the percent fluorescence recovery and were calculated by dividing the fluorescence intensity measured at 120 s after bleaching by prebleach values. (B and D) Cell extracts were probed by Western blot analysis with NS5A antiserum. The positions of bands corresponding to NS5A-GFP are indicated by arrows. In panel B, the asterisk denotes hyperphosphorylated NS5A-GFP. In panel D, the upper and lower arrows indicate the positions of hyper- and hypophosphorylated NS5A-GFP, respectively.
Preparation of cell extracts, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blot analysis.
To improve detection of the HCV NS proteins, particularly in transfections with the pCMV-JFH1Poly plasmids, cell extracts were phase separated. Briefly, a 35-mm dish of Huh-7 cells was harvested in 200 μl lysis buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.2], and 2% Triton X-114). Cell extracts were then centrifuged at 8,000 × g for 5 min, after which cellular debris was removed, and then 400 μl separation buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.2], 6% sucrose, and 0.006% Triton X-114) was added to the extracts and mixed gently. Further centrifugation at 15,000 × g for 5 min separated the extracts into two layers, with the bottom layer containing cellular membranes and any membrane-associated proteins. This membrane-containing fraction was collected, and the proteins were precipitated using 100% acetone. The precipitate was pelleted by centrifugation at 15,000 × g for 5 min and subsequently prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis (60).
RESULTS
Residues in the C terminus of NS4B are critical for HCV RNA replication.
From a previous report on predicted structural elements within NS4B, a central hydrophobic segment that is believed to contain four TMDs (TMDs 1 to 4) between amino acids 75 to 191 (Fig. 1A) was the most obvious feature within the polypeptide sequence (42). The precise junctions for the TMDs have not been determined experimentally. Upstream from the TMD region, it has been proposed that the N-terminal segment contains an amphipathic α-helix between amino acids 6 and 29 (17) and a fifth TMD termed TMX (42). A variety of predictive programs for transmembrane helices did not indicate the presence of any TMD regions beyond amino acid 191 (data not shown). However, analysis of NS4B sequences from three HCV strains (H77, Con-1, and JFH1) using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html), which predicts secondary structures in proteins, suggested the presence of two α-helices between positions 201 and 213 and positions 228 and 254 (termed helix 1 and helix 2, respectively) (Fig. 1A). Helix 1 is located within a region that is almost invariant in NS4B, whereas the amino acid sequence of helix 2 is more variable (66). From previous analysis of NS4B mutants and their ability to support HCV RNA replication, only two mutations have been examined in the C-terminal region (W252 and N254) (Fig. 1A) (38); in strain JFH1, the amino acid encoded at position 254 is threonine and not asparagine as in strain Con-1. Mutation of these amino acids either abolished (W252) or severely impaired (N254) replication (38).
FIG. 1.
Identification of amino acids in the NS4B C terminus that are critical for HCV RNA replication. (A) Schematic representation of the predicted topological arrangement of NS4B bound to the ER membrane. NS4B is predicted to contain four TMDs (numbered 1 to 4) that span the ER membrane, with the flanking N- and C-terminal ends located in the cytosol. Previously predicted features (α-helix between amino acids 6 and 29 and TMX) in the N-terminal region and the location of a nucleotide binding motif (NBM) are shown. Helices (H1 and H2), predicted by PSIPRED, that are located in the C-terminal domain are indicated. Below the cartoon, the amino acid sequence for the C-terminal region of NS4B encoded by strain JFH1 is presented. Amino acid residues that were replaced with alanine are underlined and numbered M1 to M15. Amino acids predicted to lie within helices H1 and H2 are overlined and shown in red. Asterisks denote conserved amino acids, and residues are numbered according to the N-terminal end of NS4B following cleavage from the polyprotein by the NS3/4A protease. (B) Schematic representation showing the Luc-JFH1GFP SGR encoding NS5A-GFP and the position of the C-terminal end of NS4B containing mutations M1 to M15. (C) Huh-7 cells were electroporated with in vitro-transcribed RNA derived from wt Luc-JFH1GFP, mutants M1luc to M15luc, and Luc-JFH1GND. Extracts were prepared at 4, 24, 48, and 72 h to determine luciferase activity at each time point. Assays were performed in duplicate. RLU, relative light units.
Given the low number of mutations generated in the C-terminal segment of NS4B and our prediction that two α-helices were located in this region, we performed alanine substitution mutagenesis to evaluate its importance in HCV RNA replication. Fifteen point mutations were introduced into the region between amino acids 192 and 261 (Fig. 1A). These mutations were distributed randomly along the length of the C-terminal segment, incorporating changes in the highly conserved region (amino acids 192 to 227), both α-helices, previously mutated residues at positions 252 and 254 (38), and a cysteine residue at position 257 that may be the site for palmitoylation of the protein (68). Mutations were introduced into pSGR-Luc-GFP-JFH1 (hereafter referred to as Luc-JFH1GFP) (Fig. 1B), an HCV SGR that expresses GFP-tagged NS5A to enable direct visualization of the protein in live cells (30); the mutant constructs were referred to as M1luc, M2luc, etc.
To determine whether these changes affected replication, mutant RNA was electroporated into Huh-7 cells and luciferase activity was measured at regular time intervals for up to 72 h (Fig. 1C). Luciferase activity from Luc-JFH1GFP was almost 50-fold higher at 72 h than the 4-h value, consistent with previous results (30). Of the 15 mutants, 8 replicated to similar levels as Luc-JFH1GFP over a 72-h period (M1luc, M6luc, M7luc, M9luc, M10luc, M12luc, M14luc, and M15luc). Two mutants, M3luc and M11luc, were attenuated for replication, as luciferase activity dropped 10- and 20-fold, respectively, compared to that of Luc-JFH1GFP in the first 24 h following introduction of subgenomic RNA into cells. Luciferase activities increased after this initial decline but remained lower than that of Luc-JFH1GFP by 72 h. Luciferase levels for five of the mutants (M2luc, M4luc, M5luc, M8luc, and M13luc) declined as quickly as those seen with a GND control mutant, and these mutants therefore were judged to be unable to replicate. Four of these mutations were located in the highly conserved segment between amino acids 192 and 227 (M2, M4, M5, and M8), and only one mutation (M13) was found in the less well-conserved region beyond amino acid 227 (Fig. 1A). Mutant M13 corresponds to a tryptophan (W252) that has been identified previously as essential for RNA replication (38).
Residues in the C-terminal region of NS4B are important for formation of foci.
To characterize further the behavior of the mutations that blocked replication, we tested whether each mutant could form MAFs, the presumed sites of viral RNA replication (25). Mutations M2, M4, M5, M8, and M13 were introduced into the pCMV-JFH1Poly vector, which contained the wt coding sequences for NS3-NS5B from strain JFH1 and incorporated a GFP-tagged version of NS5A (Fig. 2A). The resultant constructs were termed M2poly, M4poly, etc. Each construct was transfected into Huh-7 cells, and NS5A-GFP and NS4B were visualized (Fig. 2B). Both NS4B and NS5A-GFP, expressed from pCMV-JFH1Poly, were localized in discrete punctate foci (Fig. 2B, JFH1Poly), which were indistinguishable from MAFs visualized in cells actively replicating HCV RNA (24, 30, 59). In contrast, NS5A-GFP expressed from pCMV-JFH1Poly-Δ4B, which lacked the entire NS4B coding region (Fig. 2A), was localized almost exclusively in a pattern consistent with localization to the ER, and there was little evidence for the presence of the GFP-tagged NS5A in foci (Fig. 2B). Indeed, this pattern of NS5A-GFP distribution was similar to that observed when it was expressed in the absence of the other NS proteins (30). Analysis of the five NS4B mutants revealed three distinct phenotypic categories. First, M2Poly and M5Poly produced foci containing NS4B that were indistinguishable from those observed with wt protein, and NS5A-GFP colocalized with NS4B in these structures (Fig. 2B, +). In the second category (Fig. 2B,+/−), M4Poly and M8Poly exhibited a dual phenotype, with ∼50% of cells showing colocalization of NS4B and NS5A-GFP in foci [Fig. 2B, M4Poly(1) and M8Poly(1)], while the remainder revealed little distribution of either protein in foci [Fig. 2B, M4Poly(2) and M8Poly(2)]. M13Poly was the sole member of the third category (Fig. 2B, −), in which both NS4B and NS5A-GFP were rarely detected in foci and the proteins were predominantly localized in an ER-like pattern. Since single-fluorescence images do not give an overall representation of protein localization, we selected at least 250 cells, which had been transfected with the NS4B Poly mutants and assigned the distribution of NS5A-GFP into one of two categories, predominantly foci (Fig. 2C, left image) or predominantly ER (Fig. 2C, right image); the localization of NS5A-GFP alone was assessed since it had a pattern of distribution identical to that of NS4B in the indirect-immunofluorescence studies. In agreement with the data in Fig. 2B, the localization of NS5A-GFP was markedly different for NS4B mutants M4, M8 (approximately equal numbers of cells with and without foci), and M13 (a low proportion of foci was detected) compared to mutants M2 and M5 (predominantly foci). To eliminate the possibility that introducing mutations into NS4B had decreased stability of the protein or affected polyprotein processing, cells expressing each mutant were examined by Western blot analysis (Fig. 2D). NS4B was present at the correct size (approximately 27 kDa) in each extract, and none of the mutants were expressed to lower levels than wt protein. These results showed that amino acid residues N206 (M4), E226 (M8), and particularly W251 (M13) (Fig. 1A) were important for the ability of NS4B to generate foci and for incorporation of NS5A-GFP at these sites.
Mutations in the C terminus of NS4B influence the mobility and phosphorylation of NS5A.
FRAP is a well-defined method for studying protein mobility (23), and using this technique, we have demonstrated previously that NS5A-GFP is considerably more mobile in the absence of other NS proteins (30). These data suggested that one or more of the NS proteins present in the polyprotein alter the mobility of NS5A. To determine whether NS4B was a factor in NS5A mobility, we compared the fluorescence recoveries of NS5A-GFP expressed from pCMV-JFH1Poly and pCMV-JFH1Poly-Δ4B. In agreement with our previous data (30), NS5A-GFP expressed alone gave a higher rate of fluorescence recovery than expression of the protein from pCMV-JFH1Poly, in which it is expressed as part of the NS3-NS5B polyprotein, but was less mobile than DNase X, a control ER-targeting protein (Fig. 3A). From the results obtained with pCMV-JFH1Poly-Δ4B, fluorescence recovery for NS5A-GFP was more rapid in the absence of NS4B although not as rapid as for NS5A-GFP expressed alone (Fig. 3A). The reduced fluorescence recovery did not result from disrupted cleavage at the NS4A/5A junction site due to the removal of NS4B, since the apparent molecular weight of NS5A-GFP expressed from pCMV-JFH1Poly-Δ4B was identical to that from the wt construct pCMV-JFH1Poly (Fig. 3B). We interpreted these results to indicate that NS4B plays a major role in lowering the mobility of NS5A-GFP, perhaps through their association, although NS3, NS4A, or NS5B also may contribute partly to reducing its mobility. Given that the difference in NS5A-GFP mobility was readily measurable in the presence and absence of NS4B, we conducted FRAP studies with the five pCMV-JFH1Poly NS4B mutants (M2Poly, M4Poly, etc.) (Fig. 3C). From this analysis, M2Poly and M5Poly, both of which could form MAFs, gave rates of NS5A-GFP fluorescence recovery comparable to that obtained for wt pCMV-JFH1Poly (Fig. 3A and C). In contrast, fluorescence recovery rates for M4Poly and M8Poly were higher, while NS5A-GFP expressed from M13Poly had a mobility similar to that from pCMV-JFH1Poly-Δ4B (Fig. 3A and C). These patterns of fluorescence recovery correlate with the ability of each of the mutants to form foci and are consistent with the notion that the tethering of NS5A to RCs is linked to focus formation.
NS5A exists as two phosphorylated species of 56 kDa (hypophosphorylated NS5A) and 58 kDa (hyperphosphorylated NS5A), and NS4B is required for production of the hyperphosphorylated form (35, 48). The need for NS4B to generate hyperphosphorylated NS5A was confirmed by comparing the NS5A-GFP species expressed from pCMV-JFH1Poly and pCMV-JFH1Poly-Δ4B (Fig. 3B). With the wt construct, NS5A-GFP was detected as two bands, presumed to be hypo- and hyperphosphorylated species, whereas only the hypophosphorylated protein was found for pCMV-JFH1Poly-Δ4B (Fig. 3B). For the NS4B mutant plasmids, M2Poly and M5Poly produced both phosphorylated species of NS5A-GFP, whereas the abundance of hyperphosphorylated protein was reduced for M4Poly and M8Poly, and M13Poly generated only the hypophosphorylated form (Fig. 3D). Taken together with the FRAP analysis and examination of focus formation, our results establish that there is close correspondence between the effects of the various NS4B mutations on formation of foci and mobility and phosphorylation of NS5A, providing strong evidence that these processes are linked.
Nonreplicating NS4B mutants can be complemented in trans.
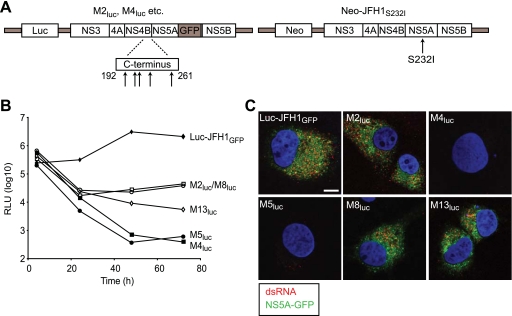
It has been demonstrated that the function of NS5A in RNA synthesis can be complemented in trans, allowing restoration of replication for subgenomic RNA carrying defective NS5A (2, 64). Based on analysis with a limited series of mutants, these studies also concluded that NS4B, as well as the other NS proteins important for replication, could not be trans-complemented. Since we had identified a group of five novel mutants in NS4B which abolished replication, we examined their ability to be rescued in replication assays. In vitro-transcribed subgenomic RNA for each mutant was electroporated into both Huh-7 cells and 2/1 cells, a Huh-7 cell line harboring an autonomously replicating wt JFH1 SGR, and luciferase activity was measured over 72 h (Fig. 4B). Since the wt SGR in 2/1 cells contains the neo gene to confer antibiotic resistance (Fig. 4A), any luciferase activity detected would result from input mutant RNA. We also included Luc-JFH1GFP and Luc-JFH1GND RNAs as positive and negative controls in the two cell lines. Both control RNAs gave similar patterns of luciferase activity in Huh-7 and 2/1 cells; enzyme activity from Luc-JFH1GFP increased by 57- and 41-fold between 4 and 48 h in Huh-7 and 2/1 cells, respectively, and then either gave a further modest rise (in 2/1 cells) or a slight decrease (in Huh-7 cells) (Fig. 4B). In contrast, luciferase values from Luc-JFH1GND RNA steadily declined from 4 h onwards; this inability to complement a GND mutation in NS5B with a wt replicon agrees with previously published reports (2, 64). For the NS4B mutants, there was no evidence for replication in Huh-7 cells, in agreement with the data in Fig. 1C (Fig. 4B, panel i). In 2/1 cells, mutants M4luc and M5luc also failed to replicate and behaved almost identically to Luc-JFH1GND (Fig. 4B, panel ii). However, M2luc and M8luc gave increased luciferase activity between 24 and 72 h, indicating that replication was at least partially restored for these mutant SGRs. M13luc also gave a slight rise in enzyme levels between 48 and 72 h, consistent with a low level of replication in 2/1 cells. To demonstrate that any relative rise in luciferase activity, particularly for M2luc and M8luc, also was reflected in increased expression of viral proteins from the mutant SGRs, 2/1 cells were examined for the presence of NS5A-GFP. The endogenous SGR in 2/1 cells produces untagged NS5A, and so detection of NS5A-GFP would indicate expression from the mutant SGRs (Fig. 4A). For wt Luc-JFH1GFP, cells expressing NS5A-GFP were readily detected (Fig. 4C), reflecting the high levels of luciferase activity produced by this replicon (Fig. 4B). As predicted from the low luciferase values, NS5A-GFP expression from Luc-JFH1GND was not detected in 2/1 cells (Fig. 4C). In the case of mutants M4luc, M5luc, and M13luc, we also could not detect NS5A-GFP expression (Fig. 4C). However, NS5A-GFP was found in a low number (up to 5%) of 2/1 cells electroporated with M2luc and M8luc RNAs (Fig. 4C). The distribution of the GFP-tagged protein colocalized with the NS5A detected in these cells using an anti-NS5A antiserum (Fig. 4D). From these results, we concluded that it was possible to trans-complement some mutations in NS4B that abolish replication.
FIG. 4.
trans-complementation of SGRs with defective NS4B in cells expressing a wt HCV replicon. (A) Schematic representation of mutant and wt (helper) SGRs. The positions of mutations at the C terminus of NS4B are indicated by arrows, and the location of GFP in the NS5A coding region for the mutant SGRs is also indicated. The helper SGR was constitutively expressed in 2/1 cells. (B) Huh-7 (i) and 2/1 (ii) cells were electroporated with in vitro-transcribed RNAs derived from wt Luc-JFH1GFP, the indicated Mluc mutants, and Luc-JFH1GND. Extracts were prepared at 4, 24, 48, and 72 h to determine luciferase activity at each time point. Assays were performed in duplicate. RLU, relative light units. (C and D) 2/1 cells were electroporated with in vitro-transcribed RNAs derived from wt Luc-JFH1GFP, the indicated Mluc mutants, and Luc-JFH1GND. At 72 h after electroporation, cells were fixed and probed with NS5A antiserum (red). Cells containing NS5A alone (indicated by arrowheads) represent expression from the wt SGR in 2/1 cells; cells containing both NS5A and NS5A-GFP (indicated by arrows) represent expression from both mutant and wt SGRs. In panel D, merged and unmerged images from cells expressing both NS5A (red) and NS5A-GFP (green) are shown. Nuclei were stained with DAPI. Scale bars represent 20 μm (C) and 10 μm (D).
Reconstitution of replication from nonreplicating NS4B and NS5A mutant SGRs.
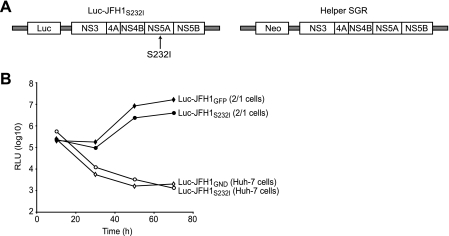
Having established that it was possible to trans-complement NS4B, we wished to extend our studies to investigate whether combining replicons with defective mutations in NS4B and NS5A could generate active RCs. From parallel studies of NS5A mutants with the JFH1 SGR, we had identified a mutation at position 232 (S232I) in the NS5A coding region that abolished replication in Huh-7 cells and behaved identically to Luc-JFH1GND (Fig. 5). In strain Con-1, mutation at this site (referred to as S2204) can either enhance or repress replication, depending on other mutations in the SGR (39). Introducing this defective replicon (termed Luc-JFH1S232I) into 2/1 cells gave luciferase values that were only fourfold lower at 72 h than those for wt Luc-JFH1GFP (Fig. 5B). Thus, replication of Luc-JFH1S232I can be restored through complementation with a wt replicon, indicating the ability to rescue deleterious mutations in NS5A, in agreement with previous reports (2, 64).
FIG. 5.
trans-complementation of a mutant NS5A SGR in cells expressing a wt HCV replicon. (A) Schematic representation of mutant (Luc-JFH1S232I) and wt (helper) SGRs. The position of the S232I mutation in NS5A is indicated. The helper SGR was constitutively expressed in 2/1 cells. (B) Huh-7 and 2/1 cells were electroporated with in vitro-transcribed RNA derived from Luc-JFH1S232I. For controls, Huh-7 and 2/1 cells were electroporated with Luc-JFH1GND and Luc-JFH1GFP RNAs, respectively. Extracts were prepared at 4, 24, 48, and 72 h to determine luciferase activity at each time point. Assays were performed in duplicate. RLU, relative light units.
To determine whether combining replicons with defective mutations in NS4B and NS5A could yield active RCs, Huh-7 cells were electroporated simultaneously with RNAs from each of the NS4B mutant replicons that were unable to replicate (M2luc, M4luc, M5luc, M8luc, and M13luc) and Neo-JFH1S232I (Fig. 6A). For these experiments, the luciferase gene in Luc-JFH1S232I was replaced with the neo gene so that any detected enzyme activity was derived from the mutant NS4B SGRs. The pattern of results obtained was identical to that for complementation in the 2/1 cells (compare Fig. 4B, panel ii, and 6B); luciferase activities for mutants M4luc and M5luc steadily decreased to low levels by 72 h, whereas values for M2luc and M8luc initially dropped between 4 and 24 h but then increased thereafter up to 72 h. Enzyme levels for M13luc did not show a similar rise between 24 and 72 h but also did not decline to the same extent as those for M4luc and M5luc. Therefore, we concluded that a low level of complementation could be achieved for M13luc. To demonstrate that HCV RNA replication did occur in cells with a combination of the Neo-JFH1S232I SGR and the M2luc, M8luc, and M13luc replicons, expression of NS5A-GFP was examined along with detection of dsRNA using J2, a monoclonal antibody that detects RNA-RNA hybrids of greater than 40 bp. Recently, we have shown that this antibody detects dsRNA in both HCV-infected cells and cells harboring SGRs at cytoplasmic sites that are consistent with the location of active RCs (59, 61). As a positive control, we showed that NS5A-GFP and dsRNA were found in cells electroporated with wt Luc-JFH1GFP RNA (Fig. 6C). Consistent with the rapid decline in luciferase activity, neither NS5A-GFP nor dsRNA was detected in cells electroporated with a combination of either M4luc or M5luc RNAs and Neo-JFH1S232I RNA (Fig. 6C). In contrast, NS5A-GFP and dsRNA were present in a low number of cells coelectroporated with Neo-JFH1S232I RNA and the M2luc and M8luc SGRs; NS5A-GFP and dsRNA also were detected occasionally with the M13luc SGR (Fig. 6C). In cells electroporated individually with these RNAs, there was no detection of either dsRNA or NS5A-GFP (data not shown). We also tested rescue of Luc-JFH1S232I by the NS4B mutant SGRs in which the luciferase reporter gene had been replaced with the neo gene (M2neo, M4neo, etc.). In these assays, replication of Luc-JFH1S232I could be detected with M2neo and M8neo and to a lesser extent with M13neo (data not shown). Hence, we conclude that the combination of two replicons, one with a mutation in NS4B and the other with a mutation in NS5A, both of which block replication, can reconstitute active RCs.
FIG. 6.
Reconstitution of active replication by combining defective SGRs in NS4B and NS5A. (A) Schematic representation of mutant SGRs in NS4B (M2luc, M4luc, etc.) and NS5A (Neo-JFH1S232I). Positions of mutations at the C terminus of NS4B in the Mluc SGRs and in NS5A in Neo-JFH1S232I are indicated. (B and C). Huh-7 cells were electroporated with combinations of in vitro-transcribed RNAs from the Mluc SGRs and from Neo-JFH1S232I. For control purposes, Huh-7 cells were electroporated with Luc-JFH1GFP and Neo-JFH1S232I RNAs. For panel B, extracts were prepared at 4, 24, 48, and 72 h to determine luciferase activity at each time point. RLU, relative light units. Assays were performed in duplicate. For panel C, cells were fixed at 72 h after electroporation and probed with monoclonal antibody J2 to detect dsRNA (red). Cells were examined also for the presence of NS5A-GFP (green). Nuclei were stained with DAPI, and the scale bar represents 10 μm.
The C-terminal domain of NS4B influences production of infectious virus.
Eight of the mutations in the C-terminal region of NS4B (M1, M6, M7, M9, M10, M12, M14, and M15) had no effect on RNA replication, while two mutations (M3 and M11) reduced replication in transient assays (Fig. 1A and C). To determine whether the C terminus of NS4B contributed to assembly and release of infectious particles, all 10 mutations were introduced into the full-length JFH1 cDNA (Fig. 7A). RNA from these mutant constructs (termed M1JFH1, M3JFH1, etc.) was electroporated into cells, and virus release was assayed by determining TCID50 values (Fig. 7B). The majority of the mutants produced virus titers comparable to those for wt JFH1, with the exception of three mutants (M3JFH1, M6JFH1, and M11JFH1) (Fig. 7B). M3JFH1 and M11JFH1 gave reduced amounts of virus production, which is likely to be a consequence of impaired RNA replication as deduced from luciferase assays (Fig. 1C, panels i and iii). However, M6JFH1 consistently produced higher viral titers than wt JFH1 at all time points, and infectious virus production increased by up to fivefold by 72 h after electroporation of RNA into cells (Fig. 7B). Higher levels of released virus were not a consequence of greater abundance of viral proteins in M6JFH1 RNA-electroporated cells compared to wt JFH1 based on Western blot analysis of NS5A (Fig. 7C, compare upper panels for JFH1 and M6JFH1); similarly, the abundance of viral RNA as determined by quantitative reverse transcription-PCR was comparable in cells electroporated with M6JFH1 and wt JFH1 RNAs (data not shown). However, NS5A levels were increased in M6JFH1-infected cells, thereby reflecting the higher titers obtained with this construct (Fig. 7C, compare lower panels). We also compared the localization and signal intensities of core, NS5A, and dsRNA in cells electroporated with M6JFH1 and JFH1 RNAs by indirect immunofluorescence, but no apparent differences were found (data not shown).
FIG. 7.
NS4B influences production of infectious HCV virions. (A) Schematic representation of JFH1 and J6-JFH1 constructs containing mutations at the C terminus of NS4B. For JFH1, the positions of 10 inserted mutations in NS4B are indicated by arrows. For J6-JFH1, the location of the M6 mutation in NS4B is shown. The gray region (core-NS2) indicates the coding sequences from strain HC-J6, which replaced the corresponding segment in strain JFH1. (B) Huh-7 cells were electroporated with wt and mutant (MJFH1) JFH1 RNAs, and medium was removed from cells at 24, 48, and 72 h after electroporation. Naïve Huh-7 cells were inoculated with medium to determine TCID50 values at 24, 48, and 72 h. Error bars indicate standard deviations. (C) Huh-7 cells were either electroporated with JFH1 and M6JFH1 RNAs or infected with 1 ml of medium removed at 24, 48, and 72 h after electroporation. For electroporated samples, extracts were prepared at 24, 48, and 72 h, and for infected samples, extracts were prepared at 72 h after infection. Extracts were probed with NS5A antiserum by Western blot analysis, and bands corresponding to NS5A are indicated by arrows. (D) Huh-7 cells were electroporated with J6-JFH1 and M6J6-JFH1 RNAs, and medium was removed from cells at 24, 48 and 72 h after electroporation. Naïve Huh-7 cells were inoculated with medium to determine TCID50 values at 24, 48, and 72 h. (E) Huh-7 cells were either electroporated with J6-JFH1 and M6J6-JFH1 RNAs or infected with 1 ml of medium removed at 24, 48, and 72 h after electroporation. Samples were prepared and extracts were analyzed as described for panel C.
To determine whether this increase in virus production was specific to JFH1, the M6 mutation was introduced into J6-JFH1, a chimeric virus in which strain JFH1 sequences from core to the loop region between TMDs 1 and 2 in NS2 (amino acids 1 to 864 in the JFH1 polyprotein sequence) are replaced with those from strain HC-J6 (67). J6-JFH1 is equivalent to a chimeric construct named Jc1, which typically gives virus titers that are about 10-fold greater than those of strain JFH1 by 72 h after RNA electroporation and characteristically releases far larger amounts of virus at 24 h than JFH1 (51). In agreement with these reported results, we found that the relative differences in TCID50 values between J6-JFH1 and JFH1 were 55-fold and 11-fold after 24 and 72 h, respectively (comparing TCID50 values in Fig. 7B with those for J6-JFH1 in Fig. 7D). Introducing the M6 mutation into J6-JFH1 (M6J6-JFH1) did not enhance virus titers above those obtained for J6-JFH1 (Fig. 7D). Indeed, the TCID50 values obtained at 72 h were intermediate between those for J6-JFH1 and M6JFH1. Moreover, the high titers typically achieved by J6-JFH1 by 24 h were not attained (Fig. 7D). The lower TCID50 values also were reflected in reduced abundance of NS5A in M6J6-JFH1- compared to J6-JFH1-infected cells (Fig. 7E). These results suggest that the M6 mutation in NS4B can enhance virus release by JFH1 but exerts a dominant effect in the context of other alterations that improve virus yields.
DISCUSSION
NS4B is a key component of the HCV replicase complex, but, apart from an ability to rearrange the ER membrane to form foci, any additional contribution of the protein to either RNA replication or other stages of the virus life cycle are not well understood. In this report, we have identified amino acids in the C-terminal region of the protein that are essential for RNA synthesis. Additional studies revealed that mutations which block RNA replication do not prevent formation of foci, indicating that NS4B provides other functions to facilitate genome synthesis. Moreover, we establish that the contribution of NS4B to the HCV life cycle includes a role in not only replication but perhaps also assembly and release of infectious virus. Finally, in contrast to previous reports (2, 64), the function of NS4B in RNA synthesis can be complemented in trans by replicons supplying a wt version of the protein, and we show that active replication can be reconstituted from two inactive replicons. Hence, our study provides novel insight into the processes governing the formation and functions of HCV replicase complexes.
The hydrophobic character of NS4B has restricted experimental studies of its structure. Therefore, identifying structural features within the protein has relied upon predictive analysis. From previous reports, the central hydrophobic region of NS4B is arranged as four TMDs, which span the ER membrane, and the N-terminal segment contains an amphipathic α-helix (17, 42). We have extended this analysis to the C-terminal domain and predicted that it contains two α-helical elements. The first helix (helix 1) lies within a highly conserved region at the C terminus (termed segment 1, amino acids 192 to 227), while the second helix (helix 2) is located in a more variable region (segment 2, amino acids 228 to 261) (Fig. 8A). Comparative studies and analysis of NS4B sequences found in HCV-infected patients (66) suggest that the N-terminal end of helix 2 may define the beginning of the variable sequence. This boundary separates the C-terminal region of NS4B into two sections of approximately equal length (36 and 34 amino acids). The mutations that we have introduced into NS4B are also evenly distributed between the two segments (M1 to M8 in segment 1 and M9 to M15 in segment 2) (Fig. 8A). Four out of the five mutations, which block replication, are located in the more highly conserved segment. Thus, segment 1 apparently makes a greater contribution to HCV RNA replication than the more variable segment 2. The only amino acid in segment 2 identified in our study that was critical for replication was the tryptophan residue at position 252 (M13), which is invariant in all genotypes. In a previous study, mutation of this amino acid also blocked replication of a strain Con-1 SGR (38). Hence, this residue is probably essential for genome synthesis by all strains of the virus.
FIG. 8.
Summary of the mutations that affect NS4B function and model for reconstitution of active replication from defective replicons. (A) Schematic representation of the C terminus of NS4B. The position and properties of each mutation that alters the behavior of NS4B are shown. NT, not tested. (B). Model for generation of sites with active RCs from SGRs with defects in NS4B and NS5A.
Several studies have demonstrated that NS4B creates foci at the ER membrane, which are the sites of viral RNA synthesis (18, 24, 59). Analysis of each of the mutations introduced into a polyprotein carrying sequences for NS3-NS5B demonstrated that inhibiting replication does not invariably prevent formation of foci. We consider that expressing NS4B in the context of the other NS proteins is preferable for examining the effects of mutations, since the foci formed by some mutants when expressed alone were much larger than those typically found by expression of the JFH1 polyprotein (data not shown); such large foci may represent aggregation of NS4B in the absence of the NS proteins. The mechanisms involved in rearranging ER membranes by NS4B to create foci are not known but are likely to involve multimerization of the protein combined with interactions with other viral NS components as well as possibly cellular factors. The lack of any apparent change to focus formation by introducing mutations at positions 196 (M2) and 211 (M5) (Fig. 8A) could be explained by characteristics of foci, which are altered but not resolved visually by light microscopy. Alternatively, the focus-forming capability of NS4B may not be affected by these mutations, but other roles for the protein in the RC could be perturbed. Given the large excess of NS proteins compared to both positive- and negative-strand RNA (52), RCs may have both direct and indirect tasks in viral RNA synthesis (e.g., a structural role to create the environment for replication). Thus, replication perhaps relies on a combination of RCs with distinct properties, which are defined by the multifunctional characteristics of components such as NS4B.
We found that mutations in NS4B that perturbed formation of foci also altered the behavior of NS5A. NS5A exists as both hypo- and hyperphosphorylated forms in cells harboring the SGR (8). The functional importance of both forms is unclear, although reducing the abundance of the hyperphosphorylated species stimulates RNA replication (3, 49) and can reduce virus assembly and release (63). Hence, NS5A hyperphosphorylation may act as a modulatory switch between RNA synthesis and later phases of the virus life cycle. Hyperphosphorylation requires expression of NS5A from a polyprotein consisting of sequences from NS3-NS5A (35, 48). NS4B plays a critical role in this process, presumably through influencing interactions of NS5A with the cellular phosphorylation machinery. Our studies demonstrated that the ability of NS4B to generate foci is coupled to NS5A hyperphosphorylation. Moreover, there was a correlation between inefficient focus formation as a result of mutations in NS4B and increased mobility of NS5A (summarized in Fig. 8A). From these data, we propose that the production of foci at the ER membrane by NS4B binds NS5A more tightly to complexes formed by the NS proteins and this environment facilitates hyperphosphorylation. Tighter binding of NS5A could aid hyperphosphorylation either through increased contact of the protein with cellular kinases that are recruited to foci or through its protection from phosphatases. The nature of the interactions that bind NS5A to NS complexes within foci is not known. Previous studies have shown that there is a network of interactions between the NS proteins, including association of NS5A with NS4B (12, 21, 36, 50). Therefore, the mutations in NS4B that alter the behavior of NS5A could result from modifications to interactions between the two proteins. Alternatively, changes to the NS4B sequence may indirectly perturb attachment of NS5A to other NS proteins. Addressing this question requires a more thorough understanding of the protein-protein contacts that occur within RCs.
In earlier studies, trans-complementation of RNA replication could be achieved only with NS5A and not with any other HCV NS protein, including NS4B (2, 64). Our analysis with NS4B mutants that block replication demonstrates that complementation of the protein is possible with mutants M2 (G196A) and M8 (E226A); partial complementation could be achieved also with mutant M13 (W252A) (Fig. 8A). However, the efficiency of complementation was less for the NS4B mutants than for the NS5A S232I mutant. Based on the luciferase values at 72 h after electroporation (Fig. 4B, panel ii, and 5B), we estimate that the NS5A S232I mutation has a complementation efficiency of about 24%, compared to approximately 1% for NS4B mutants M2 and M8; the complementation efficiency for M13 is lower still at 0.05%. Both G196 (M2) and E226 (M8) are highly conserved amino acids that are predicted to lie within unstructured regions flanking helix 1 (Fig. 8A). Interestingly, the other two mutations (M4 [N206A] and M5 [F211A]) in segment 1, which inhibit RNA replication and cannot be complemented, reside within helix 1. Previous studies have demonstrated a lack of NS4B complementation using replicons containing mutations at positions K135 and V186 (referred to as residues K1846 and V1897 by Appel et al. [2]), both of which are located within the TMD region of the protein. Therefore, structurally ordered segments of NS4B may not be accessible for complementation, whereas amino acids that lie outside of such regions could be less constrained. Based on this hypothesis, it may be possible to design other mutations in unstructured areas of NS4B that can be complemented.
Our ability to create active RCs from two inactive SGRs is an important finding and offers new insight into replication of the viral genome. It has been proposed that RCs are in a “closed” conformation, with the replicase components (apart from NS5A) acting strictly in cis to synthesize viral RNA (2). The findings from our studies indicate that such stringent constraints may not arise, since NS4B and NS5A can complement replicons defective in either protein. The efficiency of complementation was far greater for NS5A than for NS4B, suggesting that their modes of complementation may differ. In the case of NS5A, it has been proposed that it is less tightly associated with membranes than NS4B, since NS5A attachment is mediated by an amphipathic α-helix whereas NS4B contains TMDs (2, 9, 42). Thus, NS5A would be predicted to be tethered less tightly to the replicase unit and therefore may transfer between RCs at different intracellular sites. An identical scenario may not be the case for NS4B, since its distribution is largely confined to foci whereas NS5A locates to both foci and the ER membrane. Since NS4B does not readily transfer between foci (25), it is likely that complementation occurs within individual sites of viral RNA replication. Therefore, complementation of NS4B could require incorporation of more than one RNA genome during formation of foci (Fig. 8B). Such events may be rare and account for the lower efficiency of complementation by NS4B than by NS5A. Within any foci that incorporate both defective NS4B and NS5A SGRs, we presume that NS proteins are translated from both RNAs, creating a pool of viral components. These components could combine, either as complexes or as individual proteins, to reconstitute active RCs (Fig. 8B). As a consequence, both defective SGRs are mutually dependent for continued replication of viral RNA. This model could have implications for maintenance of quasispecies pools in infected patients, since mixed infection of individual hepatocytes by viruses with genomes carrying mutations that are defective in replication may permit formation of sites active in viral RNA synthesis.
The availability of a system that produces infectious HCV from tissue culture cells now permits studies of the requirements for virus assembly and release. Apart from the obvious requirement of viral structural components, it has been demonstrated recently that both NS3 and NS5A contribute to the production of infectious virus (4, 43, 63). Our results also establish a function for NS4B in modulating virus production. A single amino acid substitution (N216A [M6]) (Fig. 8A) was sufficient to increase the titer of JFH1 virus by about fivefold. One possibility is that this substitution may improve release of RNA from replication sites for packaging into capsids, and indeed, the C-terminal region of NS4B has been implicated recently in RNA binding (16). Alternatively, the alteration at amino acid position 216 may modulate the function of other replicase components, such as NS5A, to increase delivery of genomes to packaging sites. We also examined whether enhanced virus titers could be achieved by introducing the same alteration into J6-JFH1, a recombinant that generates much greater amounts of infectious virus than JFH1 (51). However, the N216A mutation in NS4B lowered virus production by J6-JFH1. These data suggest that there may be a complex interplay between NS4B, either as an individual replicase component or as part of the replicase complex, with the structural proteins. Within such interactions, NS4B may act as a modulator of virus production. A role for NS4B in controlling viral RNA synthesis has been demonstrated from studies with chimeric replicons (7), and our analysis may indicate that the protein's regulatory functions extend to later stages of the virus life cycle. A greater understanding of such potential modes of regulation is needed and could represent an attractive target for antiviral therapy.
Acknowledgments
We are grateful to Takaji Wakita for supplying JFH1 plasmid DNAs. We also thank Mark Harris and Steve Griffin for providing NS5A antiserum and Sarah Gretton for supplying NS4B antiserum. We are indebted to Graham Hope, Joyce Mitchell, Brenna Flatley, and Patricia Domingues for expert technical assistance.
This work was supported by the Medical Research Council (United Kingdom).
Footnotes
Published ahead of print on 10 December 2008.
REFERENCES
- 1.Alberti, A., L. Chemello, and L. Benvegnu. 1999. Natural history of hepatitis C. J. Hepatol 31(Suppl. 1)17-24. [DOI] [PubMed] [Google Scholar]
- 2.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel, N., T. Pietschmann, and R. Bartenschlager. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 793187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog 4e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 673835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1512-22. [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J. 2007. Allelic variation in the hepatitis C virus NS4B protein dramatically influences RNA replication. J. Virol. 815724-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 9.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2778130-8139. [DOI] [PubMed] [Google Scholar]
- 10.Deuffic-Burban, S., M. K. Mohamed, B. Larouze, F. Carrat, and A. J. Valleron. 2006. Expected increase in hepatitis C-related mortality in Egypt due to pre-2000 infections. J. Hepatol. 44455-461. [DOI] [PubMed] [Google Scholar]
- 11.Deuffic-Burban, S., T. Poynard, M. S. Sulkowski, and J. B. Wong. 2007. Estimating the future health burden of chronic hepatitis C and human immunodeficiency virus infections in the United States. J. Viral Hepat. 14107-115. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 775401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckart, M. R., M. Selby, F. Masiarz, C. Lee, K. Berger, K. Crawford, C. Kuo, G. Kuo, M. Houghton, and Q. L. Choo. 1993. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem. Biophys. Res. Commun. 192399-406. [DOI] [PubMed] [Google Scholar]
- 14.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 765974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Einav, S., M. Elazar, T. Danieli, and J. S. Glenn. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J. Virol. 7811288-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Einav, S., D. Gerber, P. D. Bryson, E. H. Sklan, M. Elazar, S. J. Maerkl, J. S. Glenn, and S. R. Quake. 2008. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat. Biotechnol. 261019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elazar, M., P. Liu, C. M. Rice, and J. S. Glenn. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 7811393-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 842761-2769. [DOI] [PubMed] [Google Scholar]
- 19.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 765326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 7512047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 783480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Global Burden of Hepatitis C Working Group. 2004. Global burden of disease (GBD) for hepatitis C. J. Clin. Pharmacol. 4420-29. [DOI] [PubMed] [Google Scholar]
- 23.Goodwin, J. S., and A. K. Kenworthy. 2005. Photobleaching approaches to investigate diffusional mobility and trafficking of Ras in living cells. Methods 37154-164. [DOI] [PubMed] [Google Scholar]
- 24.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gretton, S. N., A. I. Taylor, and J. McLauchlan. 2005. Mobility of the hepatitis C virus NS4B protein on the endoplasmic reticulum membrane and membrane-associated foci. J. Gen. Virol. 861415-1421. [DOI] [PubMed] [Google Scholar]
- 26.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 674665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 28470-81. [DOI] [PubMed] [Google Scholar]
- 28.Hussy, P., H. Langen, J. Mous, and H. Jacobsen. 1996. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 22493-104. [DOI] [PubMed] [Google Scholar]
- 29.Hutchinson, S. J., S. M. Bird, and D. J. Goldberg. 2005. Modeling the current and future disease burden of hepatitis C among injection drug users in Scotland. Hepatology 42711-723. [DOI] [PubMed] [Google Scholar]
- 30.Jones, D. M., S. N. Gretton, J. McLauchlan, and P. Targett-Adams. 2007. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J. Gen. Virol. 88470-475. [DOI] [PubMed] [Google Scholar]
- 31.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64334-339. [DOI] [PubMed] [Google Scholar]
- 32.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215160-166. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S. Y., K. W. Park, Y. J. Lee, S. H. Back, J. H. Goo, O. K. Park, S. K. Jang, and W. J. Park. 2000. In vivo determination of substrate specificity of hepatitis C virus NS3 protease: genetic assay for site-specific proteolysis. Anal. Biochem. 28442-48. [DOI] [PubMed] [Google Scholar]
- 34.Kim, Y. K., C. S. Kim, S. H. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290105-112. [DOI] [PubMed] [Google Scholar]
- 35.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 737138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, C., J. W. Wu, K. Hsiao, and M. S. Su. 1997. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J. Virol. 716465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 38.Lindstrom, H., M. Lundin, S. Haggstrom, and M. A. Persson. 2006. Mutations of the hepatitis C virus protein NS4B on either side of the ER membrane affect the efficiency of subgenomic replicons. Virus Res. 121169-178. [DOI] [PubMed] [Google Scholar]
- 39.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 773007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 41.Lundin, M., H. Lindstrom, C. Gronwall, and M. A. Persson. 2006. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J. Gen. Virol. 873263-3272. [DOI] [PubMed] [Google Scholar]
- 42.Lundin, M., M. Monne, A. Widell, G. Von Heijne, and M. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 775428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma, Y., J. Yates, Y. Liang, S. M. Lemon, and M. Yi. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 827624-7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macdonald, A., K. Crowder, A. Street, C. McCormick, K. Saksela, and M. Harris. 2003. The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signaling. J. Biol. Chem. 27817775-17784. [DOI] [PubMed] [Google Scholar]
- 45.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 213980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5453-463. [DOI] [PubMed] [Google Scholar]
- 47.Murayama, A., T. Date, K. Morikawa, D. Akazawa, M. Miyamoto, M. Kaga, K. Ishii, T. Suzuki, T. Kato, M. Mizokami, and T. Wakita. 2007. The NS3 helicase and NS5B-to-3′X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells. J. Virol. 818030-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neddermann, P., A. Clementi, and R. De Francesco. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 739984-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neddermann, P., M. Quintavalle, C. Di Pietro, A. Clementi, M. Cerretani, S. Altamura, L. Bartholomew, and R. De Francesco. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 7813306-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piccininni, S., A. Varaklioti, M. Nardelli, B. Dave, K. D. Raney, and J. E. McCarthy. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 27745670-45679. [DOI] [PubMed] [Google Scholar]
- 51.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 1037408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 7913594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 1432493-2503. [DOI] [PubMed] [Google Scholar]
- 54.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 683631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42962-973. [DOI] [PubMed] [Google Scholar]
- 56.Song, Y., P. Friebe, E. Tzima, C. Junemann, R. Bartenschlager, and M. Niepmann. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 8011579-11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stone, M., S. Jia, W. D. Heo, T. Meyer, and K. V. Konan. 2007. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 814551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzich, J. A., J. K. Tamura, F. Palmer-Hill, P. Warrener, A. Grakoui, C. M. Rice, S. M. Feinstone, and M. S. Collett. 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 676152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Targett-Adams, P., S. Boulant, and J. McLauchlan. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 822182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Targett-Adams, P., D. Chambers, S. Gledhill, R. G. Hope, J. F. Coy, A. Girod, and J. McLauchlan. 2003. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 27815998-16007. [DOI] [PubMed] [Google Scholar]
- 61.Targett-Adams, P., G. Hope, S. Boulant, and J. McLauchlan. 2008. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J. Biol. Chem. 28316850-16859. [DOI] [PubMed] [Google Scholar]
- 62.Targett-Adams, P., and J. McLauchlan. 2005. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J. Gen. Virol. 863075-3080. [DOI] [PubMed] [Google Scholar]
- 63.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog 4e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tong, X., and B. A. Malcolm. 2006. Trans-complementation of HCV replication by non-structural protein 5A. Virus Res. 115122-130. [DOI] [PubMed] [Google Scholar]
- 65.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welker, M. W., W. P. Hofmann, C. Welsch, M. von Wagner, E. Herrmann, T. Lengauer, S. Zeuzem, and C. Sarrazin. 2007. Correlation of amino acid variations within nonstructural 4B protein with initial viral kinetics during interferon-alpha-based therapy in HCV-1b-infected patients. J. Viral Hepat. 14338-349. [DOI] [PubMed] [Google Scholar]
- 67.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262250-263. [DOI] [PubMed] [Google Scholar]
- 68.Yu, G. Y., K. J. Lee, L. Gao, and M. M. Lai. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 806013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]