Abstract
A hallmark of productive infection by DNA viruses is the coupling of viral late gene expression to genome replication. Here we report the identification of open reading frame 30 (ORF30) and ORF34 as viral trans factors crucial for activating late gene transcription following viral DNA replication during lytic infection of murine gammaherpesvirus 68 (MHV-68). The mutant virus lacking either ORF30 or ORF34 underwent normal DNA replication but failed to express viral late gene transcripts, leading to nonproductive infection. In a reporter assay system, ORF30 and ORF34 were required for MHV-68 to activate the viral late gene promoters. Furthermore, studies using chromatin immunoprecipitation assays showed that the recruitment of RNA polymerase II to the viral late promoters during lytic infection was significantly reduced in the absence of ORF30 or ORF34. Together, the results suggest that ORF30 and ORF34 may play an important role in the assembly of the transcription initiation complex at the late gene promoters. Our discovery of the viral mutants that uncouple late gene transcription from DNA replication lays an important foundation to dissect the mechanism of this critical step of gene expression regulation.
Viral gene expression during productive infection by DNA viruses is temporally regulated and typically divided into early and late phases separated by viral genome replication. Late genes are expressed after the onset of viral DNA replication and, since they mainly encode structural proteins, their expression leads to the assembly and the release of infectious particles. Although late gene expression is tightly coupled to genome replication in virtually all DNA viruses, its underlying mechanisms are not fully understood. For simian virus 40, a small virus with a circular DNA genome, amplification of viral DNA is required in trans to attenuate the repressor of viral late promoters (35, 42), and the viral large T antigen also plays an essential role to activate the promoters (3, 18). Adenoviruses, with larger and linear genomes, display a cis requirement of viral DNA replication for activation of late gene transcription (33). Moreover, in adenoviruses, trans-acting factors can also be regulated by DNA replication in multiple ways. For example, binding of a cellular transcription factor to the late promoter is greatly facilitated by viral genome replication (34). Also, it has been shown that expression of a viral activator (IVa2) of late gene transcription is induced as a result of titrating a limiting inhibitory factor during viral DNA synthesis (14).
Herpesviruses, with their enormous coding capacity in genomes larger than 100 kb, are characterized by highly ordered cascades of gene expression. Upon cell entry, immediate-early (α) genes are expressed, which in turn induces transcription of early (β) genes encoding proteins involved in genome replication, followed by late (γ) gene expression. While the regulation of immediate-early and early gene expression has been studied extensively in herpesviruses, very little is known about the mechanisms regulating late gene expression. Most of our current understanding is based on the studies of herpes simplex virus (HSV). DNA replication is required in cis for activity of late promoters (17, 24). While early viral gene promoters typically consists of distal regulatory sequences upstream of the TATA box, the critical elements of late promoters have been mapped to regions near the transcription start sites (8, 10, 12, 13, 16, 25, 32, 36). These cis-acting elements include a TATA box (8, 12, 16), an initiator element at the transcription start site (32, 36), and a downstream activation sequence in the 5′-untranslated region (10, 13, 25). Multiple HSV proteins (ICP4, ICP8, and ICP27) have been shown to be necessary for efficient expression of late genes (9, 20, 28). All three of these viral proteins have been demonstrated to interact with the general transcription machinery and could thus function to facilitate the assembly of transcription preinitiation complexes (4, 41). Nevertheless, since these viral proteins play multiple roles, including regulating early gene expression and genome replication, the specific molecular mechanisms by which they control late gene expression are not fully understood. Furthermore, despite the fact that a large amount of information has been gathered, how DNA replication is coupled to late gene expression remains a mystery.
The gamma subfamily of human herpesviruses, such as Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV), is linked to a number of malignancies and lymphoproliferative diseases. Therefore, it is important to study the molecular mechanisms of gammaherpesvirus replication. Although a significant number of genes are conserved among herpesviruses, there are also subfamily specific genes that have not been previously studied and may be essential for viral lytic replication. Because EBV and KSHV cannot efficiently undergo de novo lytic infection in cell culture, we chose to conduct the functional analysis of viral open reading frames (ORFs) in another member of the gamma subfamily, murine gammaherpesvirus 68 (MHV-68), which readily infects many cell lines and undergoes lytic replication. Previously, we identified MHV-68 ORF18 and ORF24 (2, 37) as necessary for the activation of late gene transcription. In the present study, we found that two mutant MHV-68 viruses lacking either ORF30 or ORF34 displayed a similar phenotype, in which the mutants were unable to express viral late gene transcripts despite having normal levels of viral DNA replication. Using a reporter assay, we show that the viral late gene promoters are not activated in the absence of ORF30 or ORF34. Using a chromatin immunoprecipitation (ChIP) assay, we found that the binding of RNA polymerase II (RNAPol II) to the viral late gene promoters during lytic infection is diminished without ORF30 or ORF34. These results strongly support the role of ORF30 and ORF34 in stimulating late gene expression by recruiting the transcription initiation complex.
MATERIALS AND METHODS
Cells.
All of the cells used in the study were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Flp-in-T-Rex-293 cell line (Invitrogen), which contains an integrated FLP recombination target site and constitutively expresses the Tet repressor, was used to generate the complementing cell line that expresses ORF30 (FT-30) or ORF34 (FT-34) upon induction by tetracycline (1 μg/ml). The established complementing cell lines were maintained in the presence of hygromycin (150 μg/ml) and blasticidin (7.5 μg/ml).
Plasmids.
To introduce mutations into MHV-68, a shuttle plasmid based on pGS284 (kindly provided by Greg Smith at Northwestern University) was constructed. The translational stop codons in the targeted ORF were introduced by PCR. The sequences upstream of the stop codons (A fragments) were amplified by primers of AF and AR, while the downstream sequences (B fragments) were amplified by primers of BF and BR using the wild-type (WT) MHV-68 virion DNA as the template. The primers used to introduce the stop codons for ORF30 are 30SAF (5′-TCT CCT CGA GCT GTG GCA AAT GAT CCC TCT-3′), 30SAR (5′-CAG GCC TTC AAT TAA TTA ACC GGC CTG ATC AGT AAC CAG ATT C-3′), 30SBF (5′-CCG GTT AAT TAA TTG AAG GCC TGA TCT CCT GGA TGT GGT GAA T-3′), and 30SBR (5′-TCT CGC TAG CAC ATG GGT GAT TCC CTG ACA-3′); for ORF34, the primers are 34SAF (5′-GGC AGA TCT TGG TAC AGA GTA CTC ACA TG-3′), 34SAR (5′-CCA TGG TCA ATT AAT TAA CCG GAA TTG CCA AAT CCA CAC ACC-3′), 34SBF (5′-CCG GTT AAT TAA TTG ACC ATG GAA TAT GAG CTG GAG CGT GCC-3′), and 34SBR (5′-TTA CCC GGG AAT GGG TCC ACA GGA TGA AG-3′). The A and B fragments were designed to have 20- to 25-bp overlapping sequences containing restriction enzyme sites for later screening purposes. In a subsequent PCR, the A and B fragments were used as templates and amplified by primers of AF and BR. The final PCR products were digested with appropriate enzymes and cloned into pGS284. To generate complementing cell lines, the protein expression plasmids of ORF30 (pFLAG30) and ORF34 (pFLAG34) were generated. The predicted coding regions were amplified by PCR using primers and cloned into a modified expression plasmid of the tagged protein. The reporter plasmids based on pGL3basic (Promega) were generated by inserting the 1-kb region upstream of the ATG translation initiation codon of ORF26 or ORF65 amplified by PCR (primers for the ORF26 putative promoter, 5′-CTC TGC TAG CGG CAT CAA CCT CTG TGT TTA TTG-3′ and 5′-CGG CCT CGA GGA TAA TTA ATA AAC TTC AGG TTC C-3′; primers for the ORF65 promoter, 5′-CTC TGC TAG CCA GGA TGG TTC TTC ATG AAT G-3′ and 5′-CGC TCT CGA GCA CCC TTA TTG GAA CAC TTT TGA C-3′). To construct reporter plasmids containing the origin of lytic replication (ori-Lyt) of MHV-68, the putative promoters together with the luciferase coding sequence and the polyadenylation site were PCR amplified from the pGL3basic-derived reporters and then cloned into a unique NaeI site of pMOΔ16, which includes a minimal 1.2 kb of MHV-68 ori-Lyt (5). The early gene ORF57 reporter construct contains a 565-bp promoter cloned into pGL2basic (a gift from Samuel Speck) (21) and the early gene M3 reporter construct contains a 600-bp promoter cloned into pGL3basic (23).
Construction of 30S and 34S viruses.
The recombinant viral MHV-68 bacterial artificial chromosome (BAC) plasmids (30S BAC and 34S BAC) containing triple stop codons in ORF30 or ORF34 were generated by a two-step allelic exchange described by Smith and Enquist (30). The incorporation of stop codons was determined by PCR and restriction enzyme digestion by examining the insertion of new sites engineered next to stop codons. The restriction patterns of BAC plasmids isolated from the positive colonies were verified by comparing them to those of WT BAC. The corresponding viruses, 30S and 34S, were reconstituted by transfecting FT-30 or FT-34 with the recombinant 30S BAC and 34S BAC plasmids using Lipofectamine 2000 (Invitrogen). The resultant viruses were propagated on complementing cells, and their titers [50% tissue culture infective dose(s) (TCID50)] were determined by limiting dilutions on complementing cells.
Analysis of DNA, protein, and RNA.
Total DNA was isolated from cells and analyzed by Southern blot as described previously (39). The viral copy numbers were determined by quantitative real-time PCR using primers for the ORF65 (M9) gene (29). Western blots were used for detecting viral proteins with rabbit polyclonal antibodies against ORF65, ORF26, or MHV-68-infected rabbit cells. The anti-ORF26 antibodies were generated in a similar manner as anti-ORF65 antibodies (38). Briefly, the coding sequence of ORF26 was cloned into a bacterial expression vector, and the recombinant His-tagged ORF26 was purified and injected into rabbits to induce antibody production. The anti-MHV-68 polyclonal antibodies were generated by injecting rabbits with detergent-treated lysates harvested from MHV-68-infected rabbit cells. To analyze viral transcripts, total RNA was extracted from cells by Tri-Reagent (Molecular Research Center) and subjected to Northern blot analysis as described previously (39). To generate the cDNA probes for membrane arrays of viral ORFs, total RNA was labeled by reverse transcription in the presence of [α-32P]dATP using a Strip-EZ RT kit (Ambion). The hybridization steps were carried out similarly to those of the Northern blots, and the quantification of signals was performed by using a Storm imaging system (Molecular Dynamics) (23).
Dual luciferase reporter assays.
The reporter plasmids were transfected into BHK-21 cells, together with an internal control plasmid, pRLCMV (Promega), which contains a Renilla luciferase under the cytomegalovirus enhancer and immediate-early promoter. At 24 h posttransfection, the cells were split and seeded into multiple wells for infection. Infection was carried out 24 h after seeding, and cell lysates were harvested at 24 h postinfection for the dual-luciferase reporter assay (Promega). Firefly luciferase activity was normalized against Renilla activity, and the fold of activation was calculated by comparing the normalized values of infection to those obtained from uninfected samples.
ChIP assays.
ChIP was performed by using a ChIP assay kit (Millipore) according to the manufacturer's instructions, with some modifications. Briefly, 106 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and glycine was added to 0.14 M. The cells were washed and collected in cold phosphate-buffered saline. Cells were centrifuged, and the pellet was washed once with Mg-NI buffer (15 mM Tris-HCl [7.5], 5 mM MgCl2, 60 mM KCl, 0.5 mM dithiothreitol, 15 mM NaCl, 300 mM sucrose), spun down, resuspended in Mg-NI-NP40 buffer (Mg-NI buffer plus 1% NP-40), and then incubated on ice for 10 min. The nuclei from the NP40-lysed cells were spun down and resuspended in Cal-NI buffer (15 mM Tris-HCl [7.5], 1 mM CaCl2, 60 mM KCl, 0.5 mM dithiothreitol, 15 mM NaCl, 300 mM sucrose) and spun down again, and the nucleus pellet was resuspended in Ca-NI buffer. S7 nuclease (Roche) was then added (12.5 μg), and the mixture was incubated on ice for 1 h. The sample was spun down, and the pellet was resuspended in sodium dodecyl sulfate lysis buffer (Millipore) and sonicated by using a Dismembrator 100 (Fisher) with a microtip for four cycles of 10 s on and 45 s off to shear DNA to a size range of 0.2 to 0.5 kb. The sonicated chromatin was spun down at 4°C, and the supernatant was collected for immunoprecipitation. The chromatin was diluted 10 times with ChIP dilution buffer (Millipore) and precleared for 1 h with protein A-agarose beads saturated with salmon sperm DNA (Millipore). Then, 2 μg of anti-RNAPol II antibody (sc-899X; Santa Cruz Biotechnology) or the control rabbit antibody was added, and the sample was mixed overnight at 4°C in an orbital mixer. The antibody complex was collected and incubated with protein A-agarose beads at 4°C in an orbital mixer for 1 h. The beads were then washed for 5 min on a rotating platform twice with each of the buffers (Millipore) in the following order: low-salt immune complex wash buffer, high-salt immune complex wash buffer, LiCl immune complex wash buffer, and TE buffer. The complex was eluted from the beads by incubating them with 250 μl of freshly prepared elution buffer (1% sodium dodecyl sulfate, 0.1 M NaHCO3) at room temperature for 15 min on a rotating platform. The beads were spun down, the supernatant was collected, and the elution was repeated one more time. The eluted complex was reverse cross-linked through incubation at 65°C overnight, and the DNA was recovered by a PureLink PCR purification kit (Invitrogen). For real-time PCR quantification, 1% of the recovered DNA was used in each triplicate with a Platinum SYBR Green qPCR SuperMix (Invitrogen). The primers were designed to amplify the promoter regions of ORF52 (forward [5′-CCT TTC TAC TTC CTT GAC CAT TTC-3′] and reverse [5′-AGT GCC CTG GAG ACA ACC-3′]), M7 (forward [5′-TGC TCG AGT GGA GTC ATA AAC GGT GCC AA] and reverse [5′-TAT TTG AAA CAA CAG AAA ACA C]), and GAPDH (forward [5′-TAC TAG CGG TTT TAC GGG CG] and reverse [5′-TCG AAC AGG AGG AGC AGA GAG CGA-3′]). The real-time PCR was run in an MJ Opticon. After denaturation at 95°C for 2 min, 40 PCR cycles of 15 s at 95°C, 15 s at 60°C, and 30 s at 72°C were carried out, and then a melting-temperature analysis was performed. For each run and set of primers, a standard containing known copies of viral DNA or cellular DNA was included to obtain the copy number in the ChIP samples.
RESULTS
Generation of null mutants of ORF30 and ORF34.
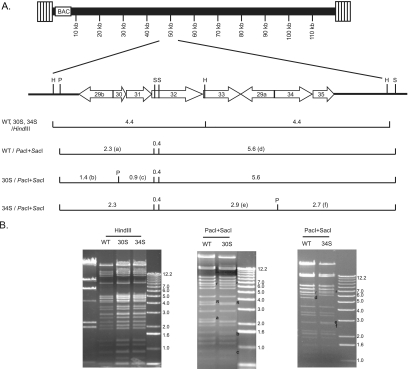
ORF34 is predicted to encode a gene product of 332 amino acids; it is one of the viral ORFs previously identified as essential for MHV-68 lytic replication by random transposon insertional mutagenesis on an infectious BAC (26, 31). ORF30, found in most gammaherpesviruses, encodes a small polypeptide of 80 amino acids, and its role in viral replication has not yet been determined. To investigate the functions of these two ORFs during viral infection, translational triple stop codons were introduced near the N terminus of the predicted coding sequence on the MHV-68 BAC by two-step recA-mediated homologous recombination (15, 30). The insertion of stop codons leads to the production of truncated gene products of 20 and 30 amino acids for ORF30 and ORF34, respectively. A novel PacI site was included in the triple stop codons for screening and verification purposes. The genomic structures and the relevant restriction enzymes sites of the resultant null mutants (30S and 34S), as well as the WT, are illustrated in Fig. 1A. The DNA restriction enzyme digestion patterns of HindIII and a combination of PacI and SacI were analyzed to confirm the integrity and correctness of the MHV-68 BACs. As predicted, in the PacI and SacI double digestion, a 2.3-kb DNA band in the WT BAC is replaced by two bands of 1.4 and 0.9 kb in the 30S BAC, and a 5.6-kb DNA band in the WT BAC is replaced by two bands of 2.9 and 2.7 kb in the 34S BAC (Fig. 1B).
FIG. 1.
Genomic structures of WT and mutant viral BACs. (A) Schematic illustrations of the BAC plasmids at the regions encoding ORF30 and ORF34. Triple stop codons and a PacI site (marked with a “P”) were introduced into ORF30 or ORF34. The SacI (S) and HindIII (H) sites are also marked. The restriction fragments differing between WT and 30S or 34S are indicated by letters in parentheses. (B) Restriction enzyme analysis of the BACs. WT, 30S, and 34S BACs were digested with either HindIII or a combination of SacI and PacI. The DNA fragments differing between WT and the two mutants are labeled with a, b, c, d, e, and f according to the illustration in panel A. The fragments labeled “R” contain 40-bp repeats, and those labeled with “r” contain 100-bp repeats. There are different numbers of repeats present on the WT and mutant BACs.
ORF30 and ORF34 are essential for productive viral infection.
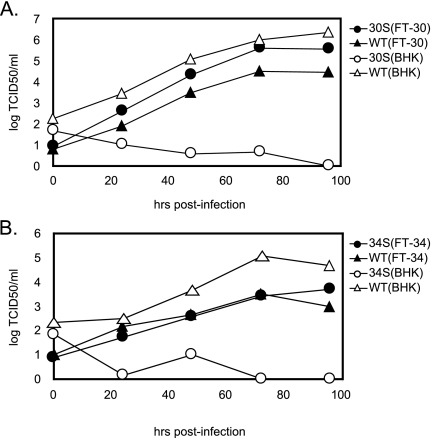
Unlike for the WT BAC, transfection of the 30S or 34S BAC plasmid into BHK-21 cells did not lead to productive viral replication. However, when provided with a protein expression plasmid containing an intact ORF30 or ORF34, the 30S and 34S mutants were capable of completing a productive life cycle (data not shown). Therefore, to rescue the 30S and 34S viruses, we transfected the mutant BAC plasmids individually into the corresponding complementing cell lines that express either ORF30 (FT-30) or ORF34 (FT-34). The viral stocks of 30S and 34S were then generated, and it was estimated that the frequency of a revertant virus regaining the WT ORF30 or ORF34 in the stocks was below the detection limit of 1 in 105 TCID50/0.1 ml. The viral growth of 30S (Fig. 2A) and 34S (Fig. 2B) was examined in BHK-21, as well as in the complementing cells. In BHK-21 cells, the mutant viruses were severely attenuated compared to the WT virus, supporting the indispensable roles of ORF30 and ORF34 during viral replication. No cytopathic effect was observed up to 14 days after infection of 30S or 34S virus, while BHK-21 cells infected with WT reached 100% cytopathic effect after 4 days. Conversely, in complementing FT-30 and FT-34 cells, 30S and 34S viruses were able to replicate comparably to WT. Taken together, the results clearly demonstrate that the defects of the mutant viruses are due to the lack of a functional ORF30 or ORF34.
FIG. 2.
Growth kinetics of WT and mutant viruses. (A) Replication of WT and 30S in BHK-21 and FT-30 cells. (B) Replication of WT and 34S in BHK-21 and FT-34 cells. The cells were infected at 0.01 TCID50 per cell of the indicated virus. The supernatants and cell lysates were harvested at multiple times after infection, and the virus titers were determined by limiting dilution on complementing FT-30 (A) or FT-34 (B) cells.
ORF30 and ORF34 are indispensable for viral DNA replication.
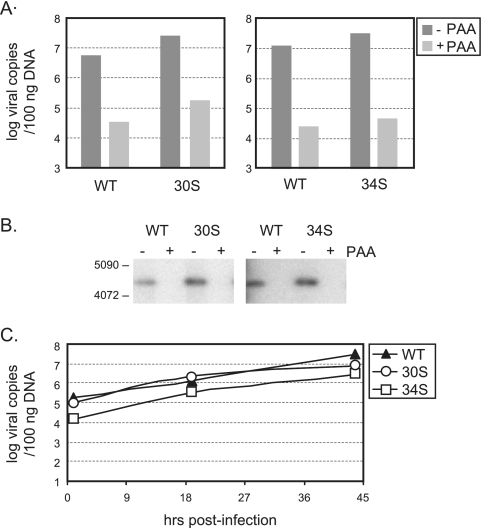
To determine which stage of viral replication requires ORF30 and ORF34, we examined the production of viral DNA. BHK-21 cells were infected with individual viruses at 2 TCID50 per cell, and the total amount of viral DNA in the infected cells at 24 h postinfection was quantified by real-time PCR. In comparison to that of WT infection, similar copy numbers of viral genomes were generated by infection of 30S or 34S (Fig. 3A). Treatment of a viral DNA polymerase inhibitor, phosphonoacetic acid (PAA), caused a 100-fold decrease in viral genome copy numbers for all infections, confirming that WT and mutant viruses produce similar levels of newly synthesized viral DNA. The real-time PCR result was further corroborated by Southern blot analysis, which showed a WT level of DNA replication by 30S and 34S viruses (Fig. 3B). In addition, we carried out a time course study for genome replication and measured the viral genome copy numbers at different times (1, 19, and 44 h after infection). The kinetics of viral genome amplification were similar between WT and mutant viruses (Fig. 3C). Together, the results indicate that ORF30 and ORF34 are not required for viral DNA replication.
FIG. 3.
Genome replication of WT and mutant viruses. (A) Quantitation of viral DNA copy numbers by real-time PCR. (B) Southern blot analysis of viral DNA. BHK-21 cells were infected at 2 TCID50 per cell without or with PAA (0.2 mg/ml) and harvested for DNA extraction at day 1 after infection. For the real-time PCR, 100 ng of total DNA was used. For the Southern blots, 1 μg of total DNA was digested with HindIII, and a 4.4-kb viral DNA fragment was detected with a probe derived from the region of ORF34. (C) Kinetics of DNA replication. Cells were infected at 2 TCID50 per cell. At 1, 19, and 44 h postinfection, total DNA was isolated, and viral DNA copy numbers were determined by real-time PCR.
The 30S and 34S viruses are deficient in the expression of late genes.
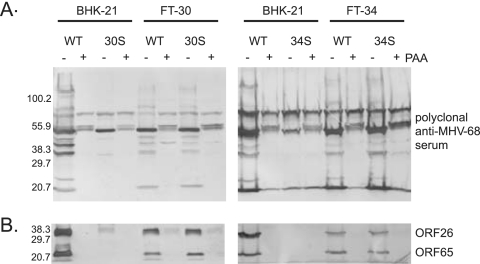
Since DNA replication of 30S and 34S viruses was not blocked, we then analyzed the expression of late viral gene products, which are defined by their dependence on viral DNA replication for expression. The polyclonal rabbit anti-MHV-68 antibodies detected multiple viral lytic proteins in WT-infected cells. Expression of a number of these lytic proteins was sensitive to PAA treatment, indicating that they are late gene products (Fig. 4A). These late viral proteins were absent from BHK-21 cells infected with 30S or 34S. We also examined the expression of two well characterized late genes, ORF26 and ORF65 (27, 29, 33-35), which encode viral capsid proteins and were readily detected in cells infected with the WT virus (Fig. 3B). However, both 30S and 34S viruses failed to produce detectable ORF26 and ORF65 gene products in BHK-21 cells. The expression of late gene products was restored in complementing cells (FT-30 and FT-34). Hence, the Western analyses demonstrate that the lack of intact ORF30 or ORF34 resulted in blocked expression of late gene products.
FIG. 4.
Expression of viral lytic proteins. Western blot analysis of viral proteins with the rabbit polyclonal antibodies against MHV-68-infected cell lysates (A) or against viral ORF26 and ORF65 (B). BHK-21, FT-30, and FT-34 cells were infected at 2 TCID50 per cell without or with PAA (0.2 mg/ml). The cell lysates were harvested at 2 days after infection for Western blots.
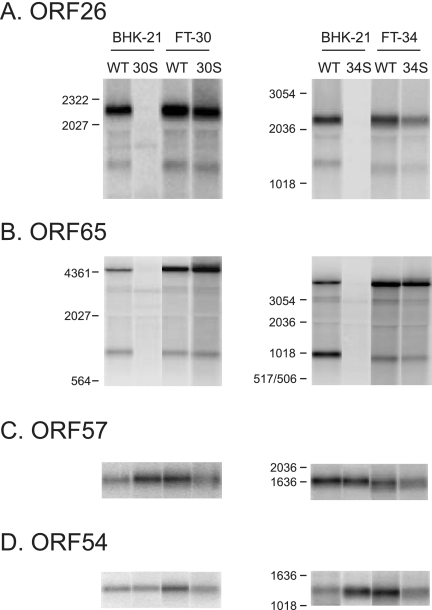
Since our data show that ORF30 and ORF34 are required for the synthesis of late viral proteins, we conducted Northern analyses to study whether late viral transcripts were also affected by the deficiency in ORF30 or ORF34. Transcription of ORF26 and ORF65 was readily detected in BHK-21 cells infected with the WT virus but not in those infected with either 30S or 34S (Fig. 5A and B). This defect is due to the lack of ORF30 or ORF34 because the expression of the capsid transcripts was recovered in complementing cells. We also examined the RNA levels of two early genes, ORF57 and ORF54 (1, 7, 22, 23), and found that the early gene transcripts were at comparable levels among cells infected with the WT, 30S, or 34S virus (Fig. 5C and D). The results indicate that ORF30 and ORF34 are required for the expression of the late gene transcripts but not for the early ones.
FIG. 5.
Expression of viral gene transcripts. Northern blot analysis of viral gene transcripts using gene-specific probes derived from the region of ORF26 (A), ORF65 (B), ORF57(C), or ORF54 (D). BHK-21, FT-30, and FT-34 cells were infected at 2 TCID50 per cell and harvested at day 2 postinfection for RNA extraction. The total RNA was then used for Northern blots.
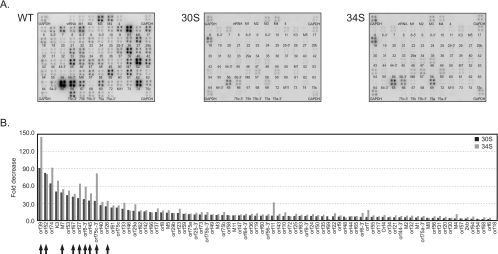
To examine transcription of other late genes, genome-wide analysis of viral gene expression was performed by using arrays containing DNA fragments derived from individual MHV-68 ORFs (23). Infection of the 30S or 34S virus showed a visible reduction of viral gene expression compared to that of the WT virus (Fig. 6A). Quantitation of the membranes revealed that the most affected ORFs (ORF8, ORF26, ORF27, ORF39, ORF45, M7, ORF52, ORF67, and ORF75c) are the genes encoding virion components (Fig. 6B, marked by arrows), which have previously been shown to be late genes (1, 7, 23). We did observe a small reduction in the expression of early genes, such as ORF57 and ORF54. This is likely due to some degrees of second-round infection for the WT at 2 days postinfection when total RNA was harvested for arrays, but not for replication-deficient 30S and 34S viruses.
FIG. 6.
Viral gene expression profiles of WT and mutant viruses. (A) MHV-68 membrane arrays probed with 32P-labeled cDNA generated from RNA of cells infected with WT, 30S, or 34S. (B) Quantitative comparisons of viral gene transcripts. BHK-21 cells were infected at 2 TCID50 per cell, and total RNA was harvested at 48 h after infection. The cDNA probe was generated by reverse transcription of RNA with oligo(dT) in the presence of [32P]dATP and was used for hybridization with membranes spotted with DNA fragments derived from viral ORFs (A). The signals of the spots were normalized against those of GAPDH at four corners of the membranes. The normalized values from the 30S array or 34S array were divided by the corresponding values from the WT array to derive the fold decrease (B). The arrows indicate examples of previously described late genes.
ORF30 and ORF34 play a critical role in the induction of late gene promoters.
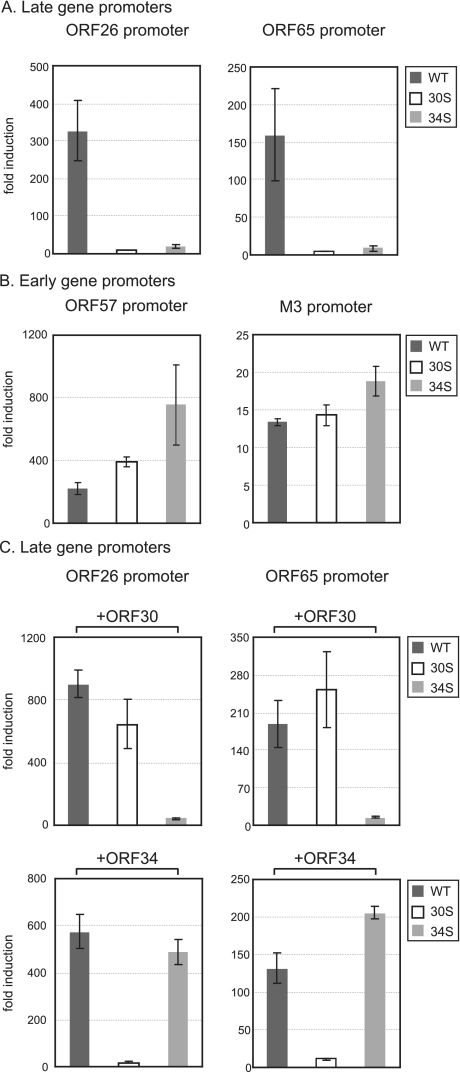
In order to further delineate the roles of ORF30 and ORF34 during transcription of late viral genes, we examined the activation of late promoters in a reporter system. We first cloned the 1-kb DNA sequence upstream of the translation start codon of ORF26 or ORF65 into a luciferase reporter plasmid (pGL3basic). Since late gene transcription depends upon DNA replication, we then moved the luciferase expression cassette driven by the putative late gene promoter into a plasmid containing the lytic replication origin (ori-Lyt) of MHV-68 (5), which allows for replication of the reporters upon infection. BHK-21 cells were transfected with the reporter constructs and then infected with individual viruses. At 24 h postinfection, the cells were harvested for measurement of luciferase activity. As shown in Fig. 7A, infection of WT activated the ORF26 promoter by 325-fold and the ORF65 promoter by 160-fold. However, these two late gene promoters were only minimally induced (<10% of the WT level) by either 30S or 34S virus (Fig. 7A). This is not a general defect of the mutant viruses in activating the viral gene promoters, since both 30S and 34S viruses were able to stimulate the two early gene promoters to a comparable level or to an even higher level than the WT virus (Fig. 7B). In addition, we also performed a rescue experiment by exogenously expressing ORF30 or ORF34 in cells. Clearly, ORF30 was able to support the 30S virus to activate the ORF26 and ORF65 promoters and, likewise, ORF34 could restore the ability of the 34S virus to induce the late gene promoters (Fig. 7C). Together, the reporter assays indicate that ORF30 and ORF34 are crucial for the virus to stimulate the late gene promoter activity.
FIG. 7.
Activation of viral promoters by WT and mutant viruses. The reporter activity of the late gene promoters (A and C) and the early gene promoters (B) without (A and B) or with (C) the exogenous expression of ORF30 or ORF34. BHK-21 cells were first transfected with individual firefly luciferase constructs driven by the promoters of ORF26 (late), ORF65 (late), ORF57 (early), and M3 (early). In all transfections, an internal control, a Renilla luciferase plasmid under the control of a constitutively active promoter, was included. In panel C, the protein expression plasmid of ORF30 or ORF34 was transfected as well. The transfected cells were then infected with individual viruses at 2 TCID50 per cell and harvested for luciferase assays at 24 h after infection. The firefly luciferase activity was normalized against that of Renilla. The fold induction by virus infection was derived by dividing the normalized values of the infected cells with those of the uninfected cells.
ORF30 and ORF34 are required for recruitment of RNAPol II to the late gene promoters.
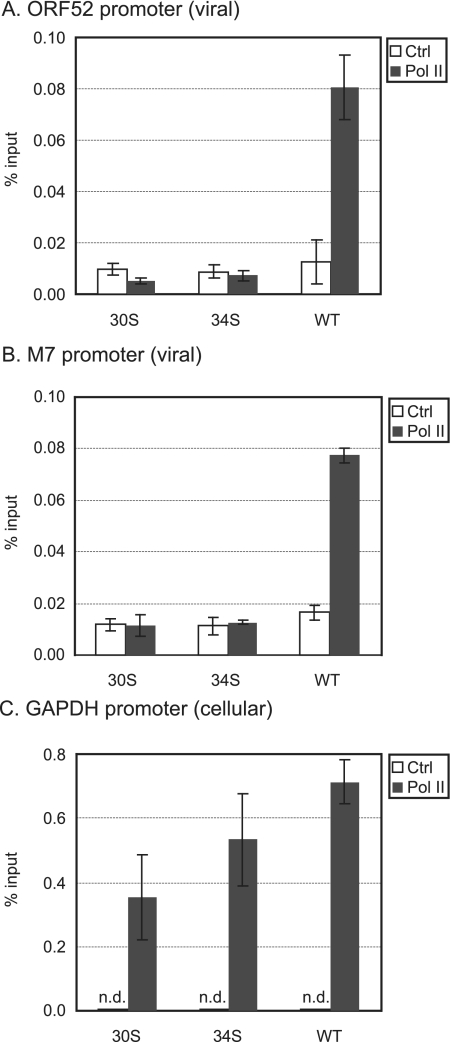
Binding of RNAPol II to promoters is a necessary step in the initiation of transcription. We therefore used ChIP assays to investigate whether the recruitment of RNAPol II to the late gene promoters was disrupted by the lack of ORF30 or ORF34. Chromatin from infected cells was immunoprecipitated with an anti-RNAPol II antibody or a control antibody. Real-time PCR was then carried out to quantify the amounts of the immunoprecipitated DNA with specific primers for the viral promoters of ORF52 and M7 or the cellular promoter of a housekeeping gene, GAPDH. We chose to examine the promoter regions of ORF52 and M7 because they are among the most highly expressed viral late genes (23), and their expression was greatly reduced in the absence of ORF30 or ORF34 (Fig. 6). As shown in Fig. 8, the anti-RNAPol II antibody significantly immunoprecipitated the late gene promoter regions in the WT-infected cells (∼6-fold enrichment for the ORF52 promoter region and ∼5-fold for the M7 promoter region). On the other hand, in either 30S- or 34S-infected cells, binding of RNAPol II to the viral late gene promoters was not detected. To confirm that ChIP was working properly among different samples, the cellular GAPDH promoter region was examined, and comparable amounts were present in the immunoprecipitated DNAs from WT-, 30S-, and 34S-infected cells (Fig. 8C). Therefore, the ChIP results indicate that ORF30 and ORF34 are essential for the virus to recruit RNAPol II to the late gene promoters.
FIG. 8.
Recruitment of RNAPol II to the late gene promoters. Quantitation of the immunoprecipitated DNA from the promoter regions of ORF52 (A), M7 (B), and GAPDH (C). 293T cells were infected with WT, 30S, or 34S and cross-linked at 18 h postinfection. ChIP assays with anti-RNAPol II antibody (indicated by Pol II) or a control rabbit antibody (Crtl) were performed. The immunoprecipitated DNA was quantitated by real-time PCR and expressed as the percentage of input DNA. The averages and standard deviations were obtained from triplicates. n.d., not detectable.
DISCUSSION
Here we studied the essential functions of ORF30 and ORF34 in MHV-68 and identified their roles at a critical point when the virus makes a progression from DNA replication to the expression of late genes. Without intact ORF30 and ORF34, viral replication is arrested after DNA synthesis, resulting in a failure to express late gene transcripts. Moreover, we showed that there was only minimal induction of late promoters by the 30S and 34S viruses. The data demonstrate that late gene expression depends not only on viral DNA replication but also on the virally encoded trans factors, ORF30 and ORF34. Unlike the viral proteins implicated thus far in regulating HSV-1 late genes, the gene products of ORF30 and ORF34 function specifically to stimulate the promoters of late genes, since their absence has little effect on either early gene expression or viral DNA replication. The homologues of MHV-68 ORF34 are found not only in other gammaherpesviruses (e.g., EBV BGLF3 and ORF34 of HVS and KSHV) but also in betaherpesviruses (e.g., HCMV UL95 and HSV-6 U67). Very little is known about the functions of these ORF34 homologues, except for the critical role of HCMV UL95 in viral replication (6, 40). The homologues of MHV-68 ORF30 are only found in other gammaherpesviruses (ORF30 of HVS and KSHV) but not in betaherpesviruses. Analysis of the coding sequences of ORF30 and ORF34 does not reveal any significant homology to cellular genes or recognizable domains, such as a DNA-binding motif. Neither protein appears to be specifically localized in the nucleus when individually expressed from a plasmid. Future experiments to explore interacting partners of ORF30 and ORF34 may provide clues to the molecular mechanisms by which these two viral ORFs activate late gene transcription.
In this report, we also found that the binding of RNAPol II to the late gene promoters depended upon ORF30 and ORF34. This result indicates a critical role of ORF30 and ORF34 in stimulating the assembly of the transcription complex at the late gene promoters. We are aware that during infection of the WT virus, the viral promoter region bound by the RNAPol II represents a small fraction of the input viral DNA in comparison to the fraction of the bound cellular promoter of a housekeeping gene, GAPDH. This may reflect the fact that only some of the viral DNA templates are transcriptionally active. We interpreted that by the time the WT-infected cells were harvested for ChIP assays, the virus had undergone DNA replication, late gene expression and formation of capsids, and thus the viral genomes were already packaged into capsids and had become inaccessible to transcription by RNAPol II. Nevertheless, there was no detectable binding of RNAPol II to the late gene promoters (ORF52 and M7 promoters) during infection of the 30S or 34S virus, despite the similar amounts of viral DNA that were in the chromatins used for immunoprecipitation. The data support the interpretation that the mutant viruses were not able to efficiently recruit the RNAPol II to the late gene promoters.
In animal cells, the formation of functional RNAPol II machinery on a core promoter is controlled by a variety of intricate mechanisms operating through vastly diverse cis-regulatory DNA sequences and protein factors. These proteins bind to promoters and enhancers, function to remodel chromatin structure, and mediate communication between sequence-specific transcriptional factors and the general transcription machinery. Based upon what is known about the mechanisms by which transcriptional cofactors regulate gene expression, we can envision several possible roles that ORF30 and ORF34 might play to activate late gene transcription. First, they might be required to create an accessible chromatin structure for the transcription machinery. For example, during lytic infection of HSV-1, histone modification associated with transcription activation has been detected at the viral promoters and coding regions (19, 27). Moreover, it has also been demonstrated that an HSV-encoded transcriptional activator, VP16, can recruit chromatin-remodeling complexes to target promoters (11). Further experiments to examine the chromatin structure of the viral genomes of 30S and 34S may shed light on this possibility. Second, ORF30 and ORF34 might function as coactivators to interface with the transcription machinery. The expression of late gene transcripts depends strictly upon viral DNA replication, indicating that a signal imparted from replication is relayed to the transcription apparatus. Because the mutant viruses lacking either ORF30 or ORF34 failed to recruit RNAPol II to the late promoters despite undergoing normal genome replication, it is possible that ORF30 and ORF34 are involved in the cooperation between the complexes that mediate viral DNA replication and gene transcription. Third, like many transcription activators, the viral proteins might recognize and bind to specific DNA sequences. Although we were not able to find any apparent DNA-binding domain in ORF30 or ORF34, there is no experimental data to formally exclude this possibility. We have some evidence that, similar to those of other herpesviruses, the late gene promoters of MHV-68 are very simple and only contain a core element without any requirement for upstream or downstream sequences. Therefore, it would seem unlikely that ORF30 and ORF34 function as sequence-specific DNA-binding activators.
With an amenable genetic system and robust replication, MHV-68 provides unique advantages for studying aspects of lytic replication of gammaherpesviruses. Our previous and current studies have used this system to identify four previously uncharacterized viral proteins—ORF18 (2), ORF24 (37), ORF30, and ORF34—as critical regulators of late gene transcription. The viral late genes, most of which encode structural proteins, are usually expressed at high levels to produce a large amount of infectious virions. Therefore, it is thought that controlling this class of genes so that they are expressed only after DNA replication is most effective for virus production. However, much work still needs to be done to understand the mechanisms by which late gene expression is controlled. Further study of these four newly identified late gene regulators may help to elucidate these mechanisms.
Acknowledgments
We thank Helen Brown, Yuri Shindo, Eric Bortz, Victoria Bender, and Joyce Wu for editing the manuscript and Vaithilingaraja Arumugaswami and Seungmin Hwang for their helpful comments. The shuttle plasmid used to construct the mutant viruses was provided by Greg Smith at Northwestern University, and the ORF57 promoter construct was provided by Samuel Speck at Emory University.
This study was partially supported by NIH grants DE15752, ED14153, and CA91791 and the Stop Cancer Foundation. T.-T.W. was supported by the Leukemia and Lymphoma Society and NIH grant DE18337.
Footnotes
Published ahead of print on 17 December 2008.
REFERENCES
- 1.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 766244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugaswami, V., T. T. Wu, D. Martinez-Guzman, Q. Jia, H. Deng, N. Reyes, and R. Sun. 2006. ORF18 is a transfactor that is essential for late gene transcription of a gammaherpesvirus. J. Virol. 809730-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady, J., J. B. Bolen, M. Radonovich, N. Salzman, and G. Khoury. 1984. Stimulation of simian virus 40 late gene expression by simian virus 40 tumor antigen. Proc. Natl. Acad. Sci. USA 812040-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrozza, M. J., and N. A. DeLuca. 1996. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol. Cell. Biol. 163085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng, H., J. T. Chu, N. H. Park, and R. Sun. 2004. Identification of cis sequences required for lytic DNA replication and packaging of murine gammaherpesvirus 68. J. Virol. 789123-9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 10014223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 8499-109. [DOI] [PubMed] [Google Scholar]
- 8.Flanagan, W. M., A. G. Papavassiliou, M. Rice, L. B. Hecht, S. Silverstein, and E. K. Wagner. 1991. Analysis of the herpes simplex virus type 1 promoter controlling the expression of UL38, a true late gene involved in capsid assembly. J. Virol. 65769-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, M., and D. M. Knipe. 1991. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J. Virol. 652666-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzowski, J. F., and E. K. Wagner. 1993. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J. Virol. 675098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera, F. J., and S. J. Triezenberg. 2004. VP16-dependent association of chromatin-modifying coactivators and under-representation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 789689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homa, F. L., J. C. Glorioso, and M. Levine. 1988. A specific 15-bp TATA box promoter element is required for expression of a herpes simplex virus type 1 late gene. Genes Dev. 240-53. [DOI] [PubMed] [Google Scholar]
- 13.Homa, F. L., T. M. Otal, J. C. Glorioso, and M. Levine. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 63652-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iftode, C., and S. J. Flint. 2004. Viral DNA synthesis-dependent titration of a cellular repressor activates transcription of the human adenovirus type 2 IVa2 gene. Proc. Natl. Acad. Sci. USA 10117831-17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia, Q., T. T. Wu, H. I. Liao, V. Chernishof, and R. Sun. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J. Virol. 786610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, P. A., and R. D. Everett. 1986. The control of herpes simplex virus type-1 late gene transcription: a “TATA-box”/cap site region is sufficient for fully efficient regulated activity. Nucleic Acids Res. 148247-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, P. A., and R. D. Everett. 1986. DNA replication is required for abundant expression of a plasmid-borne late US11 gene of herpes simplex virus type 1. Nucleic Acids Res. 143609-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller, J. M., and J. C. Alwine. 1984. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell 36381-389. [DOI] [PubMed] [Google Scholar]
- 19.Kent, J. R., P. Y. Zeng, D. Atanasiu, J. Gardner, N. W. Fraser, and S. L. Berger. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 7810178-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, D. B., S. Zabierowski, and N. A. DeLuca. 2002. The initiator element in a herpes simplex virus type 1 late-gene promoter enhances activation by ICP4, resulting in abundant late-gene expression. J. Virol. 761548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, S., I. V. Pavlova, H. W. Virgin IV, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 742029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackett, M., J. P. Stewart, S. de V. Pepper, M. Chee, S. Efstathiou, A. A. Nash, and J. R. Arrand. 1997. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J. Gen. Virol. 781425-1433. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 7710488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavromara-Nazos, P., and B. Roizman. 1987. Activation of herpes simplex virus 1 gamma 2 genes by viral DNA replication. Virology 161593-598. [DOI] [PubMed] [Google Scholar]
- 25.Mavromara-Nazos, P., and B. Roizman. 1989. Delineation of regulatory domains of early (beta) and late (gamma 2) genes by construction of chimeric genes expressed in herpes simplex virus 1 genomes. Proc. Natl. Acad. Sci. USA 864071-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moorman, N. J., C. Y. Lin, and S. H. Speck. 2004. Identification of candidate gammaherpesvirus 68 genes required for virus replication by signature-tagged transposon mutagenesis. J. Virol. 7810282-10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh, J., and N. W. Fraser. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 823530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 641704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickabaugh, T. M., H. J. Brown, D. Martinez-Guzman, T. T. Wu, L. Tong, F. Yu, S. Cole, and R. Sun. 2004. Generation of a latency-deficient gammaherpesvirus that is protective against secondary infection. J. Virol. 789215-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 736405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song, M. J., S. Hwang, W. H. Wong, T. T. Wu, S. Lee, H. I. Liao, and R. Sun. 2005. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc. Natl. Acad. Sci. USA 1023805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steffy, K. R., and J. P. Weir. 1991. Mutational analysis of two herpes simplex virus type 1 late promoters. J. Virol. 656454-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, G. P., and M. B. Mathews. 1980. DNA replication and the early to late transition in adenovirus infection. Cell 22523-533. [DOI] [PubMed] [Google Scholar]
- 34.Toth, M., W. Doerfler, and T. Shenk. 1992. Adenovirus DNA replication facilitates binding of the MLTF/USF transcription factor to the viral major late promoter within infected cells. Nucleic Acids Res. 205143-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiley, S. R., R. J. Kraus, F. Zuo, E. E. Murray, K. Loritz, and J. E. Mertz. 1993. SV40 early-to-late switch involves titration of cellular transcriptional repressors. Genes Dev. 72206-2219. [DOI] [PubMed] [Google Scholar]
- 36.Woerner, A. M., and J. P. Weir. 1998. Characterization of the initiator and downstream promoter elements of herpes simplex virus 1 late genes. Virology 249219-230. [DOI] [PubMed] [Google Scholar]
- 37.Wong, E., T. T. Wu, N. Reyes, H. Deng, and R. Sun. 2007. Murine gammaherpesvirus 68 open reading frame 24 is required for late gene expression after DNA replication. J. Virol. 816761-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 759262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 743659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 10012396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, C., and D. M. Knipe. 2002. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J. Virol. 765893-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo, F., and J. E. Mertz. 1995. Simian virus 40 late gene expression is regulated by members of the steroid/thyroid hormone receptor superfamily. Proc. Natl. Acad. Sci. USA 928586-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]