Abstract
The secreted cytokine alpha/beta interferon (IFN-α/β) binds its receptor to activate the Jak-STAT signal transduction pathway, leading to formation of the heterotrimeric IFN-stimulated gene factor 3 (ISGF3) transcription complex for induction of IFN-stimulated genes (ISGs) and establishment of an antiviral state. Many viruses have evolved countermeasures to inhibit the IFN pathway, thereby subverting the innate antiviral response. Here, we demonstrate that the mildly myocarditic reovirus type 1 Lang (T1L), but not the nonmyocarditic reovirus type 3 Dearing, represses IFN induction of a subset of ISGs and that this repressor function segregates with the T1L M1 gene. Concordantly, the T1L M1 gene product, μ2, dramatically inhibits IFN-β-induced reporter gene expression. Surprisingly, T1L infection does not degrade components of the ISGF3 complex or interfere with STAT1 or STAT2 nuclear translocation as has been observed for other viruses. Instead, infection with T1L or reassortant or recombinant viruses containing the T1L M1 gene results in accumulation of interferon regulatory factor 9 (IRF9) in the nucleus. This effect has not been previously described for any virus and suggests that μ2 modulates IRF9 interactions with STATs for both ISGF3 function and nuclear export. The M1 gene is a determinant of virus strain-specific differences in the IFN response, which are linked to virus strain-specific differences in induction of murine myocarditis. We find that virus-induced myocarditis is associated with repression of IFN function, providing new insights into the pathophysiology of this disease. Together, these data provide the first report of an increase in IRF9 nuclear accumulation associated with viral subversion of the IFN response and couple virus strain-specific differences in IFN antagonism to the pathogenesis of viral myocarditis.
The type I interferon (alpha/beta interferon [IFN-α/β]) response is one of the earliest host defenses against infection by a wide array of pathogenic viruses. Viral infection stimulates synthesis and secretion of IFN-α/β, and receptor binding activates the Janus kinases (JAK), JAK1 and Tyk2, to phosphorylate signal transducers and activators of transcription (STATs), STAT1 (STAT1α and STAT1β) and STAT2 (22, 48, 58, 66). Phosphorylated STAT1 and STAT2 form heterodimers and associate with p48/interferon regulatory factor 9 (IRF9), forming the heterotrimeric transcription factor complex IFN-stimulated gene factor 3 (ISGF3) (17, 72). ISGF3 complexes translocate to the nucleus and bind to specific regulatory DNA sequences called IFN-stimulated response elements (ISREs). ISGF3 binding to ISREs initiates the transcription of many IFN-stimulated genes (ISGs), including ISG56 (IFIT1) and transcription factor IRF7, leading to the establishment of an antiviral state (9). IRF7 forms homo- or heterodimers to further stimulate transcription of type I IFN genes, thereby amplifying the IFN response in a positive feedback loop (39, 56).
To circumvent this innate protective response, many viruses have evolved mechanisms to subvert the IFN-α/β pathway. These viral antagonistic strategies can be grouped into two categories: (i) suppression of IFN induction, where viruses actively block the host cell sensor machinery or the downstream signaling molecules (e.g., IRF3) preventing initiation of IFN transcription (4, 15, 38, 68), or (ii) suppression of IFN signaling, where viruses inhibit signaling events at or following activation of the IFN receptor complex to prevent establishment of an antiviral state. These mechanisms for suppression of IFN signaling include modulation of JAK tyrosine phosphorylation activity (5, 33), degradation or modulation of nuclear translocation of the ISGF3 complex (45, 51, 54), and interference with ISG promoter activity (51).
Mammalian orthoreoviruses (reoviruses), members of the Reoviridae family, form nonenveloped particles that contain a genome of 10 segments of double-stranded RNA (dsRNA) (57). Virtually all mammals, including humans, serve as hosts for reovirus infection (73). However, reovirus causes disease primarily in the very young (73). Newborn mice infected with reovirus sustain injury to a variety of organs, including the brain, heart, and liver (73). The IFN response plays an important role in reovirus-induced myocarditis in mice. Nonmyocarditic reovirus strains cause myocarditis in mice depleted of IFN-α/β (62). Accordingly, nonmyocarditic reovirus strains induce more IFN-β and are more sensitive to its antiviral effects than myocarditic reovirus strains in primary cardiac myocyte cultures (62). It is not known whether reovirus inhibits the IFN-α/β signaling pathway or whether this repression is associated with induction of myocarditis.
In this study, we found that reovirus represses the IFN-α/β signaling pathway. This repression is virus strain specific and is associated with virus strain-specific differences in the induction of myocarditis. IFN repression is mediated by the reovirus M1 gene, which was previously identified as a determinant of reovirus induction of and sensitivity to IFN-α/β (62). The M1 gene product, μ2, of strain type 1 Lang (T1L) was found to cause nuclear accumulation of IRF9, a novel effect associated with suppression of IFN signaling. Together, these data constitute the first report of an increase in IRF9 nuclear accumulation associated with viral subversion of the IFN response and provide evidence that virus strain-specific differences in IFN antagonism are a determinant of disease.
MATERIALS AND METHODS
Cells and viruses.
Mouse L929 cells were maintained in minimal essential medium (MEM) (SAFC Biosciences, Denver, PA) supplemented to contain 5% fetal calf serum (FCS) (Atlanta Biologicals, Atlanta, GA) and 2 mM l-glutamine (Mediatech, Inc., Herndon, VA). Human HEK293 cells (ATTC CRL-1573) were maintained in Dulbecco's modified Eagle medium (Gibco BRL, Gaithersburg, MD) supplemented to contain 10% FCS and 10 μg of gentamicin (Sigma Co.) per ml. Vero cells (ATCC CCL-81) were maintained in Dulbecco's modified Eagle medium supplemented to contain 10% FCS and 10 μg of gentamicin per ml. “Reassortant viruses” are viruses with gene segments derived from two different parental strains during a mixed infection. “Recombinant viruses” are viruses generated from plasmids by reverse genetics (30). The efficiently myocarditic reovirus reassortant 8B was derived from a mouse coinfected with nonmyocarditic reovirus type 3 Dearing (T3D) and mildly myocarditic reovirus T1L (61). 8B was used to generate the EW and DB series of reovirus reassortants (61). Other reovirus reassortants (E3, 3HA1, and 1HA3) were derived from T1L and T3D (12). The recombinant strain viruses were derived from plasmids expressing T1L and T3D genes (30; and T. Kobayashi and T. D. Dermody, unpublished data). All reoviruses were plaque purified and amplified in L929 cells, purified on a CsCl gradient (64), and stored in diluted aliquots at −80°C.
Plasmids.
The M1 and S2 genes from T1L were subcloned by PCR using PfuUltra II Fusion HS DNA polymerase (Stratagene, La Jolla, CA) and the plasmids pT7-M1-T1L and pT7-S2-T1L, respectively. The forward primer for the M1 gene of T1L (5′-CTCGAGGCCACCATGGCTTACATCGCAGT T-3′) and the reverse primer for the M1 gene of T1L (5′-CCCGGGTCACGCCAAGTCAGATCG-3′) added XhoI and SmaI sites, respectively. The forward primer for the S2 gene of T1L (5′-CTCGAGGCCACCATGGCTCGCGCTGCGTTC-3′) and the reverse primer for the S2 gene of T1L (5′-CCCGGGTCACCATTCGTCGCACTT-3′) added XhoI and SmaI sites, respectively. After gel purification using the QIAquick gel extraction kit (Qiagen, Inc., Valencia, CA), the purified PCR products were ligated into the XhoI and SmaI sites of the sequencing vector pSC-B (Stratagene), yielding pSC-B-M1-T1L or pSC-B-S2-T1L. The M1 and S2 genes of T1L were excised from pSC-B-M1-T1L and pSC-B-S2-T1L, respectively, and used to generate pCAGGs-M1-T1L and pCAGGs-S2-T1L, encoding the T1L M1 and S2 gene products, respectively. The inserted T1L M1 and S2 genes were confirmed by sequence analysis. To generate pCAGGs-M1-T3D and pCAGGs-S2-T3D, encoding the T3D M1 and S2 gene products, respectively, viral cDNA fragments were amplified by PCR using pT7-M1-T3D and pT7-S2-T3D (30) as templates, respectively. Amplified fragments were cloned into the XhoI-KpnI site of the multiple-cloning site of pCAGGs (43). The plasmid pCAGGs-ORF6-GFP was a gift from Ralph Baric (University of North Carolina, Chapel Hill, NC) and contains green fluorescent protein (GFP)-tagged severe acute respiratory syndrome (SARS) coronavirus open reading frame 6 (ORF6) inserted into pCAGGs. The ISRE-Luc reporter construct (Stratagene) encodes five tandem copies of the ISG54 ISRE upstream of firefly luciferase. phRL-TK (Promega, Madison, WI) carries a synthetic Renilla luciferase gene under the control of a herpes simplex virus thymidine kinase (TK) promoter.
Transfections and luciferase assays.
HEK293 cells were plated in 48-well clusters at 6 × 104 cells per well and allowed to adhere for 1 day prior to transfection. Using Fugene 6 (Roche) at 3 μl per reaction, cells were transfected with four plasmids: 0.02 μg of pISRE-Luc, 0.001 μg of phRL-TK, and 0.5 μg each of two pCAGGS constructs. After 24 h of incubation, the transfection medium was replaced with fresh medium (“mock treated”) or medium supplemented with 1,000 U of recombinant human IFN-β per ml (Calbiochem no. 407318). Cells were harvested 6 h later in luciferase assay lysis buffer, and lysates were tested for luciferase activity using a dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luciferase activity for each sample was normalized to the internal Renilla luciferase control, and the average of results for duplicate samples was divided by that for mock-treated samples, to express the data as average fold induction ± standard deviation.
Infections for protein harvest and immunoblot analysis.
L929 cells were plated in 24-well clusters at 5 × 105 cells per well. At 2 hours postplating, cells were washed twice with supplemented MEM and infected with reovirus T1L or T3D at a multiplicity of infection (MOI) of 25 PFU per cell in 700 μl of supplemented MEM. After incubation at 37°C for 1 h, 1 ml of supplemented MEM was added, and the cultures were harvested at the indicated times. “Mock-infected” cultures received medium only, were washed with supplemented MEM, and were harvested immediately after addition of 1 ml of supplemented MEM. Total cellular protein extracts were prepared using radioimmunoprecipitation assay lysis buffer (50 mM Tris HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA) containing a cocktail of protease and phosphatase inhibitors (Sigma Co., no. P8340 and P2850) by incubating on ice for 30 min and centrifuging at 10,000 × g for 10 min to remove cellular debris. Cytoplasmic and nuclear extracts were obtained using an NE-PER kit (Pierce, Rockford, IL) according to the manufacturer's instructions. Protein content was determined by a bicinchoninic acid protein assay (Pierce), and 20 μg of protein from each lysate was boiled for 5 min in 1× Laemmli sample buffer and subjected to 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis as described previously (76). Immunoblots were exposed to film and converted to digital format using an HP Scanjet G4050. Quantification of the digitized bands was facilitated using UN-SCAN-IT software (Silk Scientific, Orem, UT).
Antibodies for immunoblotting.
Primary antibodies were purchased from the following suppliers and used at the indicated final dilutions: mouse monoclonal anti-STAT1α/β (1:1,000) (no. 610186; BD Biosciences, San Jose, CA), rabbit polyclonal anti-STAT2 (1:1,000) (no. 07-140; Millipore Corp.), rabbit polyclonal anti-IRF9 (1:100) (sc-10793; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and goat polyclonal anti-actin conjugated to horseradish peroxidase (1:500) (sc-1615-HRP; Santa Cruz Biotechnology, Inc.). Rabbit polyclonal anti-reovirus μ2 (1:1,000 dilution) was kindly provided by Earl Brown (University of Ottawa, Canada).
Quantitative reverse transcription-PCR (qRT-PCR).
L929 cells were plated in 24-well clusters at 5 × 105 cells per well. At 2 hours postplating, cells were washed twice with supplemented MEM and infected with reovirus T1L or T3D at an MOI of 25 PFU per cell in 700 μl of supplemented MEM. After incubation at 37°C for 1 h, 1 ml of supplemented MEM was added. After 20 h of incubation, cultures were mock treated or treated with mouse IFN-α/β (Access Biomedical, San Diego, CA) at the indicated doses. At 5 h posttreatment, total RNA was harvested using an RNeasy kit (Qiagen, Inc., Valencia, CA), and contaminating genomic DNA was removed using RNase-free DNase I (Qiagen, Inc.). RNA was converted to cDNA by reverse transcription and amplified by quantitative PCR to determine the relative abundances of specific mRNA sequences by comparison to a standard curve generated from serial dilutions of a DNA standard, as described previously (76). Copy numbers for genes of interest were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression.
Indirect immunofluorescence.
Vero cells were plated in Lab-Tek II chamber slides (Nalge Nunc International Corp., Naperville, IL) at 5 × 104 cells per well. After incubation overnight, cells were mock treated or infected with T1L or T3D at an MOI of 25 PFU per cell. After 20 h of incubation, cultures were mock-treated or treated with 1,000 U of recombinant human IFN-β per ml for 30 min. For visualization of STAT1, cells were fixed with 100% ice-cold methanol for 10 min. After three washes with phosphate-buffered saline (PBS), cells were blocked at 37°C for 15 min in 5% goat serum (Sigma Co.) diluted in PBS. All primary and secondary antibodies were diluted in PBS containing 0.01% immunoglobulin G (IgG)-free, protease-free bovine serum albumin (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). After blocking, samples were incubated at room temperature for 1 h with a mix of mouse monoclonal anti-STAT1α/β (1:1,000; BD Biosciences) and rabbit anti-T1L and rabbit anti-T3D antisera (each at 1:5,000) (B. Sherry, unpublished data). After three washes with PBS, samples were incubated with a combination of Alexa 488-conjugated goat anti-mouse IgG (1:1,000; Invitrogen) and Alexa 594-conjugated goat anti-rabbit IgG (1:1,000; Invitrogen) secondary antibodies at room temperature for 30 min. For detection of STAT2, cells were fixed in 4% paraformaldehyde in PBS at room temperature for 10 min, and after removal of the paraformaldehyde, cells were permeabilized with ice-cold acetone-methanol (1:1) at −20°C for 30 min. After five washes with PBS, samples were blocked with 1% bovine serum albumin in PBS at 37°C for 15 min. Samples were incubated with a mix of rabbit anti-STAT2 (1:50) (sc-476; Santa Cruz Biotechnology, Inc.) and mouse monoclonal anti-reovirus σ1 (5C6) (74) and mouse monoclonal anti-reovirus σ3 (4F2) (70) (each at 1:1,000) at room temperature for 1 h. After three washes with PBS, cells were incubated with a combination of Alexa 488-conjugated goat anti-rabbit IgG (1:1,000) (Invitrogen) and Alexa 594-conjugated goat anti-mouse IgG (1:1,000) (Invitrogen) secondary antibodies at room temperature for 30 min. Cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) at room temperature for 10 min to visualize nuclei. Following a final wash with PBS, coverslips were mounted on slides using Prolong Gold reagent (Invitrogen). Images were obtained at a magnification of ×100 under oil immersion using a Nikon TE-200 inverted epifluorescence microscope. Representative fields of view illustrating infected and uninfected cells in the same field are shown.
Statistical analysis.
A Student two-sample t test (pooled variance) or a nonparametric Kruskal-Wallis analysis was applied (Systat 9.0). Results were considered significant at a P value of <0.05.
RESULTS
T1L represses IFN induction of a subset of ISGs.
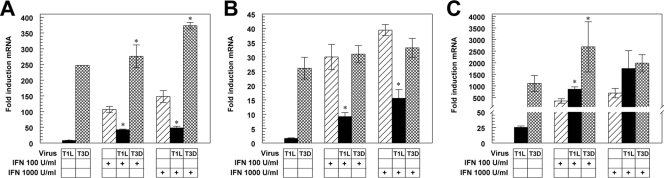
To determine whether reovirus inhibits IFN signaling, we investigated the effects of reovirus infection on IFN induction of three ISGs. L929 cells were infected with either T1L (a mildly myocarditic reovirus) or T3D (a nonmyocarditic reovirus). At 20 h postinfection, cultures were treated with IFN-α/β or buffer for 5 h, and mRNAs encoding ISGs IRF7, ISG56, and STAT1 were quantified by qRT-PCR (Fig. 1). T1L induced minimal IFN, while T3D induced high IFN levels (data not shown), as reported previously (62). As anticipated, T1L induced IRF7 and STAT1 mRNAs poorly, while T3D induced robust expression of these transcripts (Fig. 1A and B). Both strains induced ISG56, which also can be directly induced by IRF3 (19), providing a potential mechanism for increased T1L induction of this ISG (Fig. 1C) relative to the other ISGs tested and consistent with reovirus induction of this ISG in cells lacking the IFN-α/β receptor (67). IFN induced the transcription of all three genes and, as expected, induction was unchanged or enhanced when cells were infected with T3D before IFN treatment. In sharp contrast, IFN induction of IRF7 (Fig. 1A) and STAT1 (Fig. 1B) was dramatically inhibited in T1L-infected cells. However, IFN induction of ISG56 was not suppressed by T1L (Fig. 1C), consistent with an ISGF3-independent mechanism for IFN induction of this ISG. In addition, unlike STAT1 and IRF7, ISG56 contains two ISRE sequences, both of which contribute to ISG56 expression (75), suggesting that T1L is incapable of inhibiting activation of both ISRE sites simultaneously. Thus, T1L represses IFN signaling, although repression is restricted to a subset of ISGs.
FIG. 1.
T1L represses IFN induction of IRF7 and STAT1 but not ISG56. L929 cells were infected with either T1L or T3D at an MOI of 25 PFU per cell for 20 h and either mock treated or treated with IFN-α/β at the indicated dose for 5 h. RNA was quantified by qRT-PCR, and copy number was normalized to GAPDH. Fold inductions of IRF7 (A), STAT1 (B), and ISG56 (C) mRNAs are expressed relative to uninfected mock-treated cultures. Results are presented as the means of results for duplicate samples ± standard deviations from a representative of at least two independent experiments. Asterisks indicate significant differences from IFN treatment alone (Student's t test, P < 0.05).
Identification of the M1 gene of T1L as an antagonist of IFN signaling.
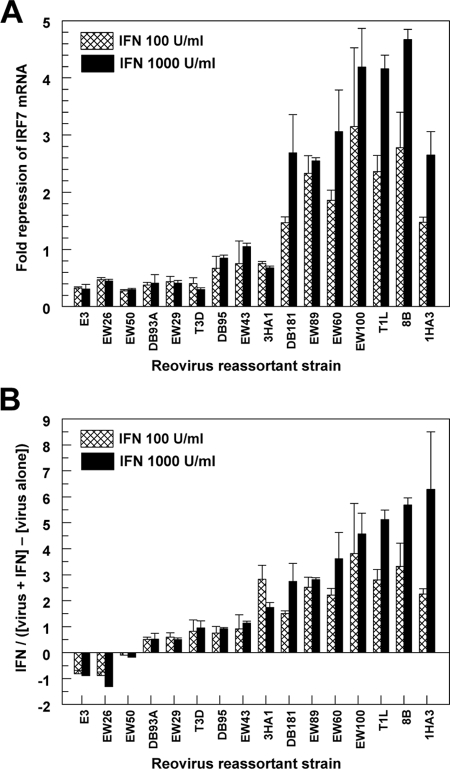
We next used reovirus reassortants containing a mixture of genes derived from T1L and T3D to identify the viral genes associated with T1L repression of IFN signaling. L929 cells were infected with a panel of reovirus reassortants for 20 h and treated with IFN-α/β for 5 h. IRF7 mRNA in the infected cultures was quantified by qRT-PCR (Fig. 2). Viral fold repression was calculated by dividing IFN induction of IRF7 mRNA in uninfected cultures by that in infected cultures (Fig. 2A). Accordingly, higher values represent viral repression of IFN-induced IRF7 mRNA. At a dose of 1,000 U/ml of IFN-α/β, viruses that repress IFN-induced IRF7 mRNA clearly segregated from those lacking this activity. A similar segregation occurred at a dose of 100 U/ml of IFN-α/β. However, some of these viruses induce IFN-β (62), and therefore these viruses would be anticipated to induce IRF7 through the native IFN response prior to addition of exogenous IFN-α/β. This extraneous IRF7 was eliminated as a confounding variable by subtracting viral induction of IRF7 in the absence of IFN-α/β treatment from viral induction of IRF7 in the presence of IFN-α/β before comparing to induction by IFN-α/β alone (Fig. 2B). A negative value indicates that the virus induced more IRF7 than IFN-α/β alone. At both doses of IFN-α/β used, the reassortant viruses distributed along a continuum of repression. However, a trend similar to that shown in Fig. 2A was maintained. Nonparametric Kruskal-Wallis analyses demonstrated that the M1 and S2 genes of T1L segregated with repression of the IFN signaling pathway using the calculation in either Fig. 2A or B (Table 1). However, only a single virus (EW43) segregated the T1L S2 gene from the T1L M1 gene. This virus did not repress IFN signaling (Fig. 2A and B), suggesting that the statistical association of the T1L S2 gene with repression of IFN signaling might reflect cosegregation with the T1L M1 gene rather than actual S2 gene product function.
FIG. 2.
Analysis of reovirus reassortants for repression of IFN-induced IRF7 mRNA. L929 cells were infected with the indicated reovirus strain at an MOI of 25 PFU per cell. At 20 h postinfection, cells were either mock treated or treated with IFN-α/β at the indicated dose for 5 h. RNA was quantified by qRT-PCR, and copy number was normalized to GAPDH. (A) Results are expressed as fold repression according to the following formula: IFN fold induction of IRF7 in mock-infected cultures divided by IFN fold induction of IRF7 in virus-infected cultures. (B) Viral direct induction of IRF7 is removed from the equation used for panel A according to the following formula: IFN fold induction of IRF7 in mock-infected cultures divided by (IFN fold induction of IRF7 in virus-infected cultures minus virus induction of IRF7). Results are presented as the means from duplicate samples + standard deviations.
TABLE 1.
Repression of IFN-induced IRF7 mRNA segregates with the T1L M1 and S2 gene segments and is associated with induction of myocarditis
| Virusb | Reovirus gene segments and proteinsa
|
Myocarditise | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Outer capsid
|
Core
|
Nonstructural
|
|||||||||
| S1 | S4 | M2 | S2c | M1d | L1 | L2 | L3 | S3 | M3 | ||
| E3 | 3 | 3 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | − |
| EW26 | 1 | 1 | 3 | 3 | 1 | 3 | 3 | 1 | 1 | 1 | − |
| EW50 | 1 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 3 | 3 | − |
| DB93A | 1 | 1 | 3 | 3 | 3 | 1 | 3 | 3 | 1 | 3 | − |
| EW29 | 3 | 1 | 1 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | − |
| T3D | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | − |
| DB95 | 3 | 3 | 3 | 3 | 3 | 1 | 3 | 3 | 1 | 3 | − |
| EW43 | 3 | 1 | 1 | 1 | 3 | 3 | 3 | 1 | 3 | 1 | − |
| 3HA1 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | ND |
| DB181 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | + |
| EW89 | 3 | 1 | 3 | 3 | 1 | 1 | 3 | 1 | 1 | 3 | + |
| EW60 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | + |
| EW100 | 3 | 1 | 3 | 1 | 1 | 3 | 3 | 1 | 3 | 3 | + |
| T1L | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | +/− |
| 8B | 3 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | + |
| 1HA3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ND |
1, derivation from T1L; 3, derivation from T3D. Results from Fig. 2 were used for a nonparametric Kruskal-Wallis analysis to identify reovirus genes associated with inhibition of IFN signaling. P values for method A and method B (Fig. 2) at each of the two IFN doses were >0.05 (not significant) for all genes other than S2 and M1.
Viruses are listed in the same order as in Fig. 2.
T1L S2 is associated with inhibition of IFN signaling. Method A, P = 0.003 (1,000 U IFN) and P = 0.012 (100 U IFN). Method B, P = 0.007 (1,000 U IFN) and P = 0.039 (100 U IFN).
T1L M1 is associated with inhibition of IFN signaling. Method A, P = 0.006 (1,000 U IFN) and P = 0.006 (100 U IFN). Method B, P = 0.025 (1,000 U IFN) and P > 0.05 (100 U IFN).
Viruses are designated as myocarditic (+) or nonmyocarditic (−) based on references 60 and 61. ND, not determined. Induction of myocarditis is associated with inhibition of IFN signaling. Method A, P = 0.004 (1,000 U IFN) and P = 0.004 (100 U IFN). Method B, P = 0.004 (1,000 U IFN) and P = 0.004 (100 U IFN).
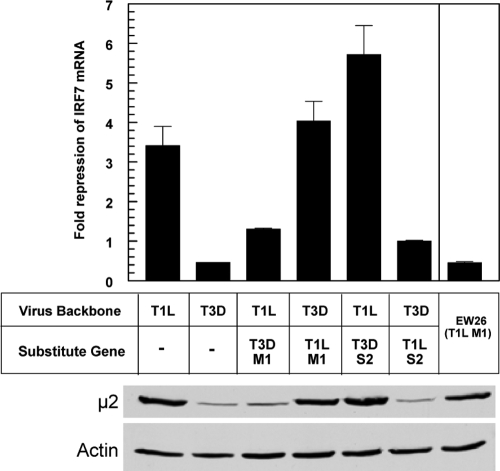
To determine whether the T1L M1 and T1L S2 genes encode repressors of IFN signaling, we engineered wild-type recombinant strains T1L and T3D and single-gene recombinant viruses by reverse genetics (30; Kobayashi and Dermody, unpublished data). L929 cells were infected with these viruses for 20 h and treated with IFN-α/β for 5 h, and IRF7 mRNA was quantified by qRT-PCR (Fig. 3). As expected, T1L repressed IFN signaling, while T3D did not. A recombinant reovirus carrying the M1 gene of T3D in the genetic background of T1L failed to inhibit IFN signaling, indicating that the T1L M1 gene is necessary for repression of IFN signaling in reovirus-infected cells. The reciprocal recombinant reovirus carrying the M1 gene of T1L in the genetic background of T3D repressed IFN signaling as effectively as T1L, indicating that the T1L M1 gene is sufficient for repression of IFN signaling in reovirus-infected cells. In contrast, recombinant reoviruses encoding heterologous S2 genes demonstrated that the T1L S2 gene is neither required nor sufficient for IFN repression. To determine μ2 expression levels in infected cells, total cell lysates were probed by immunoblotting. T1L and recombinant viruses containing a T1L M1 gene expressed μ2 at higher levels than T3D and recombinant viruses containing a T3D M1 gene, raising the possibility that repression might reflect quantitative differences in μ2 expression. However, μ2 expression in lysates from cells infected with EW26, which contains a T1L M1 gene but does not repress IFN signaling (likely reflecting a mutation or unique gene constellation), demonstrated that robust μ2 expression is insufficient to repress IFN signaling. Together, these data indicate that the T1L M1 gene, but no other T1L gene, encodes a repressor of IFN signaling.
FIG. 3.
Analysis of reovirus recombinants for repression of IFN-induced IRF7 mRNA. L929 cells were infected and treated with IFN-α/β as for Fig. 2, using recombinant reoviruses that contain single gene replacements. RNA was quantified by qRT-PCR, and copy number was normalized to GAPDH. Results are expressed as fold repression as for Fig. 2A and presented as the means of results for duplicate samples + standard deviations from a representative of two independent experiments. Calculations as for Fig. 2B generated results similar to those presented here (data not shown). Results for EW26 are from Fig. 2. The M1-encoded μ2 protein was detected by immunoblotting using rabbit polyclonal anti-μ2.
Association of antagonism of IFN signaling with induction of myocarditis.
Previous studies using a panel of reovirus reassortants identified the reovirus M1 gene as a determinant of reovirus strain-specific differences in induction of myocarditis in mice (60). In addition, reovirus strain-specific differences in induction of and sensitivity to IFN-α/β are associated with protection against myocarditis. Interestingly, these differences in the IFN response are associated with the M1, S2, and L2 genes (62). A nonparametric Kruskal-Wallis analysis here demonstrated that reovirus repression of IFN signaling is associated with viral induction of myocarditis (Table 1) (P = 0.004). Given that the M1 gene cosegregates with both reovirus repression of IFN signaling and reovirus induction of myocarditis, it is possible that these phenotypes are not causally related but instead reflect independent functions of the M1 gene product. However, the reovirus reassortant EW26, which was an “exception” for the genetic association of the M1 gene and induction of myocarditis (60), also was an “exception” for the genetic association of the M1 gene and repression of IFN signaling, suggesting that a common mutation or unique constellation of genes affects both phenotypes simultaneously. Therefore, these data strongly suggest that reovirus repression of IFN signaling is a determinant of reovirus-induced myocarditis.
Expression of the M1 gene of T1L represses IFN induction of an ISG.
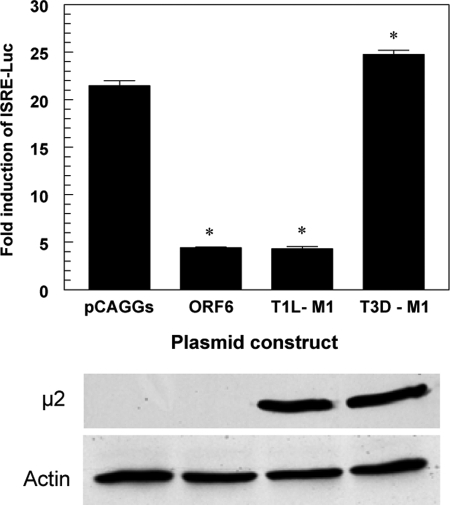
To determine whether the M1 gene of T1L functions autonomously to inhibit IFN-β signaling, we examined T1L M1 gene product μ2 inhibition of IFN-β-stimulated luciferase reporter gene expression driven by an ISRE promoter (Fig. 4). Treatment of cells with IFN-β induced ISRE reporter gene expression approximately 20-fold in cells transfected with empty vector (pCAGGs). As expected, transfection with a plasmid expressing SARS coronavirus ORF6-GFP, a known viral repressor of IFN signaling (34), significantly suppressed IFN-β induction of ISRE reporter gene expression (approximately 4.9-fold). The M1 gene of T1L inhibited IFN-β induction of ISRE reporter gene expression at a magnitude approximating that of SARS coronavirus ORF6-GFP (Fig. 4). In contrast, the M1 gene of T3D failed to repress IFN induction of the ISRE reporter gene. These results are consistent with experiments using reovirus reassortant viruses (Fig. 2; Table 1) and recombinant reoviruses (Fig. 3), in which repression of IFN-stimulated induction of IRF7 gene expression cosegregated with the M1 gene from T1L. Thus, the T1L μ2 protein is an effective inhibitor of IFN-α/β signaling that can function independently of viral infection.
FIG. 4.
The reovirus T1L M1 gene inhibits IFN induction of an ISRE promoter. HEK293 cells were transfected with an ISRE-luciferase reporter construct, a constitutively expressing Renilla luciferase construct, and the plasmids shown. At 24 hours posttransfection, cultures were either mock treated or treated with 1,000 U of IFN-β per ml for 6 h. Luciferase activity for each sample was normalized to the internal Renilla luciferase control, and the mean from duplicate samples was divided by that from the mock-treated samples to express the data as mean fold induction + standard deviation. A representative of three independent experiments is shown. Asterisks indicate significant differences from pCAGGs (Student's t test, P < 0.05). The M1-encoded μ2 protein was detected by immunoblotting using rabbit polyclonal anti-μ2.
T1L does not degrade or prevent nuclear translocation of STAT1 or STAT2.
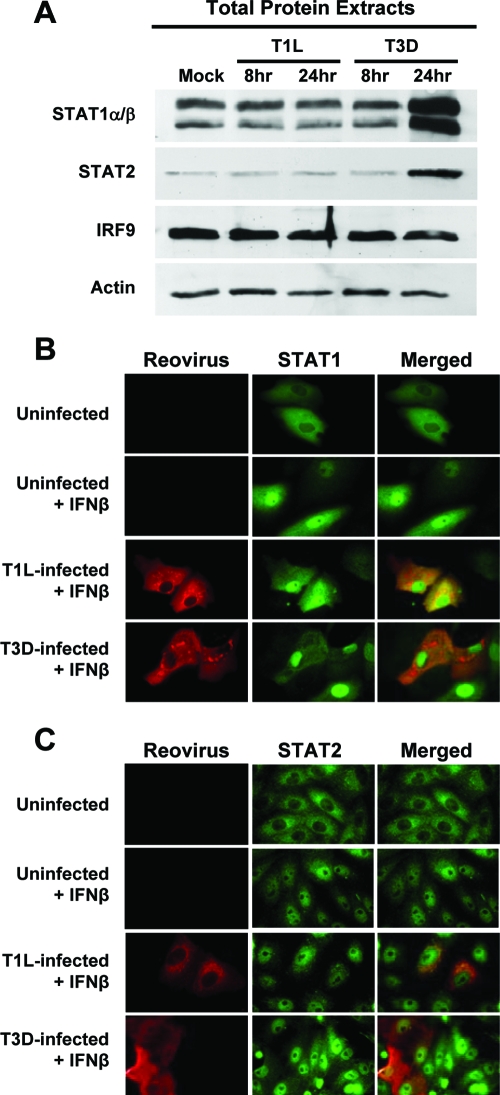
Several viruses evade the IFN response by specifically targeting components of the ISGF3 complex for proteolytic degradation (10, 13, 51). Therefore, we investigated whether the observed inhibition of IFN-induced ISG expression by T1L is attributable to degradation of STAT1, STAT2, or IRF9 protein. L929 cells were infected with T1L or T3D, and total protein extracts were harvested at various intervals postinfection (Fig. 5A). Immunoblotting for steady-state levels of STAT1, STAT2, and IRF9 revealed that neither T1L nor T3D infection resulted in degradation of ISGF3 components. The increased STAT1 and STAT2 in T3D-infected cells at 24 h postinfection most likely reflects induction of these proteins by IFN-α/β secreted during infection by this virus.
FIG. 5.
T1L does not degrade or inhibit translocation of STATs. (A) L929 cells were infected with either T1L or T3D at an MOI of 25 PFU per cell, and at the indicated times postinfection, total cellular protein was analyzed for STAT1, STAT2, and IRF9 by immunoblotting. Results are representative of two independent experiments. (B and C) Vero cells were infected with either T1L or T3D at an MOI of 25 PFU per cell and incubated for 20 h. Cells were either mock treated or treated with human IFN-β at 1,000 U per ml for 30 min. Cells were fixed and stained with antisera specific for T1L and T3D (B) (red) or monoclonal antibodies specific for reovirus σ1 and σ3 (C) (red) and either STAT1 (B) (green) or STAT2 (C) (green). A yellow color in the merged images indicates overlap of red and green pixels. Representative fields of view show infected and uninfected cells in the same field. STAT nuclear translocation was observed in every T1L- and T3D-infected cell.
In addition to targeting STATs for degradation, other viruses subvert the IFN response by modulating STAT nuclear translocation (45, 51, 53). To investigate this possibility for reovirus, Vero cells, which are defective in IFN production, were infected with T1L or T3D, incubated for 20 h, and mock treated or treated with human IFN-β for 30 min. In uninfected cultures, IFN-β rapidly stimulated both STAT1 (Fig. 5B) and STAT2 (Fig. 5C) translocation to the nucleus. Neither T1L nor T3D infection inhibited IFN-stimulated STAT1 or STAT2 nuclear translocation, indicating that this process in the IFN-mediated signal-transduction pathway is not altered by reovirus. In addition, levels of total STAT1 and STAT2 in infected cell cultures were equal to or greater than those in uninfected cultures (Fig. 5A), indicating that infection does not induce STAT degradation. Together, these data suggest that T1L does not use previously identified mechanisms involving modulation of STAT1 or STAT2 for inhibition of IFN-β signaling.
The T1L M1 gene is associated with accumulation of IRF9 in the nucleus.
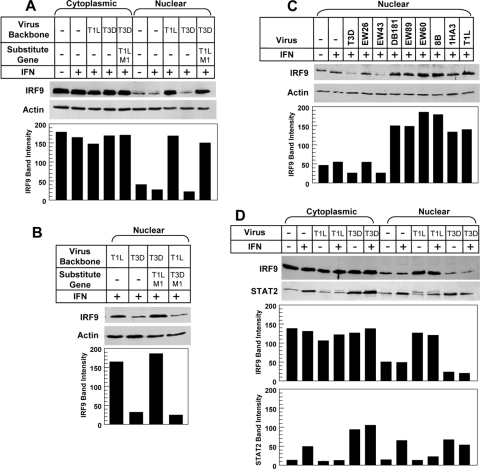
To investigate reovirus modulation of IRF9 localization in response to IFN-α/β, L929 cells were infected with the indicated virus for 20 h, treated with IFN-α/β for 5 h, and monitored for cytoplasmic and nuclear levels of IRF9 by immunoblotting (Fig. 6). Steady-state levels of IRF9 in cytoplasmic extracts were equivalent in all cases (Fig. 6A). IFN treatment alone did not increase IRF9 nuclear accumulation, presumably due to the IRF9 nuclear localization signal (NLS), which results in nuclear translocation even in the absence of IFN signaling (36). In contrast, levels of IRF9 in nuclear extracts were significantly higher in cells infected with T1L than in those infected with T3D or mock infected (Fig. 6A to D). Similar results were obtained in experiments using HEK293 cells (data not shown). A recombinant reovirus carrying the T1L M1 gene in the T3D genetic background also induced higher IRF9 levels in the nucleus than T3D, indicating that the T1L M1 gene is a genetic determinant of IRF9 nuclear accumulation (Fig. 6A and B). In contrast, the reciprocal recombinant reovirus carrying the T3D M1 gene induced low levels of IRF9 in the nucleus, comparable to those in T3D-infected cells (Fig. 6B). Thus, the T1L M1 gene is both necessary and sufficient for IRF9 accumulation in the nuclei of reovirus-infected cells.
FIG. 6.
The T1L M1 gene mediates nuclear accumulation of IRF9. L929 cell cultures were either mock treated or infected with the indicated reovirus strains at an MOI of 25 PFU per cell for 20 h and treated with 1,000 U per ml of IFN-α/β for 5 h. Cytoplasmic and nuclear protein extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nylon membrane, and immunoblotted using rabbit polyclonal anti-IRF9, -STAT2, or -actin. Gels were scanned and band intensity quantified as indicated. The band migrating above STAT2 in T1L-infected cultures in panel D was identified as a reovirus protein by mass spectrometry (data not shown).
To determine whether IRF9 nuclear accumulation is associated with inhibition of IFN signaling, reovirus reassortants that had been tested for repression of IFN induction of IRF7 (Fig. 2) were tested for modulation of IRF9 redistribution in cells. Reovirus reassortants that repress IFN signaling (DB181, EW89, EW60, 8B, and 1HA3) resulted in IRF9 accumulation in the nucleus, while those that did not repress IFN signaling (EW26 and EW43) resulted in lower nuclear levels of IRF9 (Fig. 6C). Collectively, these data indicate that the T1L M1 gene results in accumulation of IRF9 in the nuclei of reovirus-infected cells and that this nuclear retention of IRF9 is associated with repression of the IFN signaling pathway.
To determine whether exogenous IFN stimulation is required for T1L-induced nuclear accumulation of IRF9, cells were infected and then treated with IFN or control medium. T1L induced nuclear accumulation of IRF9 regardless of IFN treatment (Fig. 6D), suggesting that IFN stimulation is not required for this T1L effect. We next used immunoblotting to investigate whether T1L induces nuclear accumulation of STAT2 (Fig. 6D), since STAT2 is a binding partner for IRF9 and is involved in its nuclear-cytoplasmic shuttling (1, 36, 52). IFN treatment in the absence of viral infection induced increased STAT2 levels in the cytoplasm, consistent with STAT2 function as an ISG. IFN treatment also induced STAT2 nuclear translocation, consistent with STAT2 incorporation into ISGF3 and exposure of an NLS (1). T3D also induced STAT2 cytoplasmic expression and nuclear translocation, consistent with T3D induction of IFN (62), and these effects were maintained following IFN stimulation. In contrast, T1L failed to induce STAT2 cytoplasmic expression or nuclear translocation in the absence of IFN stimulation, consistent with the poor induction of IFN by this viral strain (62). In addition, T1L failed to induce STAT2 cytoplasmic expression upon IFN stimulation, consistent with T1L repression of IFN signaling (Fig. 1). Finally, STAT2 nuclear translocation did not increase in T1L-infected IFN-stimulated cells, despite the fact that T1L does not repress IFN-stimulated STAT2 nuclear accumulation (Fig. 5), likely reflecting failure of IFN to induce STAT2 expression as it did in mock- or T3D-infected cells.
DISCUSSION
In this study, we demonstrate that reovirus represses the IFN-α/β signaling pathway and that this repression is virus strain specific. Repression is mediated by the M1 gene product (μ2) causing nuclear accumulation of IRF9. Virus strain-specific differences in this repressor function are associated with virus strain-specific differences in induction of myocarditis, a disease modulated by the IFN response (44, 62). Together, these data provide the first report of an association between viral subversion of the IFN response and nuclear accumulation of IRF9 and link strain-specific differences in IFN antagonism to viral pathogenesis.
The inhibitory effect of T1L infection on IFN signaling cosegregated with the T1L M1 gene (Fig. 2 and 3; Table 1), and ectopic expression of the T1L M1 gene inhibited IFN-β-induced reporter gene expression (Fig. 4). Reovirus strain-specific differences in induction of and sensitivity to IFN-α/β are associated with the M1, S2, and L2 genes (62), suggesting that repression of IFN signaling either provides the underlying mechanism for these two properties or involves interactions with similar host factors. Reovirus induction of IFN-β is substantially reduced in cells derived from mice lacking the IFN-α/β receptor (B. Sherry, unpublished observations), indicating that most of the IFN-β induced by reovirus infection results from the positive amplification loop, where IFN-mediated induction of IRF7 induces additional IFN-β (56). Therefore, repression of IFN signaling would reduce reovirus induction of IFN, providing a mechanism for poor induction of IFN by T1L (62). Repression of IFN signaling also would provide resistance to the antiviral effects of IFN, providing a mechanism for the relative resistance of T1L to IFN (27, 62). Thus, T1L repression of IFN signaling provides a mechanism for strain-specific differences in both reovirus induction of and sensitivity to IFN-α/β.
Studies with pathogenic viruses have demonstrated that the type I IFN response is essential in protection against viral disease. The virulence of many viruses is enhanced in mice lacking the IFN-α/β receptor (14, 28, 32, 42, 55, 63), and absence of the IFN-α/β response can result in broader tissue tropism and lethality following infection (18, 25). Inhibition of the host IFN response is a critical determinant of viral virulence, as evidenced by the resultant attenuation when viral IFN repressor proteins are mutated (11, 24, 65, 71). Indeed, targeting these IFN repressors provides the conceptual basis for a new generation of vaccine candidates (69). Virus strain-specific differences in antagonism of the IFN response can be determinants of strain-specific differences in disease, but results are mixed. For example, while virus strain-specific differences in the NS1 protein of H1N1 influenza virus result in differences in inhibition of the IFN response (31), there is no evidence that these NS1 protein differences result in differences in influenza virus pathogenicity (46). However, studies using the NS1 gene from H5N1 influenza virus strains link NS1 effects on the IFN response with pathogenesis (59). Finally, a study using both H1N1 and H5N1 strains found that NS1 protein differences are associated with differences in pathogenicity but not through differences in modulation of the IFN response (26). Identification of an association between reovirus repression of the IFN response and reovirus-induced myocarditis here (Table 1 [EW26 in particular]) provides strong evidence that viral IFN antagonism serves as a virulence determinant in the heart.
Infection with T1L or reassortant or recombinant viruses containing the T1L M1 gene results in accumulation of IRF9 in the nucleus, an effect not previously described for any virus. In addition, nuclear accumulation of IRF9 occurs in both murine and human cell lines infected with T1L, indicating that this effect is not species specific. Paramyxovirus inhibitors of IFN are either species specific (35) or operant across a broad host range (20). The events underlying T1L M1-mediated nuclear accumulation of IRF9 are not yet understood. The M1- encoded μ2 protein (57) is an RNA-binding protein (6) present in low copy number in the virion but expressed abundantly in infected cells, where it associates with microtubules (8, 41) and contributes to formation of viral factories (47). Interestingly, despite the exclusively cytoplasmic replication strategy of reovirus, and in contrast to most reovirus proteins, μ2 also distributes to the nucleus and contains a predicted NLS (7, 47). The function of μ2 in the nucleus is unknown, but it may alter IRF9 structure or function there. In sum, none of the known properties of μ2 suggest a specific mechanism for modulation of IRF9 localization.
Nuclear-cytoplasmic shuttling of IRF9 has been well characterized (1, 36, 52). IRF9 contains an NLS but lacks a nuclear export signal, while STAT2 lacks a functional NLS but contains a nuclear export signal. They form a heterodimeric cytoplasmic complex in unstimulated cells, resulting in IRF9-mediated interaction with importins for nuclear localization. However, STAT2-mediated interactions with CRM1 in the nucleus result in nuclear export, and this dominant function determines a primarily cytoplasmic localization for both STAT2 and its partner IRF9. Upon IFN stimulation, STAT phosphorylation, and formation of ISGF3, IRF9 is again transported to the nucleus. Nuclear dephosphorylation of STAT2 results in association with CRM1, and STAT2 then escorts IRF9 back to the cytoplasm. Accordingly, we envision four possible models for μ2-mediated nuclear localization of IRF9 and repression of IFN signaling. First, nuclear accumulation of IRF9 in T1L-infected cells could reflect an upregulation of IRF9 expression, and abundant free nuclear IRF9 could compete with ISGF3 to bind but not induce expression from ISREs, thereby functioning as a dominant-negative inhibitor of IFN signaling. However, total cellular IRF9 was not increased in T1L-infected cells (Fig. 5), and qRT-PCR experiments demonstrate that T1L does not induce IRF9 mRNA between 4 and 24 h postinfection (data not shown), together indicating that T1L does not upregulate IRF9 expression. Moreover, while IRF9 does bind ISREs without inducing expression (35), IRF9 overexpression can compensate for IRF9 deficiencies with no evidence of dominant-negative function (37). Second, T1L μ2 could enhance importin function to increase IRF9 nuclear translocation, and once there, abundant free IRF9 could act as a dominant-negative inhibitor of IFN signaling. Viral modulation of importins or nuclear-cytoplasmic trafficking is well precedented, although in each case, the virus acts to impede rather than enhance normal cell function (16, 50, 53). However, this model lacks precedent for IRF9 dominant-negative function. Third, T1L μ2 could inhibit CRM1 function, thereby blocking STAT2-mediated IRF9 export. However, STAT2 nuclear localization was not increased by T1L infection (Fig. 6D), eliminating this possibility. Finally, in a fourth model, which we favor, T1L μ2 might prevent IRF9 from binding to STAT2, thereby affecting both ISGF3 function and IRF9 export. This process could occur through μ2 association with IRF9 in the cytoplasm, consistent with T1L induction of IRF9 nuclear accumulation even in the absence of IFN stimulation (Fig. 6D), or in the nucleus, consistent with μ2 nuclear localization. Alternatively, μ2 could mediate structural modifications of IRF9. Our future studies will determine whether μ2 affects the interaction of IRF9 with ISRE promoters and other components of the ISG transcriptional apparatus to attenuate ISG expression.
Previous studies of viral mechanisms for antagonizing the IFN response involving IRF9 have documented a decrease in IRF9 levels (37) or sequestration of IRF9 to prevent its translocation to the nucleus upon IFN stimulation (2) but not nuclear accumulation of IRF9 as seen here. The nonstructural protein 1 of rotavirus, another member of the Reoviridae family, modulates the IFN response by recognizing a common element of IRF proteins to mediate degradation of IRF3, IRF5, and IRF7 (3). While our results suggest that modulation of IRFs may be a common mechanism by which Reoviridae members subvert the IFN response, reovirus does not express a homolog of rotavirus nonstructural protein 1, and reovirus has not been shown to inhibit the activity of other IRF proteins.
Other viruses use multiple independent mechanisms to inhibit the IFN response. For example, the influenza virus NS1 protein (31), paramyxovirus V proteins (21, 23, 49), and herpes simplex virus type 1 (29, 40) each block both induction of IFN and IFN signaling. The reovirus S4 gene encodes a dsRNA-binding protein, σ3, which is capable of preventing activation of the antiviral ISG PKR (57). While dsRNA is not thought to be exposed in the cytoplasm during reovirus replication and σ3 has not been implicated genetically or biochemically in modulating the IFN response during reovirus infection, a role for σ3 in countering the host response remains possible. The results reported here provide the first direct evidence for a reovirus protein modulating the IFN response during infection. If σ3 also inhibits the IFN pathway by sequestering dsRNA, then reovirus, encoding only 11 proteins, has evolved at least two independent mechanisms for subverting the antiviral actions of IFN.
Acknowledgments
We thank Lianna Li, Susan Irvin, and Adolfo García-Sastre for insightful discussions and Wrennie Edwards for technical assistance.
This research was supported by Public Health Service awards T32 AI49824 and F32 AI071440 (to G.H.H.), R01 AI50080 (to T.S.D.), and R01 AI062657 (to B.S.) and by the Elizabeth B. Lamb Center for Pediatric Research.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Banninger, G., and N. C. Reich. 2004. STAT2 nuclear trafficking. J. Biol. Chem. 27939199-39206. [DOI] [PubMed] [Google Scholar]
- 2.Barnard, P., and N. A. McMillan. 1999. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology 259305-313. [DOI] [PubMed] [Google Scholar]
- 3.Barro, M., and J. T. Patton. 2007. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 814473-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 777945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 7912828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brentano, L., D. L. Noah, E. G. Brown, and B. Sherry. 1998. The reovirus protein mu2, encoded by the M1 gene, is an RNA-binding protein. J. Virol. 728354-8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broering, T. J., J. S. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein microNS forms large inclusions and colocalizes with reovirus microtubule-associated protein micro2 in transfected cells. J. Virol. 768285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho, J., M. M. Arnold, and M. L. Nibert. 2007. Silencing and complementation of reovirus core protein mu2: functional correlations with mu2-microtubule association and differences between virus- and plasmid-derived mu2. Virology 364301-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 2641415-1421. [DOI] [PubMed] [Google Scholar]
- 10.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 739928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 7713257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drayna, D., and B. N. Fields. 1982. Genetic studies on the mechanism of chemical and physical inactivation of reovirus. J. Gen. Virol. 63149-159. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, J., O. T. Lynch, Y. Suessmuth, P. Qian, C. R. Boyd, J. F. Burrows, R. Buick, N. J. Stevenson, O. Touzelet, M. Gadina, U. F. Power, and J. A. Johnston. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J. Virol. 813428-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiette, L., C. Aubert, U. Muller, S. Huang, M. Aguet, M. Brahic, and J. F. Bureau. 1995. Theiler's virus infection of 129Sv mice that lack the interferon alpha/beta or interferon gamma receptors. J. Exp. Med. 1812069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 3001145-1148. [DOI] [PubMed] [Google Scholar]
- 16.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, X. Y., D. S. Kessler, S. A. Veals, D. E. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA 878555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Sastre, A., R. K. Durbin, H. Zheng, P. Palese, R. Gertner, D. E. Levy, and J. E. Durbin. 1998. The role of interferon in influenza virus tissue tropism. J. Virol. 728550-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 765532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagmaier, K., N. Stock, S. Goodbourn, L. F. Wang, and R. Randall. 2006. A single amino acid substitution in the V protein of Nipah virus alters its ability to block interferon signalling in cells from different species. J. Gen. Virol. 873649-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 30315-32. [DOI] [PubMed] [Google Scholar]
- 22.Heim, M. H. 1999. The Jak-STAT pathway: cytokine signalling from the receptor to the nucleus. J. Recept. Signal Transduct. Res. 1975-120. [DOI] [PubMed] [Google Scholar]
- 23.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 2714621-4628. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Z., S. Krishnamurthy, A. Panda, and S. K. Samal. 2003. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 778676-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ida-Hosonuma, M., T. Iwasaki, T. Yoshikawa, N. Nagata, Y. Sato, T. Sata, M. Yoneyama, T. Fujita, C. Taya, H. Yonekawa, and S. Koike. 2005. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J. Virol. 794460-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, D., M. J. Hossain, D. Hickman, D. R. Perez, and R. A. Lamb. 2008. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. USA 1054381-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs, B. L., and R. E. Ferguson. 1991. The Lang strain of reovirus serotype 1 and the Dearing strain of reovirus serotype 3 differ in their sensitivities to beta interferon. J. Virol. 655102-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson, C., J. D. Wetzel, J. He, C. Mikacenic, T. S. Dermody, and B. L. Kelsall. 2007. Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J. Exp. Med. 2041349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, K. E., B. Song, and D. M. Knipe. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi, T., A. A. Antar, K. W. Boehme, P. Danthi, E. A. Eby, K. M. Guglielmi, G. H. Holm, E. M. Johnson, M. S. Maginnis, S. Naik, W. B. Skelton, J. D. Wetzel, G. J. Wilson, J. D. Chappell, and T. S. Dermody. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kochs, G., A. Garcia-Sastre, and L. Martinez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 817011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koerner, I., G. Kochs, U. Kalinke, S. Weiss, and P. Staeheli. 2007. Protective role of beta interferon in host defense against influenza A virus. J. Virol. 812025-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 742477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus, T. A., L. Garza, and C. M. Horvath. 2008. Enabled interferon signaling evasion in an immune-competent transgenic mouse model of parainfluenza virus 5 infection. Virology 371196-205. [DOI] [PubMed] [Google Scholar]
- 36.Lau, J. F., J. P. Parisien, and C. M. Horvath. 2000. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. USA 977278-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard, G. T., and G. C. Sen. 1996. Effects of adenovirus E1A protein on interferon-signaling. Virology 22425-33. [DOI] [PubMed] [Google Scholar]
- 38.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 1022992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 176660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melroe, G. T., N. A. DeLuca, and D. M. Knipe. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 788411-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, C. L., J. S. Parker, J. B. Dinoso, C. D. Piggott, M. J. Perron, and M. L. Nibert. 2004. Increased ubiquitination and other covariant phenotypes attributed to a strain- and temperature-dependent defect of reovirus core protein mu2. J. Virol. 7810291-10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 2641918-1921. [DOI] [PubMed] [Google Scholar]
- 43.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 44.O'Donnell, S. M., M. W. Hansberger, J. L. Connolly, J. D. Chappell, M. J. Watson, J. M. Pierce, J. D. Wetzel, W. Han, E. S. Barton, J. C. Forrest, T. Valyi-Nagy, F. E. Yull, T. S. Blackwell, J. N. Rottman, B. Sherry, and T. S. Dermody. 2005. Organ-specific roles for transcription factor NF-kappaB in reovirus-induced apoptosis and disease. J. Clin. Investig. 1152341-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 777635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pappas, C., P. V. Aguilar, C. F. Basler, A. Solorzano, H. Zeng, L. A. Perrone, P. Palese, A. Garcia-Sastre, J. M. Katz, and T. M. Tumpey. 2008. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc. Natl. Acad. Sci. USA 1053064-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 764483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pellegrini, S., and I. Dusanter-Fourt. 1997. The structure, regulation and function of the Janus kinases (JAKs) and the signal transducers and activators of transcription (STATs). Eur. J. Biochem. 248615-633. [DOI] [PubMed] [Google Scholar]
- 49.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 30333-46. [DOI] [PubMed] [Google Scholar]
- 50.Porter, F. W., Y. A. Bochkov, A. J. Albee, C. Wiese, and A. C. Palmenberg. 2006. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 10312417-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 52.Reich, N. C. 2007. STAT dynamics. Cytokine Growth Factor Rev. 18511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 805156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 7611476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryman, K. D., K. C. Meier, C. L. Gardner, P. A. Adegboyega, and W. B. Klimstra. 2007. Non-pathogenic Sindbis virus causes hemorrhagic fever in the absence of alpha/beta and gamma interferons. Virology 368273-285. [DOI] [PubMed] [Google Scholar]
- 56.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441106-110. [DOI] [PubMed] [Google Scholar]
- 57.Schiff, L. A., M. L. Nibert, and K. L. Tyler. 2007. Orthoreoviruses and their replication, p. 1853-1915. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 58.Schindler, C., and J. E. Darnell, Jr. 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64621-651. [DOI] [PubMed] [Google Scholar]
- 59.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8950-954. [DOI] [PubMed] [Google Scholar]
- 60.Sherry, B., and B. N. Fields. 1989. The reovirus M1 gene, encoding a viral core protein, is associated with the myocarditic phenotype of a reovirus variant. J. Virol. 634850-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherry, B., F. J. Schoen, E. Wenske, and B. N. Fields. 1989. Derivation and characterization of an efficiently myocarditic reovirus variant. J. Virol. 634840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherry, B., J. Torres, and M. A. Blum. 1998. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J. Virol. 721314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shresta, S., J. L. Kyle, H. M. Snider, M. Basavapatna, P. R. Beatty, and E. Harris. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 782701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith, R. E., H. J. Zweerink, and W. K. Joklik. 1969. Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39791-810. [DOI] [PubMed] [Google Scholar]
- 65.Solorzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. Garcia-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 797535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 67.Stewart, M. J., K. Smoak, M. A. Blum, and B. Sherry. 2005. Basal and reovirus-induced beta interferon (IFN-β) and IFN-β-stimulated gene expression are cell type specific in the cardiac protective response. J. Virol. 792979-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 747989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 974309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tyler, K. L., M. A. Mann, B. N. Fields, and H. W. Virgin IV. 1993. Protective antireovirus monoclonal antibodies and their effects on viral pathogenesis. J. Virol. 673446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valarcher, J. F., J. Furze, S. Wyld, R. Cook, K. K. Conzelmann, and G. Taylor. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 778426-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veals, S. A., C. Schindler, D. Leonard, X. Y. Fu, R. Aebersold, J. E. Darnell, Jr., and D. E. Levy. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 123315-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Virgin, H. W., K. L. Tyler, and T. S. Dermody. 1997. Reovirus, p. 669. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven, Philadelphia, PA.
- 74.Virgin, H. W., IV, R. Bassel-Duby, B. N. Fields, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 624594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wathelet, M. G., I. M. Clauss, C. B. Nols, J. Content, and G. A. Huez. 1987. New inducers revealed by the promoter sequence analysis of two interferon-activated human genes. Eur. J. Biochem. 169313-321. [DOI] [PubMed] [Google Scholar]
- 76.Zurney, J., K. E. Howard, and B. Sherry. 2007. Basal expression levels of IFNAR and Jak-STAT components are determinants of cell-type-specific differences in cardiac antiviral responses. J. Virol. 8113668-13680. [DOI] [PMC free article] [PubMed] [Google Scholar]