Abstract
The capsid of bacteriophage HK97 is stabilized by ∼400 covalent cross-links between subunits which form without any action by external enzymes or cofactors. Cross-linking only occurs in fully assembled particles after large-scale structural changes bring together side chains from three subunits at each cross-linking site. Isopeptide cross-links form between asparagine and lysine side chains on two subunits. The carboxylate of glutamic acid 363 (E363) from a third subunit is found ∼2.4 Å from the isopeptide bond in the partly hydrophobic pocket that contains the cross-link. It was previously reported without supporting data that changing E363 to alanine abolishes cross-linking, suggesting that E363 plays a role in cross-linking. This alanine mutant and six additional substitutions for E363 were fully characterized and the proheads produced by the mutants were tested for their ability to cross-link under a variety of conditions. Aspartic acid and histidine substitutions supported cross-linking to a significant extent, while alanine, asparagine, glutamine, and tyrosine did not, suggesting that residue 363 acts as a proton acceptor during cross-linking. These results support a chemical mechanism, not yet fully tested, that incorporates this suggestion, as well as features of the structure at the cross-link site. The chemically identical isopeptide bonds recently documented in bacterial pili have a strikingly similar chemical geometry at their cross-linking sites, suggesting a common chemical mechanism with the phage protein, but the completely different structures and folds of the two proteins argues that the phage capsid and bacterial pilus proteins have achieved shared cross-linking chemistry by convergent evolution.
A common feature of the assembly pathways of all tailed double-stranded DNA bacteriophages and herpesviruses (45) is that they package their chromosomes into unstable precursors called procapsids or proheads, which mature into virions. The virion DNA is packed to a very high density (∼0.5 g/ml) (13), so the completed DNA-filled capsids must be specifically reinforced or stabilized during maturation to withstand the very high effective internal “DNA pressure” (20, 41, 49). Virions also must resist the chemical and physical stresses of the environment outside their hosts. Phage capsids achieve this stabilization by several different mechanisms, all of which include large conformational changes in the capsid. Some phages, like P22 (14) and T7 (4), achieve stabilization by conformational changes alone. For other phages, such as T4 (43) and λ (42), full stabilization requires the binding of so-called “decoration” proteins to the outside of the capsid (44). Capsid stabilization may also occur by forming covalent cross-links between capsid protein subunits, as in phage HK97 (10, 11, 36, 51). The HK97 cross-links are isopeptide bonds between asparagine and lysine side chains on neighboring subunits that tie together structural elements on the inside of the shell to elements on the outside, thus generating a chain-mail-like network of connections among capsid subunits and making a very stable capsid (39, 51). This type of amide bond was thought to occur only in phage capsid proteins, where they occur frequently (23), but it has also been identified recently within the subunits of the pili of Streptococcus pyogenes and may be widespread in related pilus proteins (26).
HK97 is a λ-like phage of Escherichia coli (7, 25) with a long noncontractile tail and a T=7 icosahedral capsid (5). HK97 procapsids assemble from the major capsid protein, the capsid protease, and the portal protein as outlined in Fig. 1. Assembly occurs during phage infection or when the same proteins are expressed from plasmids in E. coli. After assembly of Prohead I, the protease cleaves interior domains from the major capsid protein and digests itself, yielding the procapsid, Prohead II, which contains only cleaved major capsid protein. Prohead II is the natural substrate for DNA packaging during phage infection, and it matures during packaging by a process called expansion and transforms into Head II, for which the X-ray structure has been determined (22, 51). Expansion can also be induced chemically in vitro and has been studied extensively, both biochemically and in multiple cryo-electron microscopy (cryo-EM) and X-ray structures (5, 6, 15, 16, 31-33, 39). Expansion involves extensive rearrangements of the capsid subunits which change the round, thick-walled ∼550-Å Prohead II into the angular, thin-walled ∼650Å Head II (Fig. 1). Capsid cross-links are made during expansion.
FIG. 1.
Assembly and maturation of the HK97 capsid. Three proteins are required to assemble Prohead II, the HK97 procapsid. These are the major capsid protein (gp5, 42 kDa), the protease (gp4, 25 kDa), and the portal or connector protein (gp3, 46 kDa) (12). The major capsid protein folds and assembles into hexamers and pentamers (capsomers collectively), which coassemble with the protease to form Prohead I. The protease removes 102 residues of each capsid subunit and destroys itself to create Prohead II. The portal complex normally occupies the position of one pentamer and is required for viable phage assembly, but the portal protein is optional for making particles from plasmid-expressed genes (5, 11). Prohead II expands to form the mature capsid, Head II, normally during DNA packaging, but also in vitro as described in Results.
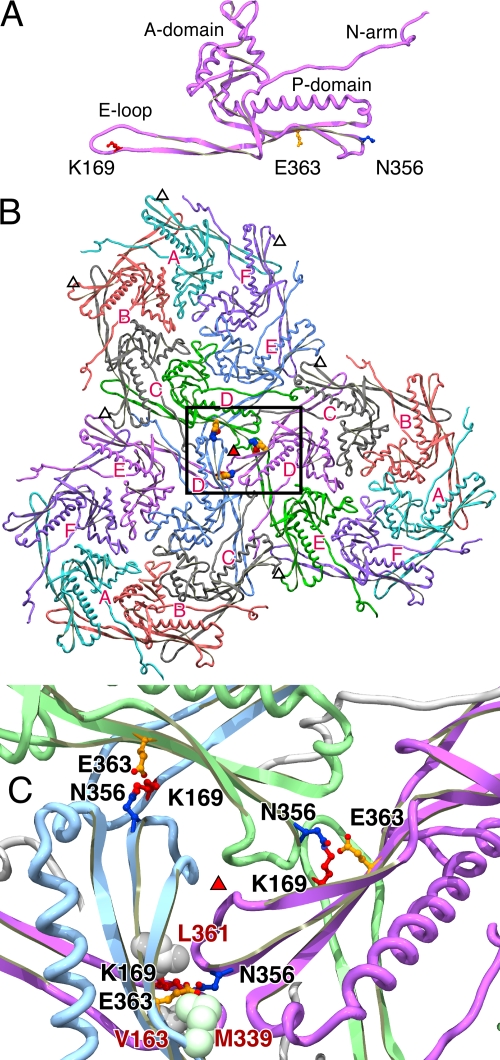
The high-resolution structure of the HK97 mature capsid provides a structural basis for understanding many aspects of assembly and cross-linking (22, 51). The subunits of the mature capsid have a continuous core with a dominant long alpha-helix and two extended arms, the N-arm and the long β-hairpin or E-loop (for Extended loop), as shown in Fig. 2A. Cross-links cannot form until a complete particle is assembled—this can be easily understood by looking at the Head II subunit by itself and the arrangement of subunits in the mature particle (Fig. 2A and B). The three residues required for cross-linking are far apart in an isolated subunit, as they are in an individual hexamer or pentamer of capsid protein from which the proheads assemble. At least three capsomers (hexamers or pentamers of capsid protein) must assemble in a manner like that shown in Fig. 2B and C in order to bring together the three residues that will participate in a given cross-link. This is not achieved directly; the capsomers initially assemble into proheads, in which the two residues that will cross-link are still separated by more than 30 Å (6). Cross-linking does not occur until the conformational rearrangements of the expansion reaction bring asparagine 356 (N356) and lysine 169 (K169) close to glutamic acid 363 (E363) in an active site-like cavity. We argue here that E363 has a catalytic role in the cross-linking reaction.
FIG. 2.
Three-dimensional models of the sites of capsid cross-linking in HK97. Views of HK97 cross-link sites based on the refined structure (PDB ID 1OHG) (22) show the proximity of the side chain of glutamate E363 to the joined residues. (A) An HK97 Head II subunit (D chain) in isolation shows the three side chains involved in capsid cross-linking: an asparagine (N356, dark blue) at the end of the P-domain, a lysine (K169, red) near the tip of the E-loop, and a glutamate (E363, orange) protruding from a β-sheet (β-J) in the P-domain. HK97 cross-links occur at sites located around the threefold and quasi-threefold axes of symmetry of the HK97 capsid, and one such trio of cross-links is shown in panels B and C. (B) Cross-links formed around a threefold axis (red-filled triangle) and their context among the three hexons that comprise a flat icosahedral face of the particle. The hexamer chains are labeled A to F, and the locations of several quasi-threefold axes are marked with open triangles. Views are from the inside of a capsid, looking outward. (C) Close-up view of the area boxed in panel B. The chains joined by the highlighted cross-links are colored identically in pairs: a lysine from a D chain is joined to an asparagine in an E chain from a neighboring hexamer, aided by a glutamate in the D chain of a neighboring hexamer. Side chain oxygens in the glutamates and cross-links are indicated in red. In the lower of the three cross-link sites, the side chains of three hydrophobic residues that surround the cross-links are shown in space-filling representations; these are residues V163, L361, and M339 and are labeled in red. Images made using SPDBV 3.7 (21) and the Persistence of Vision Raytracer (POV-Ray Mac 3.6 [http://www.povray.org/]).
In the Head II X-ray model (22), one of the cross-link residues, N356, is near the outer end of the P-domain, sandwiched within the subunit interface near junctions where three hexamers (or two hexamers and a pentamer) meet (see Fig. 2), while the other, K169, projects into the subunit interface from near the tip of the E-loop. The third residue involved in cross-linking, E363, is located on a third subunit, which lies across the local capsomer-capsomer interface from N356 (Fig. 2B and C) and one carboxylate oxygen of E363 is found only 2.5 Å away from the isopeptide bond. This arrangement is found in each of the seven unique (quasi-equivalent) cross-link sites in the Head II model (22, 51). Changing E363 to alanine (E363A) was found to abolish cross-linking (51), suggesting that E363 may have an important chemical role in the cross-linking reaction. Further support for a role of E363 in cross-linking comes from evidence that it appears to be conserved and easily identified in alignments of the HK97 capsid protein sequence with other capsid proteins in GenBank. For example, a BLAST search of the nonredundant protein database with the mature HK97 capsid protein sequence yields ∼60 non-self-matches, and 43 of these potential homologs appear to possess all three cross-linking residues (matching K169, N356, and E363 in HK97), notably including several bacteriophages for which there is experimental evidence of capsid cross-linking: D3 (19), XP10 (54), BFK20 (3), and Φ11b (2). The findings we present here suggest that both the size and the character of residue 363 can influence assembly and maturation of HK97 capsids, but only glutamate is capable of promoting efficient cross-linking. In the analogous Asn-Lys isopeptide bonds recently identified in the pili of gram-positive bacteria, a conserved glutamic (or aspartic) acid residue is also poised nearby and implicated in cross-linking, suggesting a common mechanism.
MATERIALS AND METHODS
Bacteria, plasmids, and plasmid construction.
HK97 amber mutants for complementation tests were grown on E. coli strains Ymel (supF) and LE392 (supF supE). E. coli strain BL21(DE3)/pLysS (46) was used for plasmid expression for production of proheads and for complementation tests. Plasmid pV0 (pT7-5Hd2.9) (12) was used to produce Prohead II and for construction of variants at residue E363 in gene 5. Mutations were made by using a PCR “megaprimer” method that uses one mutagenic and two flanking primers (30). Primer sequences are given 5′ to 3′. The flanking primers, AGCAGAAATCGAAAGCACAG and TCGCTGAGATAGGTGCCTC, produce a 1,341-bp product. Primer E363DNHY (CCATCCTGTGCNACGAGCGTCTGGC) was a mixed oligonucleotide to make HK97 gp5 variants E363D, E363N, E363H, and E363Y; N at the first and C at the third position changes E = GAA to the NAC set, where D = GAC, N = AAC, H = CAC, and Y = TAC. Primer E363KQ (CCATCCTGTGCMAAGAGCGTCTGGC) was a mixed oligonucleotide used to make HK97 gp5 variants E363K, E363Q; M (A or C) at first position changes E = GAA to the MAA set K = AAA and Q = CAA. The mutants were produced on a fragment that could be cut with BtgI and BamHI, and the 250-bp BtgI-BamHI fragment was purified and used to replace the corresponding wild-type segment of the parent plasmid. PCR-amplified DNA in the resulting clones was sequenced to ensure that only the desired changes were present.
Complementation tests.
Spot tests compared the efficiency of plating (EOP) of an amber mutant phage grown on a host synthesizing a mutant gp5 from a plasmid to the EOP when wild-type gp5 is provided from a control plasmid. Spot tests are variants of phage T4 techniques (1, 8). The phages were HK97 (wild-type), HK97 5am1 (amber mutant in gene 5 [34a]), and HK97 amC2 (negative control amber not complemented by plasmids used (36). Overnight cultures of BL21(DE3)/pLysS containing positive control plasmid pV0 (makes wild-type gp5 and gp4) or pV0 variant plasmids containing mutations to be tested (gene 5 mutants) were grown in LB plus 0.4% maltose and 50 μg of ampicillin/ml and 25 μg of chloramphenicol/ml. About 0.15 ml of each culture was mixed with 2.5 ml of top agar, spread onto an LB plate containing the same drugs, and allowed to solidify. Plasmid gene expression was induced by supplementing the soft agar with ∼1% lactose. Stocks of phage at ∼109 PFU/ml were serially diluted in 10-fold steps using 10 mM Tris-HCl (pH 7.5), 10 mM MgSO4, 0.01% gelatin, and 4 μl of each dilution was spotted onto plates prepared as described above and allowed to dry. After incubation overnight at 37°C, the plates were examined for clear areas indicating phage growth at the spots where phage were applied. Relative plaque counts were calculated by assigning a score of 10 for each fully cleared spot and 3 for each partially cleared spot and multiplying the scores together. For example, wild type typically yields five clear and one partially clear spot for a count of 10 × 10 × 10 × 10 × 10 × 3 = 3 × 105. The relative EOP is the plaque count on the mutant-expressing host divided by that on the wild-type-expressing host.
Preparation of Proheads.
Proheads were made using plasmids which express HK97 genes 4 and 5 under the control of the T7 φ10 promoter and purified by differential centrifugation, polyethylene glycol precipitation, and velocity sedimentation in glycerol gradients, as described previously (11, 34). Plasmid pV0 (pT7-5Hd2.9) (12) was used to produce Prohead II. Prohead I was made using a protease-knockout plasmid pVB [pT7-5Hd2.9(fsBstB1)] (12). Cells were held under inducing conditions for ∼16 h at 28°C and used immediately.
Prohead I purification steps prior to chromatography were done using TKG buffer (20 mM Tris-HCl [pH 7.5], 100 mM potassium glutamate). Proheads were further purified by adsorption to Poros HQ20 quaternary-amine ion-exchange columns (ABI, Foster City, CA) in 20 mM tris(hydroxymethyl)amino-methane hydrochloride (Tris-HCl)-bis-tris-propane buffer (Sigma) at pH 7.5 with 20 mM NaCl and elution with a linear gradient of NaCl to 0.5 M at 13 to 33 ml/min using a BioCAD chromatography system (ABI). Prohead samples were concentrated by ultracentrifugation, resuspended in a small volume, and dialyzed into a buffer appropriate for the next step.
Some mutant prohead-producing strains yield a mixture of Prohead I and Prohead II. Accordingly, Prohead II was further purified for some mutants by dissociating the unwanted Prohead I using glucose (34). Such Prohead-I-containing mixtures were treated at ∼4 mg/ml with ∼40% glucose and 10 to 20 mM Tris-HCl (pH 9.5) at room temperature overnight (34). After treatment, the remaining proheads (Prohead II, mostly) were separated from capsomers by pelleting via ultracentrifugation and then purified by using Poros HQ20 chromatography as described above.
Agarose gel analysis of HK97 head-related structures.
Native-agarose gels were run using TAMg buffer (40 mM Tris base, 20 mM acetic acid [pH 8.1], 1 mM magnesium sulfate) as described previously (11). Samples were prepared for electrophoresis by adding a 1/9 volume of 50% (vol/vol) glycerol, 0.025% bromophenol blue, and 0.025% xylene cyanol XFF. After electrophoresis, the gels were fixed and washed in 95% ethanol, dried (initially with no heat), stained in 0.05% Coomassie brilliant blue R250 in 50% (vol/vol) methanol-10% acetic acid, and destained in 10% acetic acid.
SDS-polyacrylamide gel analysis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis methods were modified from those of Laemmli (29) and used a low-cross-linker acrylamide mix (9) containing 33.5% (wt/vol) acrylamide and 0.3% (wt/vol) methylene bisacrylamide to enhance the resolution of large oligomers. Samples were heated in boiling water for 2.5 min after mixing them with concentrated SDS sample buffer to obtain 0.0625 M Tris-HCl (pH 6.8), 2% SDS, 5% (vol/vol) 2-mercaptoethanol, 10% (vol/vol) glycerol, and ∼0.01% bromphenol blue. To avoid artifactual cross-linking of HK97 capsid proteins, most samples were rapidly denatured using trichloroacetic acid (TCA) precipitation before being solubilized in SDS (11). Samples were rapidly mixed into ice-cold 10% TCA, left on ice for 10 to 30 min, centrifuged to collect the precipitated protein, which was then washed with cold acetone and recentrifuged, dried under vacuum, suspended in SDS sample buffer, and heated.
Expansion and cross-linking reactions.
SDS-glycerol expansion was induced by mixing samples with SDS sample buffer, followed by incubation at room temperature for 1 h or overnight. Reactions were stopped by heating in boiling water for 2.5 min. Low-pH/high-salt expansion was induced by diluting Prohead II to 2 mg/ml in expansion buffer (50 mM sodium acetate [pH 4.1], 400 mM KCl), and these reactions were stopped at the times indicated in the figures and legends by two- or fourfold dilution into 100 mM Tris-HCl (pH 8.0) for agarose gel analysis or by TCA precipitation for SDS gels. Dimethyl formamide (DMF) expansion was induced by diluting Prohead II samples ∼10-fold to ∼2 mg/ml into 40% (vol/vol) DMF in TAMg (agarose gel) buffer. After 1 h, the samples were diluted 10-fold into TAMg buffer without DMF and allowed to incubate until samples were taken for analysis, as indicated. Samples were loaded directly onto agarose gels after the addition of dyes and glycerol or TCA precipitated to stop all further reactions for SDS gels at the times indicated in the figures and legends.
Expansion rates were calculated by a simple exponential curve fitting using Kaleidagraph 4 (Synergy Software, Reading, PA) from densitometry data obtained from stained agarose gels using an Aristo V810 grid light box (Aristo Grid Lamp Products, Waterbury, CT) and the EDAS 290 camera system and 1D Image Analysis Software from Kodak (Rochester, NY).
Electron microscopy.
Prohead samples were adsorbed to carbon films and examined in a Zeiss EM902 electron microscope at 80 kV after negative staining with aqueous 0.5% uranyl acetate. Prohead samples were diluted 1/50 to 1/200 with TKG buffer; adsorbed to glow-discharged, carbon-coated Parlodion films on 400 mesh grids for 1 to 2 min; washed with 3 to 5 drops of water; and stained for 30 s. Grids were examined using a liquid nitrogen-cooled anticontaminator during operation. Images were recorded on Kodak 4489 film at a magnification of approximately ×30,000.
RESULTS
Most substitutions at glutamic acid 363 allow particle assembly.
The HK97 cross-linking reaction is not a conventional enzymatic reaction—it takes place only within a particle with ∼400 subunits and only during or after the transformation to mature capsid, so it was important to determine whether substitutions for E363 support formation of a functional particle, Prohead II, before testing the cross-linking phenotypes of mutants. E363 in the HK97 major capsid protein was changed in plasmid clones to alanine, aspartic acid, histidine, lysine, tyrosine, glutamine, and asparagine by mutating codon 363 of HK97 gene 5 using a PCR-based strategy. Each variant was named for its substitution, for example, E363A. The mutations were created in a plasmid that produces the HK97 major capsid protein and protease and thus makes wild-type Prohead II at high efficiency when expression is induced (11). Complementation tests were done to compare the biological function of each of the mutants to wild type (39). None of the mutant proteins was able to support production of viable phage particles in the complementation tests (Table 1). In addition, the production of the mutant proteins interfered with the growth of wild-type phage.
TABLE 1.
Complementation tests for function of mutant proteins substituted at E363 in the HK97 major capsid protein
| Plasmid | Relative EOPa for phage
|
|
|---|---|---|
| Amber mutant in MCP | Wild typeb | |
| Wild type | 1.0 | 1.0 |
| E363A | 0.0001 | 0.03 |
| E363D | 0.0001 | 0.01 |
| E363H | 0.0001 | 0.03 |
| E363K | 0.0001 | 0.03 |
| E363N | 0.0001 | 0.03 |
| E363Q | 0.0003 | 0.1 |
| E363Y | 0.0003 | 0.01 |
| Vector | 0.001 | 1.0 |
The relative EOPs of a capsid protein amber mutant and wild-type phages on strains expressing E363 mutant proteins were measured as described in Materials and Methods. MCP, major capsid protein.
Poor growth of wild-type phage on strains producing mutant proteins show the dominant-negative effects of the mutant proteins on phage production.
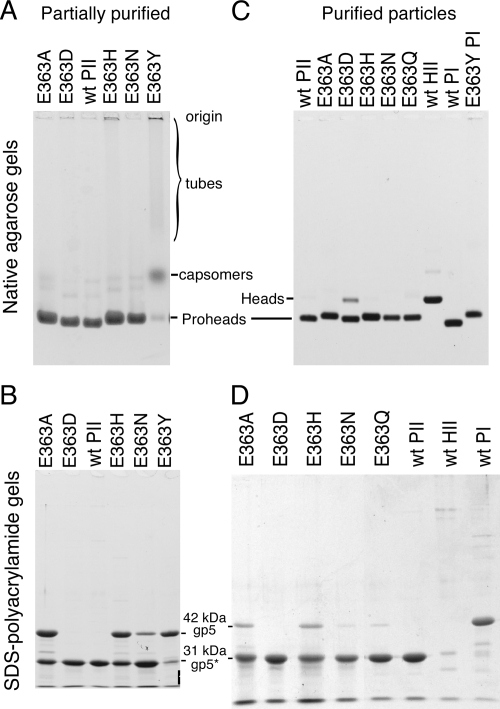
We purified prohead-sized particles from induced cultures of cells containing the mutant plasmids (11, 34) and analyzed the products by using native and denaturing gel electrophoresis and electron microscopy. HK97 proheads make a characteristic band in nondenaturing agarose gels, but Prohead I overlaps Prohead II (Fig. 3), so SDS-polyacrylamide gels were used to distinguish Prohead I, which is composed of the 42-kDa major capsid protein (gp5), from Prohead II, which contains only the 31-kDa (gp5*) cleaved form of the major capsid protein. By these criteria, five of the mutants made Prohead II, as shown in Fig. 3, but some assembly defects were observed, the most common being accumulation of Prohead I. Except for E363D, the mutant proheads had lower mobility in agarose gels due to the effect of the mutations on the charge of the particles.
FIG. 3.
Analysis of E363 mutant proheads on agarose and SDS gels. (A) Agarose gel of partially purified particles. Prohead I and Prohead II migrate similarly, but mutants vary in mobility due to changes in charge. Expanded particles (Head II) have lower mobility; tubes appear as material stuck in sample wells or as smears (see lanes for E363H and E363Y). (B) SDS-polyacrylamide gel of samples shown in panel B. A 31-kDa band of cleaved gp5* indicates that Prohead II is present. A 42-kDa band of uncleaved gp5 may derive from Prohead I particles, but the tubular structures made by mutants are also made of uncleaved protein. (C) Further purification of mutant proheads reduces the tubular forms, as shown by the lack of smears in the agarose gel shown, leaving mostly proheads. (D) An SDS gel shows that additional purification steps have decreased the portion of Prohead I.
The most conservative replacement, the aspartic acid substitution E363D, behaved the most like wild type of all of the mutants. E363D produced Prohead II exclusively, as indicated by a prohead band in agarose gels (Fig. 3A) and the presence of only the 31-kDa cleaved version of the major capsid protein in an SDS gel (Fig. 3B). The E363D particles also had wild-type morphology when examined in the electron microscope (Fig. 4A). Mutants E363A, E363N, E363H, and E363Q all produced a prohead band in agarose gels (Fig. 3), but the presence of both cleaved and uncleaved major capsid protein bands in the SDS gel lanes (Fig. 3B and D) showed that each of these four mutants accumulated some Prohead I in addition to Prohead II. The recovery of both Prohead I and Prohead II suggests that proteolytic processing is altered by these four mutations, which was not expected because the changes affect a part of the protein far from the sites of protease action. We argue below that incomplete proteolysis could be an indirect effect of slow assembly in these mutants.
FIG. 4.
Morphology of assembled and misassembled E363 mutant proteins. Samples of partially purified wild-type and mutant proheads were examined by electron microscopy using negative stain as described under Materials and Methods. (A) Mutant E363D proheads appeared to be uniform and similar to wild-type particles (not shown). (B) Mutant E363H makes both round, prohead-like particles and long tubes of capsid protein. (C) Mutant E363Y makes mostly long tubes of capsid protein.
The E363H mutant produced a substantial amount of correctly assembled Prohead II (Fig. 3), along with some Prohead I, but it also produced long tubes of capsid protein that were visible by electron microscopy as shown in Fig. 4B. This is a more drastic assembly defect than seen in the E363D, E363Q, E363A, or E363N mutants. The E363Y mutant produced small amounts of Prohead I and unassembled capsomers, but the bulk of the capsid protein was assembled into very long tubes (Fig. 4C) that were progressively lost during purification, since the protocol is optimized to purify prohead-sized particles. It was possible to obtain a small amount of pure E363Y Prohead I by using ion-exchange chromatography (Fig. 3C). On agarose gels, the E363Y tubes appear as a broad, slowly migrating smear plus material trapped in the wells (Fig. 3A); the E363H tubes appear as a similar smear and well-entrapped material. The lysine mutant E363K produced no assembled proheads (data not shown) and was not examined further. The mixtures of Prohead II and Prohead I obtained from four of the mutants were treated with glucose and high salt to dissociate Prohead I (34) and repurified. For E363N and E363Q, this produced relatively pure Prohead II (Fig. 3D).
E363 mutant proheads are capable of expansion.
In wild-type HK97 particles, cross-links appear during the expansion transformation as the active sites for cross-linking are formed (15). Because cross-linking is so tightly coupled to expansion, we had to be sure that all of the E363 mutant proheads were able to expand before we could confidently interpret our cross-linking experiments. Figure 5 shows an agarose gel of the set of mutant proheads after treatment with DMF at 36% (vol/vol) and pH 8. All of the tested particles expanded by the criterion of producing a slower-migrating band. In the cases of E363H and E363Y, the Prohead I present in the purified mutant preparations also expanded, producing a low-mobility band identified in Fig. 5. Expanded Prohead I is not on the normal HK97 maturation pathway, but we have described this particle before; it looks similar to expanded HK97 Head II in a cryo-EM reconstruction and is only partially cross-linked, possibly because uncleaved capsid protein prevents complete expansion (40).
FIG. 5.
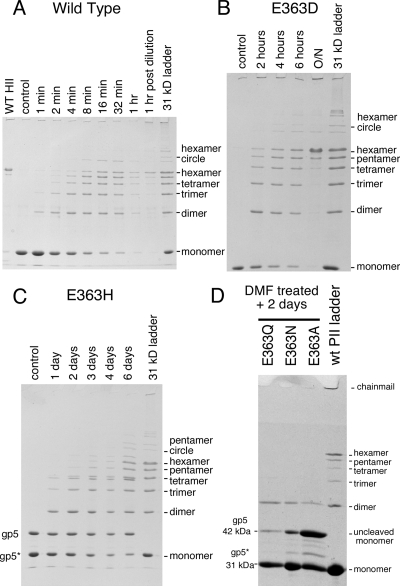
Expansion of mutant proheads. (A) Expansion of mutants induced by DMF. Wild-type and mutant Prohead II particles were diluted into TAMg agarose gel buffer (wild-type control) or TAMg containing 40% (vol/vol) DMF, left overnight, run in a 0.9% agarose gel, and stained. The low-mobility Head bands in the gel indicate that all DMF-treated Prohead II particles expanded into heads. E363H and E363Y Prohead I expanded to yield the lower-mobility expanded Prohead I band as indicated; wild-type Prohead I and the Prohead I present in the E363A sample failed to expand under the same conditions. (B) Expansion of E363 mutant proheads at pH 4.1. Proheads were treated at pH 4.1 in 0.4 M KCl; at the given times, reactions were stopped by dilution into a neutralizing buffer and run in an agarose gel. The fraction of particles expanded as measured from gel bands by densitometry was plotted versus time. The expansion rates (where the fraction expanded α1 − e−kt and t = time in minutes in the gel assay) were as follows: wild-type, k ≈ 1.0 × 10−2 min−1; E363Q, 1.2 × 10−2 min−1; E363H, 1.3 × 10−2 min−1; E363A, 3.6 × 10−3 min−1; E363N, ∼3.0 × 10−4 min−1; and E363D, ∼3.4 × 10−4 min−1.
We also tested the expansion of the E363 mutants under one of the extensively used conditions known to induce expansion in vitro, low pH in high salt (15, 31, 32). Wild-type and mutant Prohead II samples were treated at pH 4.1 in 0.4 M KCl for various times, diluted to neutralize and stop expansion, and analyzed on agarose gels to check for conversion from Prohead II to Head. The data for all of the mutants are shown graphically in Fig. 5, which shows that all mutants were capable of expansion, although the rate of expansion varied over a wide range for the mutants under these conditions. There was no obvious relationship between the rates of expansion at low pH, as measured by this assay, and the cross-linking properties of the amino acid substitutions revealed in other assays.
Cross-linking of E363 mutants: three different assays.
The ability of the E363 mutants to promote cross-link formation was examined by using three different treatments that induce the expansion and cross-linking of wild-type proheads. The conditions were as follows: (i) SDS-glycerol gel sample buffer, (ii) pH 4 in high salt, and (iii) 36% DMF at pH 8. Each of these conditions triggers the HK97 expansion transformation that is necessary for cross-links to form. Although these three conditions are quite different from each other, the results obtained generally agree well across the three conditions (and with others not shown here), and this increases our confidence that the results reflect fundamental properties of the proheads. The results are summarized in Table 2 and described in detail below.
TABLE 2.
In vitro capsid protein cross-linking in HK97 major capsid protein mutants
| Prohead | Cross-linking under various conditionsa
|
|||
|---|---|---|---|---|
| SDS-glycerol | High salt (pH 4.1)
|
DMF, 1 h, diluted 4 to 8 days | ||
| At pH 4 | Neutralized 3 to 6 days | |||
| Wild type | +++ | ++ | ++++ | ++++ |
| E363D | + | − | +++ | ++++ |
| E363H | + | − | +++ | +++ |
| E363Q | Trimer | − | Trimer | Trimer |
| E363N | Dimer | − | Dimer | Trimer |
| E363A | − | − | − | Dimer |
Purified mutant Prohead II particles were induced to expand and cross-link by various regimes described in the text and in Materials and Methods. Cross-linking result: ++++, nearly complete; +++, extended ladder, little monomer left; +, ladder present; -, no cross-linking. “Trimer” refers to trimer plus dimer; “dimer” indicates dimer, but no trimer.
SDS-induced cross-linking.
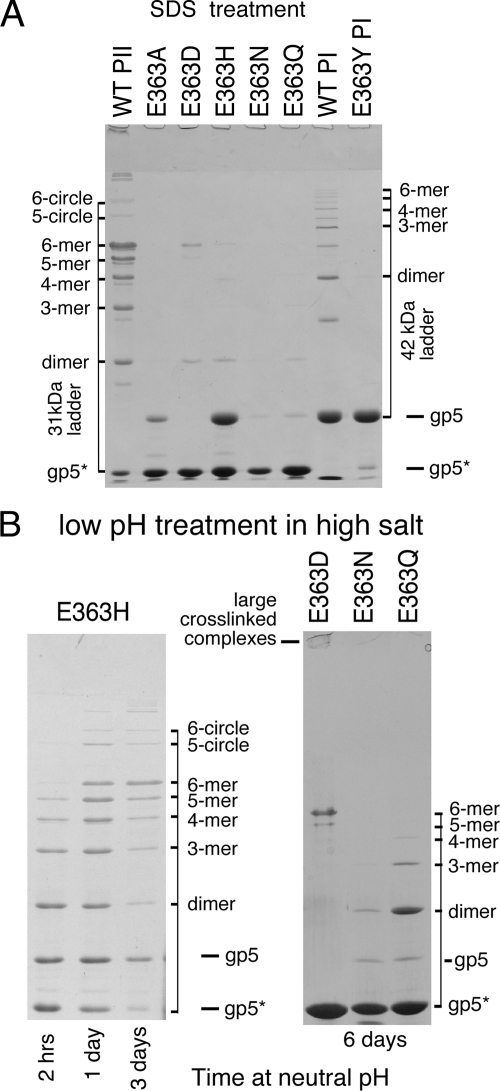
In the simple SDS-gel-buffer-induced cross-linking test (11), HK97 proheads are mixed with SDS gel sample buffer (containing glycerol and 2-mercaptoethanol), which causes wild-type proheads to begin to expand and cross-link spontaneously. After a few minutes of incubation with SDS gel buffer, Prohead I and Prohead II each normally produce a characteristic ladder of bands of cross-linked oligomers when subsequently analyzed on an SDS gel. A 1-h exposure to SDS-gel-buffer produced no evidence of cross-linking for any of the E363 mutant proheads (not shown). However, after overnight incubation (Fig. 6A), the E363D variant showed a small amount of cross-linked dimer, pentamer, and hexamer, E363H showed traces of dimer through hexamer, E363Q showed traces of dimer and trimer, E363N showed a trace of dimer, while E363A showed no cross-linking at all in this test.
FIG. 6.
Cross-linking induced by SDS-glycerol and low pH in high salt. (A) Wild-type (WT) and mutant Prohead II particles were mixed with SDS-glycerol gel sample buffer, left overnight, and then denatured at 100°C for 2.5 min before running on a 10% SDS-polyacrylamide gel. Treatment of wild-type proheads produced particles with a broad range of cross-link species for both Prohead II and Prohead I, as indicated; mutants E363D, E363H, E363Q, and E363N all show capsid protein dimers and some have larger oligomers. (B, left). E363H proheads were diluted 10-fold into expansion buffer (50 mM sodium acetate [pH 4.1], 400 mM KCl), left for 1 h, and then neutralized by diluting twofold into 0.1 M Tris-HCl (pH 8.0). Samples were TCA precipitated, denatured in SDS, and run on a SDS-10% polyacrylamide gel. The sample had both Prohead I and Prohead II, which expanded and cross-linked, yielding oligomers of 31- and 42-kDa monomers. (B, right) E363D, E363N, and E363Q Prohead II particles (∼20 mg/ml) were diluted 10-fold into 50 mM sodium acetate (pH 4.1)-400 mM KCl, left overnight, and then neutralized by diluting twofold into 0.1 M Tris-HCl (pH 8.0). After neutralization, samples were TCA precipitated at the times indicated in the figure and run on a SDS-10% polyacrylamide gel.
For the E363Y mutant, we were not able to prepare Prohead II, but we were able to purify Prohead I. We exposed the E363Y Prohead I particles to SDS-gel-buffer expansion conditions (Fig. 6A) and saw no evidence of cross-linking; under these conditions, the wild-type Prohead I clearly showed a ladder of cross-linked products, demonstrating that expansion was induced. Prohead I does not normally expand in vivo, but wild-type Prohead I can undergo a substantial amount of cross-linking in vitro, both after treatment with SDS gel buffer (11) (Fig. 6A) or after low-pH-urea treatment (40).
Low-pH-induced cross-linking.
Acidification of Prohead II has been used extensively for studying HK97 capsid maturation (15, 31, 32, 50) and was used in the previous section to examine rates of expansion (Fig. 5). When we examined samples from the same acid treatment experiments on SDS gels, no cross-linking at all was detected for any of the E363 mutants as summarized in Table 2 (gel data not shown). On the other hand, when we examined mutant particles that had been briefly exposed to acid pH and subsequently neutralized, we found that many were able to cross-link (Table 2 and Fig. 6B). We found that mutants E363D and E363H were both readily able to form a significant complement of cross-links. The E363Q and E363N mutants were also able to form cross-links but only to a rather limited extent (Fig. 6B). The E363D proheads exhibited all-or-nothing behavior, in that only a small fraction of the E363D Prohead II expanded under the low-pH treatment, while those E363D proheads that did expand appeared to be able to cross-link completely, since only monomers and the few oligomers normally found in fully cross-linked particles (pentamer, hexamer, and larger oligomers) were observed except at early time points. The acid-treated E363H mutant proheads expanded rapidly but cross-linked slowly, acquiring progressively more cross-links when sampled at 1 and 3 days after neutralization (Fig. 6B). After acid treatment followed by incubation at neutral pH for several days, E363Q was able to form a small number of cross-linked dimers and trimers, whereas E363N produced only a small amount of cross-linked dimer. E363A showed no evidence for cross-linking in these assays (not shown).
DMF-induced cross-linking.
DMF is able to induce expansion rapidly for wild-type Prohead II and all of the mutants (see Fig. 5) using a simple protocol that does not use drastic pH changes or denaturants. We used DMF to induce expansion of the set of mutants and tested for cross-linking, in some cases for rather long time periods after DMF treatment. Proheads were first diluted 10-fold into buffered 40% DMF at 23°C to induce expansion. After 1 h, samples were diluted an additional 10-fold into buffer without DMF and tested for cross-linking at intervals (Table 2 and Fig. 7). Under these conditions, wild-type Prohead II cross-links rapidly, as shown in Fig. 7A; cross-linking is 50% complete within minutes, nearly complete at the end of the 1 h of DMF treatment, and complete soon after dilution from DMF. The D and H mutants clearly achieved extensive cross-linking, although the rate for each was much slower than for the wild type. The D mutant took hours to show substantial cross-linking and overnight to go to completion (Fig. 7B), while the H mutant actually took days to show substantial cross-linking and weeks of incubation to approach completion (Fig. 7C). Ten weeks after DMF treatment, the E363H sample appeared almost completely cross-linked (data not shown). The E363Q, E363N, and E363A particles all formed a small number of cross-links after several days of incubation, but only E363Q and E363N were able to form oligomers greater than the dimer, and they did not proceed past the trimer stage in these experiments.
FIG. 7.
DMF induces extensive cross-linking in mutant proheads containing proton-accepting substitutions for E363. Wild-type (WT), E363D, and E363H Prohead II particles were diluted into agarose gel buffer containing 40% DMF to induce expansion. For the wild type, samples were taken at the times indicated, TCA precipitated, and analyzed for cross-linking on SDS-polyacrylamide gels. After 1 h exposure to 36% DMF, the samples were diluted 10-fold into TAMg agarose gel buffer. (A) The wild type cross-linked significantly within minutes, even in the presence of DMF; samples were taken from 1 to 60 min before dilution from DMF and 1 h after. (B) E363D particles cross-linked over a period of hours after dilution from DMF. (C) E363H particles cross-linked over a period of days after dilution from DMF. (D) Low levels of cross-linking are seen in E363Q, E363N, and E363A mutants when the panel of mutant proheads were analyzed after 2 days; all showed cross-linked dimers; some trimers were seen in samples taken after longer incubations (not shown).
We can make semiquantitative estimates of the relative rates of cross-linking from the experiments shown in Fig. 7 by evaluating the “ladders” of bands of cross-linked capsid protein in these SDS gels as measures of the extent of cross-linking. For example, the 8-min ladder in the wild-type cross-linking experiment (Fig. 7A) is similar to the 4-h ladder in the E363D cross-linking experiment (Fig. 7B). This suggests an ∼30-fold difference in cross-linking rate, but this probably is an underestimate because the wild-type cross-linking took place in 36% DMF, while the E363D cross-linking happened after dilution to 4% DMF, and we know that high concentrations of organic solvents reduce the rate of cross-linking by severalfold (not shown). The 4-h ladder in the E363D cross-linking experiment is similar to the 4-day ladder for the E363H mutant (Fig. 7C), implying another ∼24-fold decrease in rate for the H mutant relative to the D mutant. In summary, our cross-linking experiments show that among the substitutions examined, the extent (and by implication, the rate) of cross-linking is always in the same order, namely, E ≫ D > H ≫ Q > N > A.
The results of the in vitro cross-linking experiments described above confirmed that replacing E363 with another acidic or mildly acid residue does not abolish the ability of HK97 to carry out cross-linking, although the rates of cross-link formation by these variants are very much reduced. The other three substitutions only exhibited very low levels of cross-linking. The alanine substitution showed the least cross-linking activity, which appears to be a result of the background or uncatalyzed cross-linking reaction.
DISCUSSION
Rapid cross-linking is essential for producing viable phage.
The cross-linking experiments described above show that HK97 proheads with mutations at residue E363 cannot cross-link to any substantial extent, with the exception of the aspartate and histidine mutants, which cross-link at much lower rates than wild type. However, complementation tests of the entire set of E363 mutants (Table 1) showed that every one of the substitution mutants, including the E363D and E363H mutants, are defective and even interfere with the growth of wild-type phage in a dominant-negative manner. Even though we have shown that the wild type, the aspartate mutant, and the histidine mutant are all capable of cross-linking completely in vitro, only the wild-type protein is capable of producing viable phages in vivo. The relative order of their reaction rates for the three cross-linking-competent proteins, i.e., E ≫ D > H, is preserved for all of the conditions we tested, although the absolute rates were quite varied. We argue below that these relative differences in cross-linking rate reflect real differences in the abilities of the different amino acid side chains to promote cross-linking. The dominant-negative character of the mutant proteins in the complementation tests further emphasizes that having a full or nearly full capability to cross-link rapidly is essential for viability. Thus, it appears that successful phage production requires not only that cross-links form but that cross-linking proceeds rapidly and promptly goes to completion, which only happens with the wild-type protein. We have previously shown that cross-linking starts early in the expansion process and continues during expansion (15). This leads to the picture that cross-linking starts early during DNA packaging and is sufficiently complete by the time packaging is done to protect the capsid from physical disruption due to the “DNA pressure” in the tightly packed capsid.
Assembly defects in E363 mutants are not the cause of defective cross-linking.
Six mutants at position 363 in the HK97 major capsid protein produced prohead-like particles. However, some mutations resulted in defective assembly, including accumulation of tubes or Prohead I. The cross-linking defects we observed could be a result of defective assembly or independent consequences of the mutations. Since cross-linking takes place at capsomer interfaces (Fig. 2), there is an inherent possibility that mutations in cross-linking residues may affect assembly. This is demonstrated by another substitution at a cross-linking residue, N356D (see Fig. 2), which makes only hexamers and pentamers that do not assemble and cannot cross-link (12).
In the mature capsid (Fig. 2), E363 projects from a β strand of one hexamer (or pentamer) toward the adjacent capsomer (hexamer or pentamer), where it has polar interactions with the cross-link (the former N356) and with another residue (R194) on the same subunit (22). In Prohead II (PDB IDs 1IF0 [6], 2GP1, and 3E8K [18]), the relative position of E363 is conserved, in that it remains at the interface between capsomers and preserves its polar interactions during expansion, including those with N356 and R194, which help to hold the capsid together as it expands (18). The relative location of E363 is also likely to be preserved between Prohead II and Prohead I, because their shells are so similar in cryo-EM reconstructions (6, 40). If this were true, it would explain how the negatively charged aspartic acid in the N356D mutant might prevent assembly by repelling the approach of a similarly charged E363 residue. Thus, it appears that E363 lies at the interfaces involved in assembly and plays one role there, as well as participating in cross-linking. This helps explain why the character of E363 substitutions roughly correlates with their assembly properties. The E363 substitutions with relatively small side chains (A, D, Q, and N) assembled into normal-appearing Prohead II, but the substitutions that had bulky-ring side chains (H and Y) also made tubes of capsid protein, because normal assembly is somehow thwarted by these larger side chains. The neutral (A, Q, N, and Y) or slightly positive (H) E363 substitutions accumulated Prohead I, but most produced Prohead II as well. We suggest that this “cleavage defect” may be a consequence of slow assembly due to weakened ionic interactions between mutant capsomers, which allows much of the protease to self-destruct before it is captured by assembly.
Since HK97 prohead assembly occurred with both large or small substitutions and with negatively charged or neutral or slightly positive substitutions at E363, we argue that the capsidlike structures made by these E363 mutants, once assembled, must be very similar to normal, wild-type capsids. In addition, these mutant proheads expanded under the same conditions as wild-type particles, further supporting the interpretation that the mutants behave very much like the wild type in the transformations required for cross-linking and making it reasonable to draw conclusions from our experiments about how the chemical nature of residue 363 influences cross-linking.
Cross-linking active site and possible mechanisms.
In the mature Head II structural model (22, 51), each cross-link is found in a tightly packed region bordered by two adjacent capsomers, one providing the N356 cross-link donor subunit and a second providing the E363-containing subunit (Fig. 2). The active site is capped by an E-loop carrying the cross-link acceptor, K169 from the subunit adjacent to the E363 donor subunit. This environment has several polar residues (including lysine, arginine, glutamate, and aspartate [data not shown, but see reference 22]), many of which are involved in polar interactions that tie the glutamate-bearing and asparagine-bearing subunits together across the capsomer boundaries. However, three hydrophobic residues also pack close to the asparagine-lysine cross-link and E363. A view of how the side chains of these three hydrophobic residues cluster around the isopeptide cross-link in Head II is shown Fig. 2C. Two of these, Met 339 (M339) and Leu 361 (L361), lie on either side of E363 on the same subunit. L361 is packed close to the cross-linking K169, which is flanked by the third hydrophobic side chain, Val 163 (V163), on the same E-loop. Thus, it appears that K169 and E363 are brought together within a relatively hydrophobic environment whenever the active site is formed. K169 and V163 are also part of the lid for the enclosed cavity that is created when the tip of the E-loop moves to lie against the capsid surface during maturation.
We have suggested one possible reaction mechanism before, that cross-linking proceeds via a nucleophilic displacement of the asparaginyl amide by the lysine side chain as outlined in Fig. 8 (22). This mechanism requires that the lysine side chain be uncharged, which is not likely when the E-loop is in the up position of Prohead II and the lysine side chain is exposed to solvent. The model predicts that when the lid of the reaction pocket is closed as the lysine side chain enters, a proton is transferred from the lysine to the glutamate, removing the positive charge from the lysine so that K169 can attack the asparagine carbonyl carbon. In aqueous solution, such a proton transfer would be strongly disfavored because the pKa values of both glutamate and lysine favor their charged forms at intermediate pH levels; however, we propose that the proton transfer would remove the energetic cost of burying both charged groups in the hydrophobic reaction pocket. The protonated glutamic acid is also positioned so that it could polarize the asparagine carbonyl carbon, making it more susceptible to attack by the nucleophilic uncharged amino group of K169.
FIG. 8.
Proposed mechanism of cross-linking. The proposed role of E363 in cross-linking (22). E363 extracts a proton from K169+ to produce a neutral amine, K169, a nucleophile that can attack the carbonyl carbon of N356 to produce an isopeptide cross-link between two polypeptide chains, with the loss of ammonia.
A variation on this proposed mechanism invokes the conversion of the N356 side chain to a succinimide, which would be more susceptible to attack by lysine. We have tested for the presence of succinimides by precipitating proteins from HK97 cross-linking reactions and treating the products with hydroxylamine anhydride to cleave any available succinimides (28), but we have thus far found no compelling evidence for a succinimide at position 356 in the HK97 subunit (data not shown). One other analogous reaction has been reported, in the tetrapeptide VNGA derived from a rabbit liver enzyme (35), which occurs via an attack by the N terminus of the peptide on the adjacent asparagine side chain to produce a cyclic peptide (35). However, it is not clear how the mechanism of this unusual peptide cyclization is related to the one used for HK97 cross-linking.
HK97 cross-linking: a proton acceptor appears to be required.
The results of cross-linking assays for the set of substitution mutants are consistent with the hypothesis that it is the ability of the side chain at position 363 to act as a proton acceptor (i.e., general base) that is crucial to promote cross-linking during HK97 capsid maturation, since the most effective substitutions for E363 were E363D and E363H, both of which share the potential to act as a proton acceptor. The large difference in the rate of cross-linking observed between the most conservative substitution, aspartate, and the wild type, glutamate, which differ only by one methylene group, further suggests that correct positioning of the carboxylate is crucial, as might be expected for a residue central to the chemical mechanism. An analogous substitution of glutamate by aspartate in the active site of the enzyme triose phosphate isomerase was found to decrease the enzymes' catalytic activity by 1,000-fold (27, 37).
Three other mutants, the tyrosine, lysine, and alanine substitutions, are not expected to have any ability to act as proton acceptors. The tyrosine mutant showed no ability to cross-link in our test using Prohead I. The lysine mutant appeared to be defective in folding or assembly, so we were unable to assess its effect on cross-linking. The side chain of the alanine mutant is so much smaller than the glutamate side chain that we assume the space occupied by the glutamate side chain in the wild-type structure is filled by water. Therefore, we suggest that the low level of cross-linking observed in the alanine mutant is likely to be the result of water acting as the proton acceptor, a situation that in many enzymes reduces the enzymatic activity more that a millionfold (27). The relatively simple catalytic mechanism we propose (proton transfer) is thus compatible with a very low level of cross-linking in the alanine mutant, as observed.
Two additional substitutions, asparagine and glutamine, have side chains that also should not serve as proton acceptors, but we find that both supported cross-linking that was slightly more extensive than that seen with the alanine mutant. These results seem inconsistent with a proton acceptor role for residue 363, but glutamine and asparagine are both known to be highly susceptible to deamidation (38). Deamidation of glutamine produces glutamic acid, which is the wild-type residue and so would support one cross-link for each deamidation. (17, 48, 52). So it seems plausible to us that the small increases above background levels in the extent of cross-linking for the Gln and Asn mutants are due to low-level side chain deamidation of these residues which can yield carboxylate-bearing side chains.
Similarities of phage capsid cross-linking and pilin cross-linking.
HK97-like asparagine-to-lysine isopeptide cross-links have also recently been shown to occur in the pili of the human pathogen S. pyogenes M1, and there are indications that the occurrence of such cross-links is widespread (26) in gram-positive organisms. Gram-positive bacterial pili are composed of long strings of pilin subunits—which in the case of the M1 strain of S. pyogenes are ca. 2 to 3 nm wide and ca. 10 nm long—that are covalently attached end-to-end to each other and to the cell surface by isopeptide bonds whose formation is catalyzed by enzymes called sortases (47, 53). In addition to these interchain, sortase-mediated isopeptide bonds, the pilin subunits themselves have now been shown to have two intrachain internal isopeptide bonds that are chemically analogous to the HK97 capsid cross-links and appear to have an analogous role in stabilizing the pilin structure (26, 53). In addition to reporting that the pilin subunits under study contained isopeptide bonds, the authors of that study were able to identify homologous isopeptide bonds in several structures already deposited in the Protein Data Bank (26). The local environments in which the pilin-type isopeptide bonds are found all have features in common with the surroundings in which the HK97 cross-links lie; the lysine-asparagine cross-links are within an enclosed pocket with surrounding hydrophobic residues (including several conserved tyrosines and phenylalanines in the case of the pilins), and there is an adjacent conserved glutamic acid side chain (or aspartic acid in some pilins). Figure 9 shows a comparison of X-ray models of the “active sites” of isopeptide cross-linking in HK97 heads and S. pyogenes pilin protein, with a superposition of the structures based on an alignment of the side chains of the active site residues. Although the three active-site residues are nearly superimposable, the rest of the three-dimensional structures are completely different. In the case of the HK97 capsid, the three residues are donated from three different polypeptide chains, while the pilin reaction uses residues from three strands of the same beta-sheet containing domain. This suggests that whatever the mechanism for isopeptide bond formation, the phage capsid and pilin proteins have evolved a similar one, but they have arrived at it independently. We expect that asparagine-lysine isopeptide cross-links may exist in many additional natural systems, but the limitations of standard methods for biochemical and structural analysis make it difficult to discern when they are present.
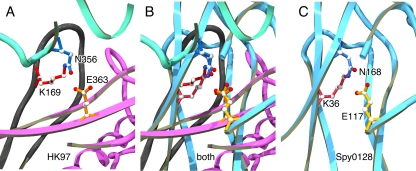
FIG. 9.
The configurations of cross-linking triads in HK97 capsids and the N-terminal domain of S. pyogenes pilin are similar. Panels A and C show close-up views of aligned cross-linking triads from one of the HK97 cross-link sites (from PDB ID 1OHG) (A) and the N-terminal cross-link site in the pilin Spy0128 from S. pyogenes M1 strain (PDB ID 3B2M) (C). (B) Overlay of the panels A and C. Parts of the X-ray models close to the cross-link triads are shown in ribbon mode, using a different color for each subunit. The cross-link side chains are in ball-and-stick mode identified by their amino acid codes and residue numbers; carbon atoms are shown in white, nitrogen in blue and oxygen in red. The two molecules were aligned by using the least squares fitting functions in the program O (Lsq_explicit and Lsq_molecule) (24) using one atom from each triad side chain, the γ carbons of their glutamates (CD) and asparagines (CG) and the ɛ carbons (CE) of their lysines. For these three atoms the final average root mean square deviation was 0.22 Å, while the isopeptide nitrogens are 1.3 Å apart.
Acknowledgments
This study was supported by National Institutes of Health grant GM47795 to R.W.H.
We thank our many collaborators for the ideas and enthusiasm generated by our interactions, including J. Johnson for discussions of reaction mechanisms and L. Liljas and C. Helgstrand for their analysis of the environment surrounding the active site.
Footnotes
Published ahead of print on 17 December 2008.
REFERENCES
- 1.Benzer, S. 1955. Fine structure of a genetic region in bacteriophage. Proc. Natl. Acad. Sci. USA 41344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borriss, M., T. Lombardot, F. O. Glockner, D. Becher, D. Albrecht, and T. Schweder. 2006. Genome and proteome characterization of the psychrophilic Flavobacterium bacteriophage 11b. Extremophiles 1195-104. [DOI] [PubMed] [Google Scholar]
- 3.Bukovska, G., L. Klucar, C. Vlcek, J. Adamovic, J. Turna, and J. Timko. 2006. Complete nucleotide sequence and genome analysis of bacteriophage BFK20—a lytic phage of the industrial producer Brevibacterium flavum. Virology 34857-71. [DOI] [PubMed] [Google Scholar]
- 4.Cerritelli, M. E., J. F. Conway, N. Cheng, B. L. Trus, and A. C. Steven. 2003. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv. Protein Chem. 64301-323. [DOI] [PubMed] [Google Scholar]
- 5.Conway, J. F., R. L. Duda, N. Cheng, R. W. Hendrix, and A. C. Steven. 1995. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J. Mol. Biol. 25386-99. [DOI] [PubMed] [Google Scholar]
- 6.Conway, J. F., W. R. Wikoff, N. Cheng, R. L. Duda, R. W. Hendrix, J. E. Johnson, and A. C. Steven. 2001. Virus maturation involving large subunit rotations and local refolding. Science 292744-748. [DOI] [PubMed] [Google Scholar]
- 7.Dhillon, E. K., T. S. Dhillon, A. N. Lai, and S. Linn. 1980. Host range, immunity and antigenic properties of lambdoid coliphage HK97. J. Gen. Virol. 50217-220. [DOI] [PubMed] [Google Scholar]
- 8.Doermann, A. H., and L. Boehner. 1970. The identification of complex genotypes in bacteriophage T4. I. Methods Genet. 66417-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreyfuss, G., S. A. Adam, and Y. D. Choi. 1984. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol. Cell. Biol. 4415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duda, R. L. 1998. Protein chainmail: catenated protein in viral capsids. Cell 9455-60. [DOI] [PubMed] [Google Scholar]
- 11.Duda, R. L., J. Hempel, H. Michel, J. Shabanowitz, D. Hunt, and R. W. Hendrix. 1995. Structural transitions during bacteriophage HK97 head assembly. J. Mol. Biol. 247618-635. [DOI] [PubMed] [Google Scholar]
- 12.Duda, R. L., K. Martincic, and R. W. Hendrix. 1995. Genetic basis of bacteriophage HK97 prohead assembly. J. Mol. Biol. 247636-647. [DOI] [PubMed] [Google Scholar]
- 13.Earnshaw, W. C., and S. R. Casjens. 1980. DNA packaging by the double-stranded DNA bacteriophages. Cell 21319-331. [DOI] [PubMed] [Google Scholar]
- 14.Galisteo, M. L., and J. King. 1993. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys. J. 65227-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gan, L., J. F. Conway, B. A. Firek, N. Cheng, R. W. Hendrix, A. C. Steven, J. E. Johnson, and R. L. Duda. 2004. Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Mol. Cell 14559-569. [DOI] [PubMed] [Google Scholar]
- 16.Gan, L., J. A. Speir, J. F. Conway, G. Lander, N. Cheng, B. A. Firek, R. W. Hendrix, R. L. Duda, L. Liljas, and J. E. Johnson. 2006. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure 141655-1665. [DOI] [PubMed] [Google Scholar]
- 17.Geiger, T., and S. Clarke. 1987. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides: succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 262785-794. [PubMed] [Google Scholar]
- 18.Gertsman, I., L. Gan, M. Guttman, J. A. Speir, R. L. Duda, R. W. Hendrix, E. A. Komives, and J. E. Johnson. An unexpected twist in viral capsid maturation. Nature, in press. [DOI] [PMC free article] [PubMed]
- 19.Gilakjan, Z. A., and A. M. Kropinski. 1999. Cloning and analysis of the capsid morphogenesis genes of Pseudomonas aeruginosa bacteriophage D3: another example of protein chain mail? J. Bacteriol. 1817221-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grayson, P., and I. J. Molineux. 2007. Is phage DNA ‘injected’ into cells: biologists and physicists can agree. Curr. Opin. Microbiol. 10401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 22.Helgstrand, C., W. R. Wikoff, R. L. Duda, R. W. Hendrix, J. E. Johnson, and L. Liljas. 2003. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 Å resolution. J. Mol. Biol. 334885-899. [DOI] [PubMed] [Google Scholar]
- 23.Hendrix, R. W. 2005. Bacteriophage HK97: assembly of the capsid and evolutionary connections. Adv. Virus Res. 641-14. [DOI] [PubMed] [Google Scholar]
- 24.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt. 2)110-119. [DOI] [PubMed] [Google Scholar]
- 25.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 29927-51. [DOI] [PubMed] [Google Scholar]
- 26.Kang, H. J., F. Coulibaly, F. Clow, T. Proft, and E. N. Baker. 2007. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 3181625-1628. [DOI] [PubMed] [Google Scholar]
- 27.Knowles, J. R. 1991. Enzyme catalysis: not different, just better. Nature 350121-124. [DOI] [PubMed] [Google Scholar]
- 28.Kwong, M. Y., and R. J. Harris. 1994. Identification of succinimide sites in proteins by N-terminal sequence analysis after alkaline hydroxylamine cleavage. Protein Sci. 3147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 30.Landt, O., H. P. Grunert, and U. Hahn. 1990. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96125-128. [DOI] [PubMed] [Google Scholar]
- 31.Lata, R., J. F. Conway, N. Cheng, R. L. Duda, R. W. Hendrix, W. R. Wikoff, J. E. Johnson, H. Tsuruta, and A. C. Steven. 2000. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell 100253-263. [DOI] [PubMed] [Google Scholar]
- 32.Lee, K. K., L. Gan, H. Tsuruta, R. W. Hendrix, R. L. Duda, and J. E. Johnson. 2004. Evidence that a local refolding event triggers maturation of HK97 bacteriophage capsid. J. Mol. Biol. 340419-433. [DOI] [PubMed] [Google Scholar]
- 33.Lee, K. K., H. Tsuruta, R. W. Hendrix, R. L. Duda, and J. E. Johnson. 2005. Cooperative reorganization of a 420 subunit virus capsid. J. Mol. Biol. 352723-735. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y., J. F. Conway, N. Cheng, A. C. Steven, R. W. Hendrix, and R. L. Duda. 2005. Control of virus assembly: HK97 “whiffleball” mutant capsids without pentons. J. Mol. Biol. 348167-182. [DOI] [PubMed] [Google Scholar]
- 34a.Li, Y. 2001. Genetic and functional analysis of the major capsid protein of bacteriophage HK97. Ph.D. thesis. University of Pittsburgh, Pittsburgh, PA.
- 35.Lura, R., and V. Schirch. 1988. Role of peptide conformation in the rate and mechanism of deamidation of asparaginyl residues. Biochemistry 277671-7677. [DOI] [PubMed] [Google Scholar]
- 36.Popa, M. P., T. A. McKelvey, J. Hempel, and R. W. Hendrix. 1991. Bacteriophage HK97 structure: wholesale covalent cross-linking between the major head shell subunits. J. Virol. 653227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raines, R. T., E. L. Sutton, D. R. Straus, W. Gilbert, and J. R. Knowles. 1986. Reaction energetics of a mutant triosephosphate isomerase in which the active-site glutamate has been changed to aspartate. Biochemistry 257142-7154. [DOI] [PubMed] [Google Scholar]
- 38.Robinson, N. E., and A. B. Robinson. 2004. Molecular clocks: deamidation of asparaginyl and glutaminyl residues in peptides and proteins. Althouse Press, Cave Junction, OR.
- 39.Ross, P. D., N. Cheng, J. F. Conway, B. A. Firek, R. W. Hendrix, R. L. Duda, and A. C. Steven. 2005. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. EMBO J. 241352-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross, P. D., J. F. Conway, N. Cheng, L. Dierkes, B. A. Firek, R. W. Hendrix, A. C. Steven, and R. L. Duda. 2006. A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J. Mol. Biol. 364512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 413748-752. [DOI] [PubMed] [Google Scholar]
- 42.Sternberg, N., and R. Weisberg. 1977. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 117733-759. [DOI] [PubMed] [Google Scholar]
- 43.Steven, A. C., H. L. Greenstone, F. P. Booy, L. W. Black, and P. D. Ross. 1992. Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, co-operativity, and super-stabilization by soc binding. J. Mol. Biol. 228870-884. [DOI] [PubMed] [Google Scholar]
- 44.Steven, A. C., J. B. Heymann, N. Cheng, B. L. Trus, and J. F. Conway. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 46.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 47.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in gram-positive bacteria. Trends Microbiol. 12228-234. [DOI] [PubMed] [Google Scholar]
- 48.Tyler-Cross, R., and V. Schirch. 1991. Effects of amino acid sequence, buffers, and ionic strength on the rate and mechanism of deamidation of asparagine residues in small peptides. J. Biol. Chem. 26622549-22556. [PubMed] [Google Scholar]
- 49.Tzlil, S., J. T. Kindt, W. M. Gelbart, and A. Ben-Shaul. 2003. Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 841616-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wikoff, W. R., J. F. Conway, J. Tang, K. K. Lee, L. Gan, N. Cheng, R. L. Duda, R. W. Hendrix, A. C. Steven, and J. E. Johnson. 2006. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J. Struct. Biol. 153300-306. [DOI] [PubMed] [Google Scholar]
- 51.Wikoff, W. R., L. Liljas, R. L. Duda, H. Tsuruta, R. W. Hendrix, and J. E. Johnson. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 2892129-2133. [DOI] [PubMed] [Google Scholar]
- 52.Xie, M., J. Aube, R. T. Borchardt, M. Morton, E. M. Topp, D. Vander Velde, and R. L. Schowen. 2000. Reactivity toward deamidation of asparagine residues in beta-turn structures. J. Pept. Res. 56165-171. [DOI] [PubMed] [Google Scholar]
- 53.Yeates, T. O., and R. T. Clubb. 2007. Biochemistry How some pili pull. Science 3181558-1559. [DOI] [PubMed] [Google Scholar]
- 54.Yuzenkova, J., S. Nechaev, J. Berlin, D. Rogulja, K. Kuznedelov, R. Inman, A. Mushegian, and K. Severinov. 2003. Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J. Mol. Biol. 330735-748. [DOI] [PubMed] [Google Scholar]