Abstract
We propose that a nucleotide template-based mechanism facilitates the acquisition of the K65R mutation in subtype C human immunodeficiency virus type 1 (HIV-1). Different patterns of DNA synthesis were observed using DNA templates from viruses of subtype B or C origin. When subtype C reverse transcriptase (RT) was employed to synthesize DNA from subtype C DNA templates, preferential pausing was seen at the nucleotide position responsible for the AAG-to-AGG K65R mutation. This did not occur when the subtype B RT and template were used. Template factors can therefore increase the probability of K65R development in subtype C HIV-1.
Antiretroviral drugs (ARVs) work by inhibiting the replication of human immunodeficiency virus type 1 (HIV-1). However, little information is available as to whether subtype diversity may affect responsiveness to therapy, and in general, the same treatment recommendations are applied to HIV-1 infections regardless of subtype (2, 9, 20, 21). Indeed, many reports have shown ARVs to be effective in the management of non-B infections. However, clinical studies in Western countries usually involve a follow-up of 2 to 4 years, in contrast to only 6 to 18 months in resource-poor settings (12). The conclusion that few subtype-specific differences may exist in regard to drug resistance mutations and responsiveness to therapy may be premature (2, 6, 9, 12, 13, 16).
Our laboratory has reported that the selection of the K65R resistance mutation by tenofovir occurs much faster in subtype C than in subtype B HIV-1 in cell culture (3). Recent findings also suggest that there may be an increased risk of K65R selection in subtype C infections after treatment failure (7; M. Hosseinipour, J. J. van Oosterhout, R. Weigel, J. Nelson, S. Fiscus, J. Eron, and J. Kumwenda, Resistance profile of patients failing first line ART in Malawi when using clinical and immunologic monitoring [TUAB0105], 17th International AIDS Conference, Mexico City, Mexico, 2008). The viral subtype should not directly impact treatment efficacy in the short term but may affect the durability of various treatment regimens in the event that some degree of viral replication and mutagenesis ultimately occur.
Recently we showed that there are few differences between the reverse transcriptases (RTs) of subtypes B and C on an enzymatic level (8). We hypothesized that some subtype differences may be governed by sequence polymorphisms. HIV-1 has high adenine content (∼35%), with many adenine homopolymer tracts, at the end of which RT exhibits characteristic pausing (11, 14, 18, 23). Such pause sites have been proposed to increase rates of strand transfer, recombination, misalignment, and misincorporation events that can contribute to the development of silent polymorphisms and drug resistance mutations (1, 4, 10, 14, 18, 24).
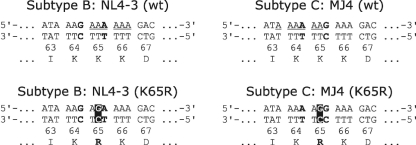
The K65R resistance mutation results from an AAA → AGA or AAG → AGG transition in HIV-1 subtypes B and C, respectively (Fig. 1). In both cases, the adenine located at the central position in the triplet codon mutates into a guanine; however, the nucleotide sequences of residues 64 and 65 are different in subtypes B and C. Based on codon usage, these polymorphisms should not yield any variation between the two subtypes. To better understand why K65R might be selected faster in subtype C under drug pressure, we now studied the acquisition of this mutation based on differences in template nucleotide sequences.
FIG. 1.
Comparison of genomic sequences of subtype B and C HIV-1. The double-stranded DNA of the pol gene that spans codons 63 to 67 is indicated. Bases that differ between the two subtypes are in bold. The site of acquisition of the K65R mutation due to an A-to-G change is highlighted. The adenine stretch, which ends at the bold G in subtype C, is underscored, whereas the stretch is shifted toward codon 66 in subtype B.
(Work by D.C. was performed in partial fulfillment of the requirements for a Ph.D. degree from the Faculty of Graduate Studies and Research, McGill University, Montréal, Québec, Canada.)
Reverse transcription involves the copying of positive-strand [(+)strand] RNA into negative-strand [(−)strand] DNA, followed by a first strand transfer and RNA degradation by the RNase H activity of RT. A second step involves the synthesis of (+)strand DNA after a second strand transfer to yield double-stranded DNA, which becomes integrated into the host cell genome. We designed specific templates to mimic (−)strand DNA synthesis from the (+)strand RNA template and (+)strand DNA synthesis from the (-)strand DNA template in the context of HIV-1 subtypes B and C. (See the supplemental material for methods and template sequences.)
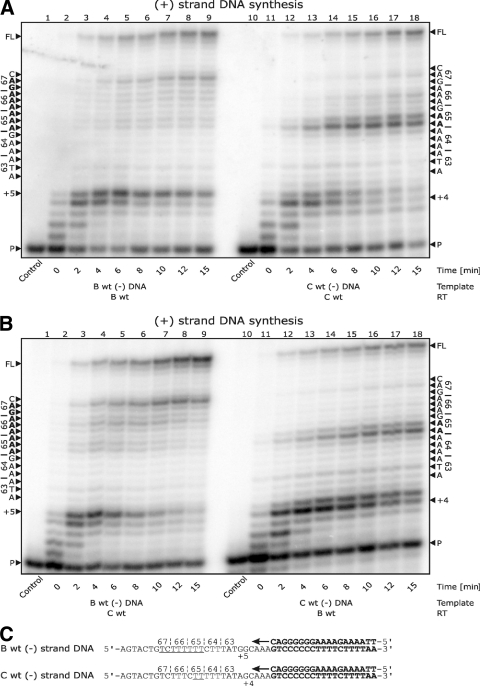
First, we compared (+)strand DNA synthesis from (−)strand DNA templates of either subtype B or C (Fig. 2). Time course experiments performed with a wild-type recombinant subtype C RT enzyme revealed pausing with the subtype C template at the last two adenine residues of the newly synthesized adenine stretch (Fig. 2A). The strongest pausing occurred at the first base of the coding sequence for residue 65, showing that RT is impaired in its ability to synthesize DNA at the exact nucleotide position responsible for the K65R mutation. As a control, we also used a recombinant subtype B RT enzyme and obtained similar results (Fig. 2B). Next, we assessed the combination of a subtype B (−)strand DNA template and subtype B RT; pausing occurred with a lesser intensity at a site immediately following the adenine stretch (Fig. 2A). In addition, a ladder of pausing sites of increasing intensities was observed throughout the adenine stretch in the pol coding sequence responsible for positions 65 to 67, regardless of whether RT of subtype B or C was employed (Fig. 2A and B). The pausing sites detected at the +5 and +4 positions are attributable to early stages of the initiation of (+)strand DNA synthesis in subtypes B and C, respectively.
FIG. 2.
(+)Strand DNA synthesis from the (−)strand DNA template of the position 65 region of pol. (A) Lanes 1 through 9 depict (+)strand DNA synthesis with subtype B wild-type RT on a subtype B template. A ladder of pausing is seen throughout the 65, 66, and 67 codons. Pausing at position +5 is attributable to early-stage initiation events. Lanes 10 through 18 depict (+)strand DNA synthesis with subtype C wild-type RT on a subtype C template. Strong pausing is seen at residue 65 and is associated with the more rapid development of the K65R mutation in subtype C. Pausing at position +4 is attributable to early-stage initiation events. (B) Lanes 1 through 9 depict (+)strand DNA synthesis with subtype C wild-type RT on a subtype B template, and lanes 10 through 18 depict (+)strand DNA synthesis with subtype B wild-type RT on a subtype C template, both showing similar results to those in panel A. The pausing events are independent of the RT enzymes and depend on the sequences used. (C) Depiction of the templates and primers used. Primers and primer annealing regions of the templates are in bold, and the regions at which pausing is seen on both templates are underlined.
These results demonstrate that the observed pausing is a nucleotide template-specific effect that is independent of the subtype of the RT enzyme used. Although adenine stretches are difficult for RT to synthesize (11, 14, 18, 23), there appear to be differences between such stretches in subtype B versus subtype C templates, such that the latter demonstrate more-accentuated pausing. Although not yet demonstrated, the strong pausing site with the subtype C template may result in dislocation mutagenesis, during which correct incorporation into a misaligned template primer followed by realignment could create the mismatch (23). Such a mechanism is consistent with the observed pausing. Both the homopolymeric nature of the nucleotide sequence of codons 64 and 65 of subtype C and the fact that the new base is the same as that found at the flank of the 5′ homopolymeric (−)strand DNA sequence help to explain why the K65R resistance mutation pathway may be more readily selected in subtype C.
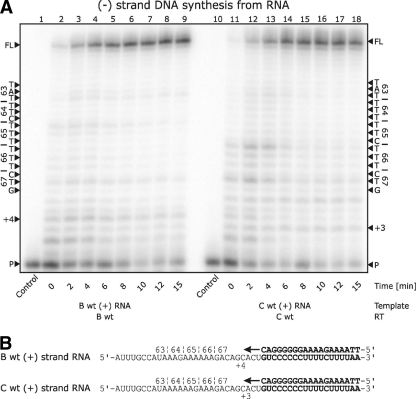
We also studied subtype B and C (+)strand RNA templates and tested them with their respective RT enzymes (Fig. 3). The results revealed that pausing at early stages of the initiation of (−)strand DNA synthesis occurred at the +4 and +3 positions in subtypes B and C, respectively. Although slight pausing was also seen at codon 66 of the subtype C sequence, corresponding to the end of a short homopolymer stretch, it was quickly alleviated as the reaction proceeded beyond 4 min, making it unlikely that a base substitution event during synthesis of (−)strand DNA from (+)strand RNA would occur.
FIG. 3.
(−)Strand DNA synthesis from the (+)strand RNA of the position 65 region of pol. (A) Lanes 1 through 9 depict (−)strand DNA synthesis with subtype B wild-type RT on a subtype B template, and lanes 10 through 18 depict (−)strand DNA synthesis with subtype C wild-type RT on a subtype C template. The data show no pausing for either enzymes or templates in the 63-to-67 region. Minor pausing attributable to initiation is seen at the +3 and +4 positions. (B) Depiction of the templates and primers used in the reaction.
These findings with DNA and RNA templates suggest that adenine/thymine-rich sequences may result in a dislocation at pausing sites which would allow for misaligned DNA synthesis and for a base substitution. In subtype C, the K65R mutation, due to an AAG → AGG transition during (+)strand DNA synthesis from the (−)strand DNA template, is likely to occur at elevated rates, whereas no increased mutation probabilities are expected during the synthesis of (−)strand DNA from the (+)strand RNA template.
The mechanism that we describe is based on the increased probability of a base substitution of HIV-1 RT in a subtype-specific nucleotide sequence, which results in the preferred selection of the K65R resistance mutation pathway in HIV-1 subtype C. This mutation can be readily maintained by any drug against which K65R confers resistance. Although the drug pressure that selects for K65R is similar in subtypes B and C, there is a fundamental difference between the two subtypes based on the propensity of the two respective templates to mutate at position 65. Previous studies have shown that the accuracy of DNA replication depends on the type of RT enzyme used and on the template sequence (11, 14, 19, 23). Even though the RTs used in our study are from different subtypes, little variation was seen at an enzymatic level (8). Therefore, variations at a genomic level of the pol nucleotide sequence, especially in homopolymers of adenine or thymine (11, 14, 18, 23), are probably responsible. Such nucleotide regions often exhibit pausing sites that may be linked to rates of base substitutions.
Clinical data available to date show noticeable differences in rates of acquisition of K65R between different subtypes (7) but not to the extent predicted by our proposed model. Although K65R confers moderate resistance against most approved nucleoside and nucleotide reverse transcriptase inhibitors (17, 22), high-level resistance may be accompanied by a loss of viral fitness (5). Furthermore, the use of an effective triple regimen should result in long-term suppression of the viral load and prevent the outgrowth of any mutated species, as has been shown in numerous clinical trials, including some in which subtype C-infected patients have been enrolled (6, 15, 16).
Supplementary Material
Acknowledgments
This research was supported by grants to M.A.W. from the Canadian Institutes of Health Research (CIHR) and the International Partnership on Microbicides (IPM). D.C. is the recipient of a CIHR M.D./Ph.D. fellowship award.
We thank Susan P. Colby-Germinario and Cesar Collazos for technical assistance.
Footnotes
Published ahead of print on 10 December 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bebenek, K., J. Abbotts, S. H. Wilson, and T. A. Kunkel. 1993. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 26810324-10334. [PubMed] [Google Scholar]
- 2.Brenner, B. G. 2007. Resistance and viral subtypes: how important are the differences and why do they occur? Curr. Opin. HIV AIDS 294-102. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, B. G., M. Oliveira, F. Doualla-Bell, D. D. Moisi, M. Ntemgwa, F. Frankel, M. Essex, and M. A. Wainberg. 2006. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS 20F9-F13. [DOI] [PubMed] [Google Scholar]
- 4.Buiser, R. G., R. A. Bambara, and P. J. Fay. 1993. Pausing by retroviral DNA polymerases promotes strand transfer from internal regions of RNA donor templates to homopolymeric acceptor templates. Biochim. Biophys. Acta 121620-30. [DOI] [PubMed] [Google Scholar]
- 5.Cong, M. E., W. Heneine, and J. G. Garcia-Lerma. 2007. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J. Virol. 813037-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DART. 2006. Virological response to a triple nucleoside/nucleotide analogue regimen over 48 weeks in HIV-1-infected adults in Africa. AIDS 20:1391-1399. [DOI] [PubMed] [Google Scholar]
- 7.Doualla-Bell, F., A. Avalos, B. Brenner, T. Gaolathe, M. Mine, S. Gaseitsiwe, M. Oliveira, D. Moisi, N. Ndwapi, H. Moffat, M. Essex, and M. A. Wainberg. 2006. High prevalence of the K65R mutation in human immunodeficiency virus type 1 subtype C isolates from infected patients in Botswana treated with didanosine-based regimens. Antimicrob. Agents Chemother. 504182-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel, F. A., C. F. Invernizzi, M. Oliveira, and M. A. Wainberg. 2007. Diminished efficiency of HIV-1 reverse transcriptase containing the K65R and M184V drug resistance mutations. AIDS 21665-675. [DOI] [PubMed] [Google Scholar]
- 9.Geretti, A. M. 2006. HIV-1 subtypes: epidemiology and significance for HIV management. Curr. Opin. Infect. Dis. 191-7. [DOI] [PubMed] [Google Scholar]
- 10.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 2501227-1233. [DOI] [PubMed] [Google Scholar]
- 11.Huber, H. E., J. M. McCoy, J. S. Seehra, and C. C. Richardson. 1989. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J. Biol. Chem. 2644669-4678. [PubMed] [Google Scholar]
- 12.Kantor, R. 2006. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr. Opin. Infect. Dis. 19594-606. [DOI] [PubMed] [Google Scholar]
- 13.Kantor, R., D. A. Katzenstein, B. Efron, A. P. Carvalho, B. Wynhoven, P. Cane, J. Clarke, S. Sirivichayakul, M. A. Soares, J. Snoeck, C. Pillay, H. Rudich, R. Rodrigues, A. Holguin, K. Ariyoshi, M. B. Bouzas, P. Cahn, W. Sugiura, V. Soriano, L. F. Brigido, Z. Grossman, L. Morris, A. M. Vandamme, A. Tanuri, P. Phanuphak, J. N. Weber, D. Pillay, P. R. Harrigan, R. Camacho, J. M. Schapiro, and R. W. Shafer. 2005. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klarmann, G. J., C. A. Schauber, and B. D. Preston. 1993. Template- directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J. Biol. Chem. 2689793-9802. [PubMed] [Google Scholar]
- 15.Margot, N. A., B. Lu, A. Cheng, and M. D. Miller. 2006. Resistance development over 144 weeks in treatment-naive patients receiving tenofovir disoproxil fumarate or stavudine with lamivudine and efavirenz in Study 903. HIV Med. 7442-450. [DOI] [PubMed] [Google Scholar]
- 16.Miller, M. D., N. Margot, D. McColl, and A. K. Cheng. 2007. K65R development among subtype C HIV-1-infected patients in tenofovir DF clinical trials. AIDS 21265-266. [DOI] [PubMed] [Google Scholar]
- 17.Parikh, U. M., D. L. Koontz, C. K. Chu, R. F. Schinazi, and J. W. Mellors. 2005. In vitro activity of structurally diverse nucleoside analogs against human immunodeficiency virus type 1 with the K65R mutation in reverse transcriptase. Antimicrob. Agents Chemother. 491139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson, J. T., D. G. Nickens, and D. H. Burke. 2006. HIV-1 reverse transcriptase pausing at bulky 2′ adducts is relieved by deletion of the RNase H domain. RNA Biol. 3163-169. [DOI] [PubMed] [Google Scholar]
- 19.Ricchetti, M., and H. Buc. 1990. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 91583-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomson, M. M., and R. Najera. 2005. Molecular epidemiology of HIV-1 variants in the global AIDS pandemic: an update. AIDS Rev. 7210-224. [PubMed] [Google Scholar]
- 21.Vergne, L., J. Snoeck, A. Aghokeng, B. Maes, D. Valea, E. Delaporte, A. M. Vandamme, M. Peeters, and K. Van Laethem. 2006. Genotypic drug resistance interpretation algorithms display high levels of discordance when applied to non-B strains from HIV-1 naive and treated patients. FEMS Immunol. Med. Microbiol. 4653-62. [DOI] [PubMed] [Google Scholar]
- 22.Wainberg, M. A., M. D. Miller, Y. Quan, H. Salomon, A. S. Mulato, P. D. Lamy, N. A. Margot, K. E. Anton, and J. M. Cherrington. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir. Ther. 487-94. [DOI] [PubMed] [Google Scholar]
- 23.Williams, K. J., L. A. Loeb, and M. Fry. 1990. Synthesis of DNA by human immunodeficiency virus reverse transcriptase is preferentially blocked at template oligo(deoxyadenosine) tracts. J. Biol. Chem. 26518682-18689. [PubMed] [Google Scholar]
- 24.Wu, W., B. M. Blumberg, P. J. Fay, and R. A. Bambara. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J. Biol. Chem. 270325-332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.