Abstract
Vaccinia virus is the prototypical member of the family Poxviridae. Three morphologically distinct forms are produced during infection: intracellular mature virions (IMV), intracellular enveloped virions (IEV), and extracellular enveloped virions (EEV). Two viral proteins, F12 and A36, are found exclusively on IEV but not on IMV and EEV. Analysis of membranes from infected cells showed that F12 was only associated with membranes and is not an integral membrane protein. A yeast two-hybrid assay revealed an interaction between amino acids 351 to 458 of F12 and amino acids 91 to 111 of A36. We generated a recombinant vaccinia virus that expresses an F12, which lacks residues 351 to 458. Characterization of this recombinant revealed a small-plaque phenotype and a subsequent defect in virus release similar to a recombinant virus that had F12L deleted. In addition, F12 lacking residues 351 to 458 was unable to associate with membranes in infected cells. These results suggest that F12 associates with IEV through an interaction with A36 and that this interaction is critical for the function of F12 during viral egress.
Vaccinia virus (VV) is a member of the Poxviridae family of viruses and replicates entirely in the cytoplasm of infected cells. During morphogenesis, the first infectious forms produced are called intracellular mature virions (IMV) (8, 12). A subset of IMV are transported along microtubules to the site of wrapping at the trans-Golgi network (6, 11) and early endosome (17), where they acquire two additional membranes and are termed intracellular enveloped virions (IEV). IEV are transported along microtubules to the cell periphery, where their outermost membrane fuses with the plasma membrane, releasing infectious virions onto the cell surface (9, 13). Surface-associated viral particles are termed cell-associated enveloped virions (CEV). Actin tails polymerized beneath CEV propel virions away from infected cells and toward adjacent cells, helping to facilitate cell-to-cell spread (7, 21). Some virions are released from the cell surface and are termed extracellular enveloped virions (EEV), which are involved in long-range virus dissemination (10).
There are seven known, IEV-specific, viral proteins: A33, A34, A36, A56, B5, F12, and F13 (reviewed by Smith et al. [15]). F12 and A36 are found exclusively on the outermost envelope of IEV and are not associated with CEV and EEV (18, 24). A36 has been reported to be involved in the transport of virions to the cell surface via an interaction with the molecular motor kinesin (20), as well as actin tail formation beneath CEV (5). F12 is a 65-kDa protein that is expressed predominantly late during infection (25). Deletion of the gene causes a small plaque phenotype on cell monolayers, indicating that F12 has a role in infectious enveloped virion production and/or release (13, 25). F12 is not predicted to contain a signal sequence or a transmembrane domain, and its method of association with the outermost IEV membrane is unknown. In this report, we show that F12 is associated with membranes through an interaction with A36. Furthermore, residues 351 to 458 of F12 were shown to be sufficient for interaction with A36 and association with membranes. Deletion of this region of F12 resulted in a small-plaque phenotype, less infectious CEV/EEV production and, consequently, less actin tail formation.
MATERIALS AND METHODS
Cells and viruses.
Monolayers of HeLa and BS-C-1 cells were maintained as described previously (3). The recombinant virus vB5R-GFP was a gift from Bernard Moss and has been previously described (22). Plaque assays and growth curves were performed as previously described (22).
Primer pair CGAATTCAGGTCAGCTAAGACTAG and CAGATCTGATGGCGTTCTATACGTT was used to amplify the 500-bp region upstream of the F12L open reading frame (ORF) (left flank) and primer pair CAGATCTCAGTTCAGTTATAAAGAA and ACTCGAGAGCTCCTTTGGTGAA was used to amplify the 500-bp region downstream of the F12L ORF (right flank). The two fragments were joined by ligation into pBMW-118 (19), which contains a gene coding for HcRed (BD Biosciences Clontech) and a xanthine-guanine phosphoribosyl-transferase selection cassette, to create pΔF12L-118. The left flank and coding sequence of F12L was amplified with the primer pair CGAATTCAGGTCAGCTAAGACTAG and CGGATCCTTAAGCGTAATCAGGCACGTCGTAAGGGTATAATTTTACCATCTGACTCATG, which added a 5′ EcoRI site (underlined) and a 3′ coding sequence for the hemagglutinin epitope (HA; italicized), followed by a BamHI site (underlined). The right flank was amplified by using the primer pair CGGATCCTAAAATTATAAAAAGTGAAAAACAATATTATTTTTATC and ACTCGAGAGCTCCTTTGGTGAA, which added corresponding 5′ BamHI and 3′ XhoI sites (underlined), respectively. The two fragments were joined by ligation into pBMW-118 to create pF12L-HA-118. The internal deletion of F12L-HA was generated by two-step PCR using pF12L-HA-118 as a template and primer pairs CGAATTCAGGTCAGCTAAGACTAG/GTTTTGAAGAAATGATTAAATCGAATGGTCGGCTCTC and CATTCGATTTAATCATTTCTTCAAAACAATACCC/ACTCGAGAGCTCCTTTGGTGAA to amplify the left flank plus the coding sequence of residues 1 to 350 and the coding sequence for residues 459 to 636 including the HA tag and the right flank, respectively. These two fragments were joined by a second PCR. The resulting fragment was cloned by using the TOPO TA cloning kit (Invitrogen) and sequenced. The fragment was subsequently cloned into pBMW-118 to create pF12LΔ351-458-HA-118.
The generation of the recombinant viruses vB5R-GFP/ΔF12L and vB5R-GFP/F12LΔ351-458-HA was performed by transient-dominant selection as previously described (4, 23). Briefly, HeLa cells infected with vB5R-GFP were transfected 2 h postinfection (p.i.) with either pΔF12L-118 or pF12LΔ351-458-HA-118 by using Lipofectamine (Invitrogen) according to the manufacturer's instructions. The following day, cells were harvested by scraping, lysed by freeze-thawing, and single-crossover recombinants were selected for by infecting BS-C-1 cells in the presence of xanthine (250 μg/ml), hypoxanthine (15 μg/ml), and mycophenolic acid (25 μg/ml). Plaques that fluoresced both green and red were picked and plaque purified three more times in the presence of selection. Subsequent plaque purification was performed four additional times in the absence of selection, with non-red plaques being picked each time. The recombinant viruses produced were amplified to high titer, and insertion of the recombinant DNA into the genome was verified via PCR and sequencing.
vB5R-GFP/F12L-HA was generated as follows. HeLa cells were infected with vB5R-GFP/ΔF12L and transfected with pF12L-HA-118 by using Lipofectamine (Invitrogen) according to the manufacturer's instructions. At 24 h p.i., cells were harvested and lysed by freeze-thawing, and the lysate was used to infect BS-C-1 cells. Plaques that fluoresced green were purified a total of four times on BS-C-1 cells. The recombinant virus was amplified and verified by PCR and sequencing.
Yeast two-hybrid constructs and assay.
Plasmids expressing the GAL4 DNA-binding domain (BD) fused to the cytoplasmic domains of A33, A34, B5, and various regions of A36 (A331-40-BD, A341-20-BD, B5300-317-BD, A3624-221-BD, A3624-80-BD, A3624-90-BD, A3624-111-BD, A3661-111-BD, A3671-111-BD, A3681-111-BD, and A3691-111-BD, respectively) were a gift from Bernard Moss and have been previously described (23). Plasmids that encode various regions of F12 fused to the GAL4 activation domain (AD) in pGADT7 (BD Biosciences Clontech) were constructed by amplifying the desired regions of F12L by PCR so that a 5′ NdeI and a 3′ EcoRI restriction endonuclease site was added. Amplified fragments were initially inserted into pGEM-T (Promega) and verified by sequencing. They were subsequently inserted in pGADT7 by using standard molecular biology techniques. The plasmids were named pGAD-F12L1-121, pGAD-F12L125-348, pGAD-F12L351-458, and pGAD-F12L456-635. Plasmids were transformed into yeast strain AH109 and assayed for interaction as previously described (23).
In vitro interaction assay.
The purification of GST-A36R24-111 has been previously described (20). In vitro transcription and translation of pGAD-F12L1-121 and pGAD-F12L351-458 in the presence of [35S]methionine-cysteine was performed by using the TNT quick-coupled transcription/translation system (Promega). Equal amounts of the products were subsequently incubated with purified glutathione S-transferase (GST) alone or GST-A3624-111 that had been bound to glutathione-Sepharose 4B beads (Amersham Pharmacia) in lysis buffer (25 mM Tris [pH 8], 0.5 M NaCl, 10 mM EDTA) as previously described (16). The beads were washed and resuspended in protein loading buffer. Protein complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and visualized via phosphorimaging. The intensity of the resulting bands was quantified with Kodak 1D image analysis software. The amount of GST and GST-A3624-111 used in the reaction was visualized by staining with SimplyBlue SafeStain (Invitrogen).
Immunofluorescence analysis.
HeLa cells were grown on coverslips and infected with vB5R-GFP, vB5R-GFP/F12L-HA, vB5R-GFP/F12LΔ351-458-HA, or vB5R-GFP/ΔF12L at a multiplicity of infection (MOI) of 1. Coverslips were harvested 24 h p.i., fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde, and permeabilized with PBS containing 10% fetal bovine serum and 0.1% saponin. Fixed cells were stained with anti-HA monoclonal antibody (MAb; Covance), followed by Texas Red-conjugated anti-mouse MAb (Jackson Immunoresearch Laboratories). Coverslips were washed three times in PBS and mounted onto slides with Mowiol that contained 1 μg of DAPI (4′,6′-diamidino-2-phenylindole; EM Sciences) to visualize DNA. To stain actin, permeabilized cells were incubated with 1 unit of Alexa Fluor 633 phalloidin (Molecular Probes) for 20 min at room temperature. To stain B5-GFP on the cell surface, fixed cells were stained without permeabilization using MAb 19C2 (11), followed by Texas Red-conjugated anti-rat MAb (Jackson Immunoresearch Laboratories). Cells were washed, incubated with 50 μg of Hoechst dye/ml, and mounted onto slides with Mowiol. Images were captured by using a Leica DMIRB inverted fluorescence microscope as described previously (19) and overlaid using Adobe PhotoShop software.
Coimmunoprecipitation.
Plasmids pA36R-V5, pA36R1-80-V5, and pA36R1-111-V5 were gifts of Bernard Moss and have been described previously (23). HeLa cells were infected with either vB5R-GFP, vB5R-GFP/F12L-HA, or vB5R-GFP/F12LΔ351-458-HA at an MOI of 5 in the presence or absence of 0.1 μg of the drug rifampin (Sigma-Aldrich)/ml. Cells were transfected at 2 h p.i., with either pA36R-V5, A36R1-80-V5, or A36R1-111-V5 using Lipofectamine (Invitrogen) according to the manufacturer's instructions. At 24 h p.i., cells were harvested by scraping, lysed in radioimmunoprecipitation assay buffer (half-strength PBS, 1% Triton X-100, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, and 0.2 mM phenylmethylsulfonyl fluoride), and centrifuged for 10 min at 14,000 × g to pellet nuclei. Supernatants were precleared with protein G- agarose (Calbiochem) for 2 h at 4°C and incubated with anti-HA MAb (Santa Cruz Biotechnology), followed by protein G-agarose (Calbiochem). Immune complexes were pelleted, washed three times in radioimmunoprecipitation assay buffer, boiled in protein loading buffer, and resolved by SDS-PAGE. Proteins were transferred to nitrocellulose membranes and incubated with either an anti-V5 horseradish peroxidase (HRP)-conjugated MAb (Invitrogen) or an anti-HA MAb (Roche), followed by an HRP-conjugated anti-rat MAb (Jackson Immunoresearch Laboratories). Bound antibodies were detected by using chemiluminescence analysis (Pierce).
Alkaline carbonate extraction.
HeLa cells were infected with either vB5R-GFP or vB5R-GFP/F12L-HA at an MOI of 10. At 24 h p.i., the cells were harvested by scraping, washed three times with PBS, and washed once with 100 mM NaCl. Cells were resuspended in 100 mM sodium carbonate (pH 11.5), homogenized by douncing, and incubated on ice for 30 min. The homogenate was centrifuged at 200,000 × g for 60 min at 4°C to pellet the membranes. Supernatants were removed and saved for analysis. Pellets were resuspended in an equal volume of 100 mM sodium carbonate (pH 11.5). Both the pellets and the supernatants were analyzed by Western blotting for the presence of F12-HA using an anti-HA MAb (Roche Diagnostics), B5-GFP using an anti-GFP MAb (Covance), and actin using an anti-actin MAb (Rockland).
Triton X-114 partitioning and membrane isolation.
Triton X-114 partitioning was performed as previously described (1). Briefly, BS-C-1 cells were infected at an MOI of 10 with either vB5R-GFP or vB5R-GFP/F12L-HA. At 24 h p.i., the medium was removed, and the cells were lysed in cold 10 mM Tris-HCl (pH 7.4) containing 2% Triton X-114 and 150 mM NaCl for 15 min on ice. The lysates were centrifuged for 7 min at 14,000 × g to remove nuclei and other insoluble material. Supernatants were layered onto a 6% sucrose cushion containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.06% Triton X-114 and warmed to 37°C until clouding occurred. The samples were centrifuged at 1,000 × g for 10 min, and the aqueous and detergent phases were extracted and analyzed by Western blotting with an anti-HA MAb (Roche Diagnostics) to detect F12-HA, anti-GFP MAb (Covance) to detect B5-GFP, and anti-actin MAb (Rockland) to detect actin as described above.
To isolate membranes, HeLa cells were infected with either vB5R-GFP, vB5R-GFP/F12L-HA, or vB5R-GFP/F12LΔ351-458-HA at an MOI of 10. At 24 h p.i., the medium was removed, and the cells were incubated in homogenization buffer (0.25 mM sucrose, 10 mM HEPES, 1 mM EDTA [pH 7.4]) containing protease inhibitor cocktail tablets (Roche Diagnostics) on ice for 20 min. Cells were disrupted by passage through a 27G syringe needle. The resulting lysates were centrifuged at 2,400 × g for 5 min to remove nuclei and insoluble material. Postnuclear lysates were centrifuged at 200,000 × g for 30 min at 4°C to pellet membranes. Supernatants were removed for analysis, and membrane pellets were suspended in an equal volume of PBS. Membrane pellets and supernatants were analyzed by Western blotting for the presence of F12-HA using an anti-HA MAb (Roche Diagnostics), B5-GFP using an anti-GFP MAb (Covance), and actin using an anti-actin MAb (Rockland).
RESULTS
F12 is not an integral membrane protein.
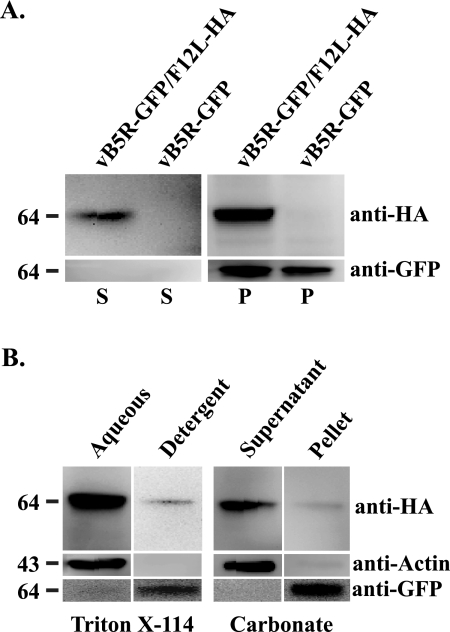
F12 is a 65-kDa protein that was shown to localize to IEV (18). Initially, we wanted to determine whether F12 is a membrane-associated or an integral membrane protein. To do this, we constructed a recombinant vaccinia virus that replaced the normal copy of F12L with one that contained the coding sequence for a C-terminal HA epitope tag. An identical construct was previously shown to functionally replace the normal F12 (25). Much like vB5R-GFP (22), our new recombinant, vB5R-GFP/F12L-HA, expressed a B5-GFP fusion protein to visualize and monitor morphogenesis at a subcellular level. Lysates were prepared from cells infected with either vB5R-GFP or vB5R-GFP/F12L-HA, and membranes in the lysates were pelleted by ultracentrifugation. The resulting supernatant and pellets were analyzed for the presence of F12-HA using Western blots. The anti-HA MAb specifically reacted with a ∼64-kDa band that was present predominantly in the pellet from cells infected with vB5R-GFP/F12L-HA, which was absent in preparations from cells infected with vB5R-GFP (Fig. 1A). Furthermore, the integral membrane protein B5-GFP was found almost entirely in the pellet from both infections. These results demonstrate that F12 is associated with membranes.
FIG. 1.
Membrane association of F12. Cells infected with either vB5R-GFP or vB5R-GFP/F12-HA were lysed. (A) Lysates were centrifuged to pellet membranes, and the resulting pellets (P) and supernatants (S) were separated by SDS-PAGE and analyzed by Western blotting with anti-HA MAb and anti-GFP MAb. (B) Cells were infected with vB5R-GFP/F12-HA and lysed 24 h p.i. The postnuclear supernatants were subjected to either Triton X-114 partitioning (B, left) or Na2CO3 extraction (B, right). For Triton X-114 partitioning, aqueous and detergent phases were separated by low-speed centrifugation, and each phase was analyzed by SDS-PAGE/Western blotting with the indicated antibodies. Na2CO3-treated membranes were pelleted, and the resulting pellet and supernatant were analyzed by SDS-PAGE/Western blotting with the indicated antibodies. The positions and masses (in kilodaltons) of markers are indicated on the left of each blot.
The amino acid sequence of F12 is not predicted to contain a transmembrane domain. It has been reported to contain two putative myristoylation sites at residues 129 and 551 (14) that may mediate its association with viral membranes. We used two standard assays, Triton X-114 partitioning and alkaline carbonate treatment, to determine whether F12 is a peripheral or an integral membrane protein. Lysates from cells infected with either vB5R-GFP or vB5R-GFP/F12L-HA were partitioned into two phases, aqueous and detergent, using Triton X-114 detergent, and each phase was analyzed for F12, actin and B5-GFP by using Western blotting. Both F12-HA and the soluble protein actin were predominantly in the aqueous phase, whereas the integral membrane protein B5-GFP was found in the detergent phase (Fig. 1B), indicating that the majority of F12 does not have an integral association with membranes. Likewise, treatment of membranes with sodium carbonate extracted F12-HA so that it did not pellet with membranes, while B5-GFP was not extracted (Fig. 1B). Both of these results indicate that the majority of F12 has a peripheral association with membranes. It is not an integral membrane protein nor does it associate with membranes through lipidation.
Residues 351 to 458 of F12 interact with the cytoplasmic domain of A36 in a yeast two-hybrid assay.
The previous results indicated that F12 is not an integral membrane protein. We hypothesized that F12 associates with membranes through an interaction with another IEV-specific, membrane-associated viral protein. To investigate this possibility, we used the yeast two-hybrid assay to look for interactions between F12 and the cytoplasmic domain of A33, A34, A36, and B5. In accordance with previous results, initial assays that used the full-length F12 fused to the Gal4 AD failed to detect an interaction (Table 1) (23). This could be due to the inefficient expression of vaccinia virus proteins from the nucleus. In an effort to circumvent this problem, we tested truncations of F12 and found that residues 351 to 458 of F12 interacted with the cytoplasmic tail of A36 but not A33, A34, or B5 (Table 1). In order to further define the region of A36 responsible for interaction with F12, we tested a series of previously constructed A36 truncations (23). The results of these assays showed that residues 91 to 111 of A36 are sufficient for interaction with residues 351 to 458 of F12 (Table 2).
TABLE 1.
Interaction of F12 with the cytoplasmic domain of various IEV proteins in a yeast two-hybrid assaya
| AD fusion | BD fusion | Growth on QDO |
|---|---|---|
| F12 (l-121) | A33 (1-40) | − |
| A34 (l-20) | − | |
| A36 (24-221) | − | |
| B5 (300-317) | − | |
| F12 (125-348) | A33 (1-40) | − |
| A34 (l-20) | − | |
| A36 (24-221) | − | |
| B5 (300-317) | − | |
| F12 (351-458) | A33 (1-40) | − |
| A34 (l-20) | − | |
| A36 (24-221) | + | |
| B5 (300-317) | − | |
| F12 (456-635) | A33 (1-40) | − |
| A34 (l-20) | − | |
| A36 (24-221) | − | |
| B5 (300-317) | − |
QDO, quadruple-dropout medium. The amino acid ranges are indicated in parentheses.
TABLE 2.
Interaction of F12351-458 with various regions of A36 in a yeast two-hybrid assaya
| AD fusion | BD fusion | Growth on QDO |
|---|---|---|
| F12 (351-458) | A36 (24-80) | − |
| F12 (351-458) | A36 (24-90) | − |
| F12 (351-458) | A36 (24-111) | + |
| F12 (351-458) | A36 (61-111) | + |
| F12 (351-458) | A36 (71-111) | + |
| F12 (351-458) | A36 (81-111) | + |
| F12 (351-458) | A36 (91-111) | + |
QDO, quadruple-dropout medium. The amino acid ranges are indicated in parentheses.
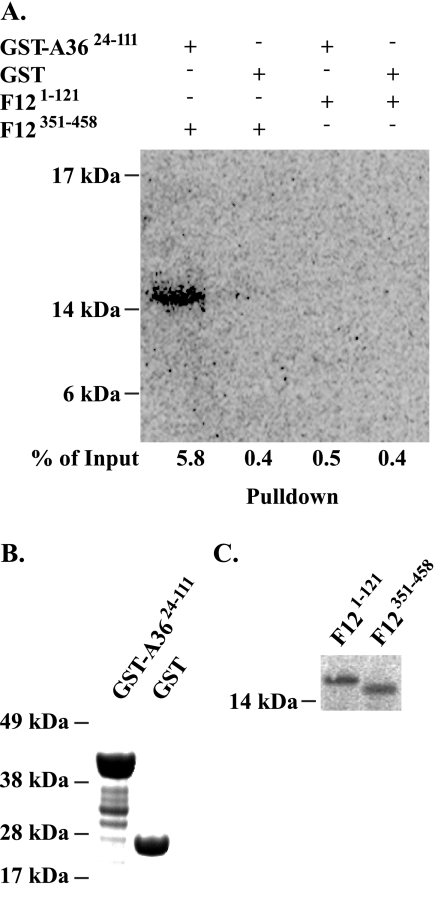
To confirm the yeast two-hybrid results, we carried out an in vitro, GST pull-down assay. In agreement with previous results, one-step purification of either GST or GST-A3624-111 from Escherichia coli resulted in prominent bands of the predicted size for GST-A3624-111 after SDS-PAGE (Fig. 2B) (20). Equal amounts of either GST or GST-A3624-111 were determined by protein concentration assay and bound to glutathione-Sepharose beads. Bound beads were incubated with 35S-labeled F12351-458 or F121-121 as a negative control (Fig. 2C). After extensive washing, complexes were eluted from the beads by boiling in SDS sample buffer and resolved by SDS-PAGE. Radiolabeled proteins were imaged with a phosphorimager. GST-A3624-111 only bound F12351-458 and not F121-121 (Fig. 2A). Importantly, neither bound an equivalent amount of GST, indicating that the interaction between F12 and A3624-111 is specific for residues 351 to 458 of F12.
FIG. 2.
In vitro interaction. In vitro 35S-labeled F121-121 and F12351-458 proteins were incubated with either GST-A3624-111 or GST alone bound to glutathione-Sepharose beads. (A) Glutathione complexes were separated by SDS-PAGE, blotted to nitrocellulose, and analyzed by phosphorimaging. Equal volumes of input GST-A3624-111 and GST (B) or 35S-labeled F12351-458 and F121-121 (C) were analyzed by SDS-PAGE. GST proteins were detected by using SimplyBlue SafeStain (Invitrogen). Radiolabeled proteins were detected by using phosphorimaging. The percentage of input in panel A was calculated by measuring band intensities of the input and resulting complexes from a single blot and corrected for volume changes that occurred during processing of the samples. For presentation purposes, a darker exposure is shown in panel A. The positions and masses (in kilodaltons) of markers are indicated on the left.
Interaction of F12 and A36 in infected cells.
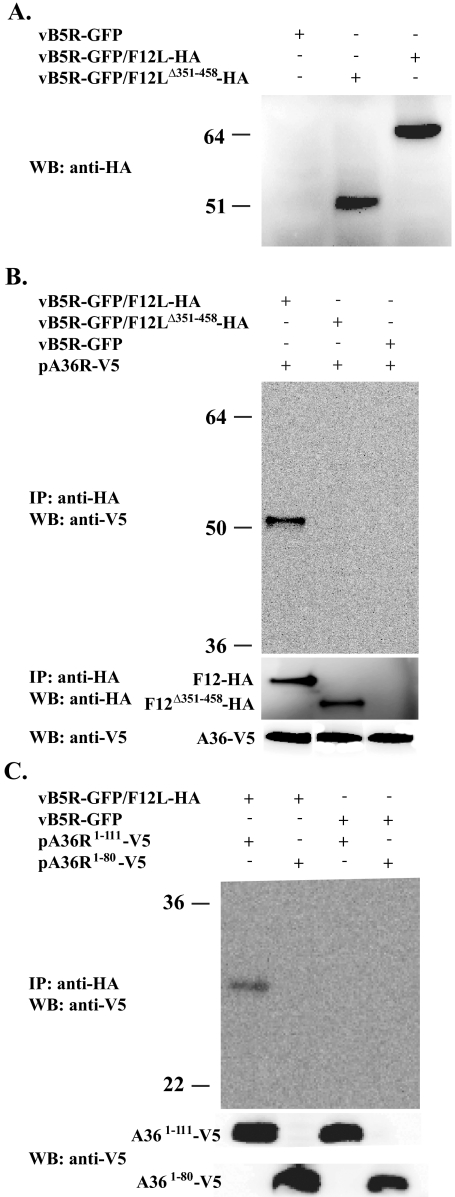
We next wanted to determine whether F12 and A36 could interact in infected cells. We constructed two additional recombinant vaccinia viruses: vB5R-GFP/F12LΔ351-458-HA and vB5R-GFP/ΔF12L. Both viruses express B5-GFP to visualize morphogenesis. The recombinant vB5R-GFP/ΔF12L has the entire F12L ORF removed, while vB5R-GFP/F12LΔ351-458-HA expresses an F12 that lacks residues 351 to 458 and has a C-terminal HA epitope tag. To ensure that equal amounts of F12-HA and F12Δ351-458-HA were expressed, lysates from cells infected with the recombinant viruses were analyzed by Western blotting. Anti-HA MAb recognized proteins of the predicted size for F12-HA and F12Δ351-458-HA (64 and 51 kDa, respectively) indicating that both HA-tagged proteins were stable and expressed at similar levels (Fig. 3A).
FIG. 3.
Coimmunoprecipitation. (A) Lysates from cells infected with the indicated viruses were analyzed by Western blotting using an anti-HA MAb. HeLa cells were infected with the indicated viruses followed by transfection with the indicated plasmids. Lysates from the infected/transfected cells were immunoprecipitated with anti-HA MAb. Immune complexes were separated by SDS-PAGE, blotted to nitrocellulose, and probed with either an anti-V5 HRP-conjugated MAb (B and C) or an anti-HA MAb (B middle). The bottom panels show the expression of A36-V5 proteins. The positions and masses (in kilodaltons) of markers are indicated on the left.
Yeast two-hybrid and in vitro pull-down assays indicated that residues 351 to 458 of F12 interact with residues 91 to 111 of A36. We predicted that the interaction between F12 and A36 is responsible for the localization of F12 to IEV. To test this prediction, we confirmed the interaction between these proteins during infection. HeLa cells infected with vB5R-GFP, vB5R-GFP/F12L-HA, or vB5R-GFP/F12LΔ351-458-HA were transfected with pA36R-V5, which expresses A36 with a V5 epitope tag. Lysates from cells infected with the recombinants were immunoprecipitated with an anti-HA MAb, and A36 was detected by Western blotting with anti-V5 HRP-conjugated MAb. In cells that expressed A36-V5 and F12-HA, A36-V5 was brought down by anti-HA MAb (Fig. 3B), which confirmed that F12 and A36 interact in infected cells. Importantly, A36-V5 was not immunoprecipitated in cells expressing F12Δ351-458-HA, confirming that residues 351 to 458 of F12 are necessary for the interaction. In addition, we performed immunoprecipitation to ensure that the anti-HA MAb immunoprecipitated equivalent amounts of F12-HA and F12Δ351-458-HA (Fig. 3B).
To test whether residues 91 to 111 of A36 are involved in the interaction, we repeated the coimmunoprecipitation using two other plasmids that express A36-V5 truncations. Plasmids pA36R1-111-V5 and pA36R1-80-V5 express V5 epitope tagged versions of A36 that either contain or lack residues 91 to 111, respectively. In cells expressing F12-HA, only A361-111-V5 was brought down by the anti-HA antibody (Fig. 3C), confirming that residues 91 to 111 of A36 are required for interaction with F12 during infection.
Growth and spread of recombinant viruses.
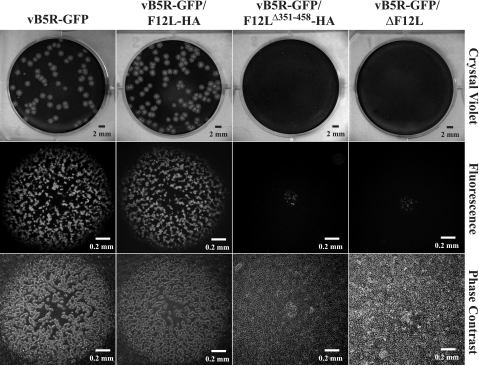
Plaque size is directly related to the amount of infectious CEV/EEV produced. Mutations that affect CEV/EEV production typically have a reduced plaque size. To look at the effect disruption of the F12:A36 interaction has on CEV/EEV production, we looked at the size of the plaques produced by our recombinants on monolayers of BS-C-1 cells. As had been reported previously (25), the addition of an HA epitope tag to the C terminus of F12 had no significant effect on the size of plaques formed by vB5R-GFP/F12L-HA (Fig. 4). In contrast, vB5R-GFP/F12LΔ351-458-HA produced small plaques that were similar in size to vB5R-GFP/ΔF12L (Fig. 4), suggesting that these two recombinants were defective in infectious extracellular virus production.
FIG. 4.
Plaque phenotypes. Monolayers of BS-C-1 cells were infected with the indicated recombinant viruses. At 72 h p.i., individual plaques were imaged by fluorescence (middle) and phase-contrast microscopy (bottom). After microscopy, cells were stained with crystal violet, and the entire well was imaged (top).
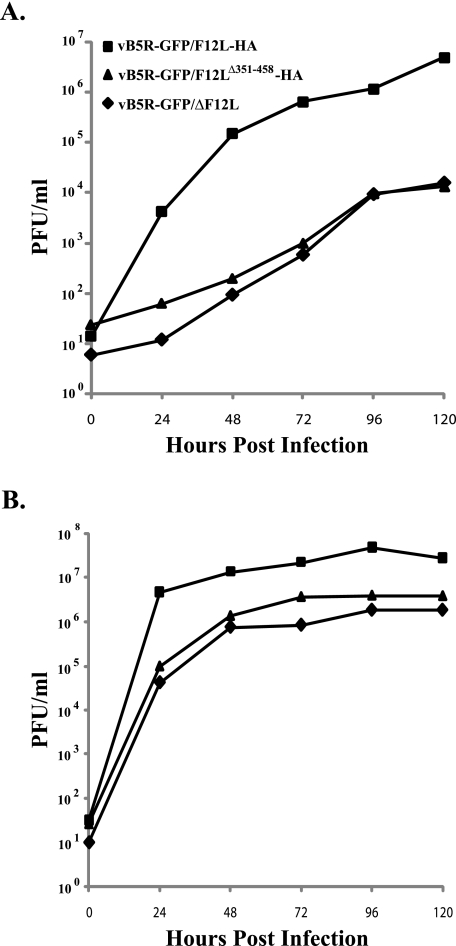
Low-MOI growth curves were performed to assess the ability of our recombinants to spread cell to cell, which is directly related to infectious CEV/EEV production. Cells infected with vB5R-GFP/F12LΔ351-458-HA and vB5R-GFP/ΔF12L released similar amounts of infectious virions that were ∼2.5 logs less than vB5R-GFP/F12L-HA (Fig. 5A). The amount of cell- associated infectious virus was also significantly less for vB5R-GFP/F12LΔ351-458-HA and vB5R-GFP/ΔF12L than for vB5R-GFP/F12L-HA (Fig. 5B). In contrast, there was little difference in cell-associated virus over the same time period for all three recombinants when a high MOI was used, indicating that IMV production was unaffected (data not shown). These results indicate that the interaction of F12 with A36 is required for efficient CEV/EEV release.
FIG. 5.
Low-MOI growth curve. BS-C-1 cells were infected at an MOI of 0.01 with the indicated viruses. At the indicated times, titers of the virions released into the medium (A) and associated with cells (B) were determined by plaque assay on monolayers of BS-C-1 cells.
Fluorescence microscopy of cells infected with recombinant viruses.
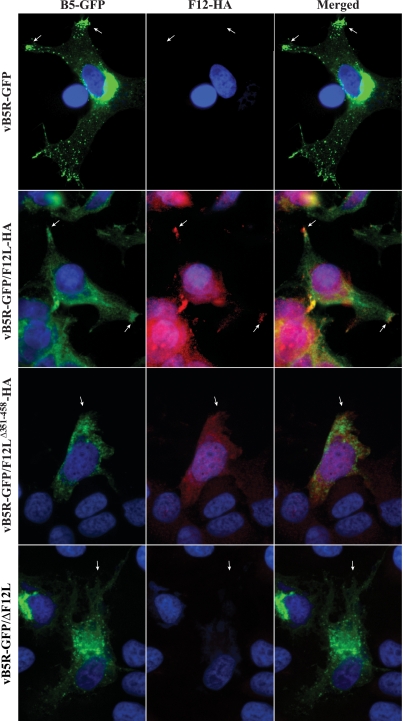
Previous reports have shown that cells infected with vB5R-GFP have a reproducible fluorescent pattern. Typically, there are three hallmarks of this pattern: an accumulation of B5-GFP in the juxtanuclear region, virion-sized particles labeled with B5-GFP, and an accumulation of signal at the peripheral extensions, or vertices, of the cell (Fig. 6, arrows). Mutations that disrupt CEV/IEV production typically result in a change in this pattern. To determine whether disruption of the F12-A36 interaction has an effect on enveloped virus formation, we examined cells infected with our recombinant viruses by using immunofluorescence microscopy. Examination of the B5-GFP staining pattern in cells infected with vB5R-GFP and vB5R-GFP/F12L-HA showed identical subcellular fluorescent patterns with GFP signal in the juxtanuclear region (site of wrapping), as well as at the cell vertices (Fig. 6, arrows). In addition, B5-GFP staining can be seen throughout the cytoplasm. B5-GFP staining can also be seen in cells infected with vB5R-GFP/F12LΔ351-458-HA and vB5R-GFP/ΔF12L (Fig. 6). However, there is an absence of signal accumulation at the vertices (Fig. 6, arrows). In addition, we looked at the localization of F12 in these cells. As had been reported previously (18), F12 localizes to the cell periphery in infected cells (Fig. 6) and appears to colocalize with B5-GFP in the vertices (Fig. 6, arrows). In contrast, only diffuse F12-HA staining was seen in cells infected with vB5R-GFP/F12Δ351-458-HA (Fig. 6), indicating that there is a defect in morphogenesis and that F12 may be mistargeted when it is unable to interact with A36.
FIG. 6.
Localization of B5-GFP and F12-HA in infected cells. HeLa cells infected with indicated viruses were permeabilized and stained with an anti-HA MAb followed by a Texas Red-conjugated anti-mouse MAb (red). Cells were imaged by fluorescence microscopy. Green represents B5-GFP fluorescence, blue shows DAPI staining, and yellow represents the overlap of green and red fluorescence. Arrows point to the vertices of the cells.
vB5R-GFP/F12LΔ351-458-HA is defective in IEV release.
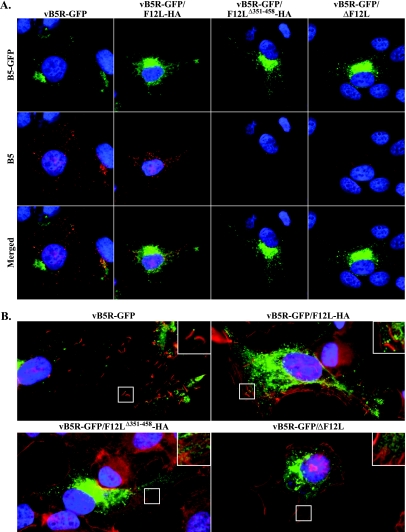
The deletion of F12 has been reported to affect the release of IEV from infected cells (18, 25). During a normal infection, B5 is deposited into the plasma membrane as IEV are released from the cell. Mutations that reduce CEV/EEV production subsequently reduces the amount of B5 on the cell surface. To determine whether our recombinants have a similar defect, we stained the surface of cells infected with our recombinant viruses for B5. Cells infected with vB5R-GFP and vB5R-GFP/F12L-HA had significant B5 staining on the surface (Fig. 7A). In contrast, the amount of B5 was severely reduced or absent in cells infected with vB5R-GFP/F12LΔ351-458-HA and vB5R-GFP/ΔF12L (Fig. 7A), suggesting a defect in IEV release.
FIG. 7.
(A) Surface staining of B5 on infected cells. HeLa cells were infected with the indicated viruses. At 24 h p.i., unpermeabilized cells were stained with an anti-B5 MAb, followed by a Texas Red-conjugated anti-rat MAb (red), and imaged by fluorescence microscopy. Green represents B5-GFP fluorescence, blue is DAPI staining, and yellow represents overlap of green and red fluorescence. (B) HeLa cells infected with indicated viruses were permeabilized, stained with Alexa Fluor 633 phalloidin (red), and imaged by fluorescence microscopy. Green represents B5-GFP fluorescence, and blue is DAPI staining. Boxed regions are enlarged to show structural details.
In addition to B5 surface staining, actin tail formation serves as an indicator of CEV production because CEV on the surface of cells form actin tails to facilitate cell-to-cell movement. To confirm that the reduction of B5 on the cell surface corresponded to a reduction of CEV, we measured actin tail production in cells infected with our recombinant viruses by staining infected cells with Alexa Fluor 633 conjugated to phalloidin. Numerous actin tails were easily identified in cells infected with either vB5R-GFP or vB5R-GFP/F12L-HA (Fig. 7B and insets). In contrast, we were unable to find actin tails in cells infected with vB5R-GFP/F12LΔ351-458-HA and vB5R-GFP/ΔF12L (Fig. 7B and insets).
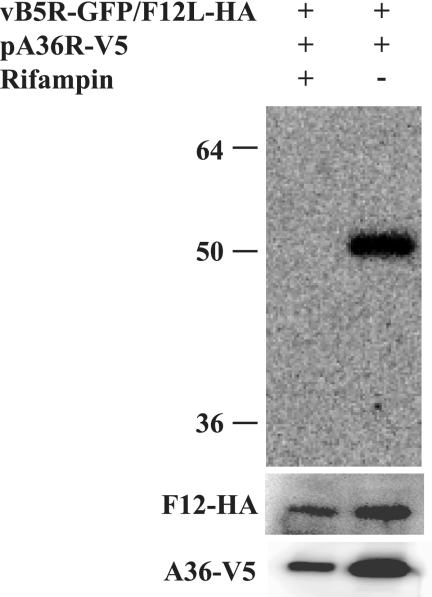
F12 and A36 fail to interact in the presence of rifampin.
Our immunofluorescence results correlate well with previous studies and demonstrate that F12 predominantly localizes to the cell periphery and not at the site of wrapping (18). The defect in localization suggests that virion morphogenesis may be required for F12 to interact with A36. To test this possibility, we repeated the coimmunoprecipitation experiments in the presence or absence of the drug rifampin, which inhibits virion morphogenesis (2). We were only able to detect an interaction between F12 and A36 in the absence of rifampin (Fig. 8). Importantly, our F12 and A36 constructs were expressed in the presence or absence of the drug, indicating that IMV, and subsequently IEV, formation is required for F12 to interact with A36.
FIG. 8.
Coimmunoprecipitation in the presence of rifampin. HeLa cells were infected with vB5R-GFP/F12-HA, followed by transfection with pA36R-V5, in the presence or absence of 0.1 μg of rifampin/ml. Lysates were immunoprecipitated with anti-HA MAb. Immune complexes were separated by SDS-PAGE and analyzed by Western blotting with anti-V5 HRP-conjugated MAb. The bottom panels show expression of F12-HA and A36-V5. The positions and masses (in kilodaltons) of markers are indicated on the left.
Residues 351 to 458 of F12 are required for the association of F12 with membranes.
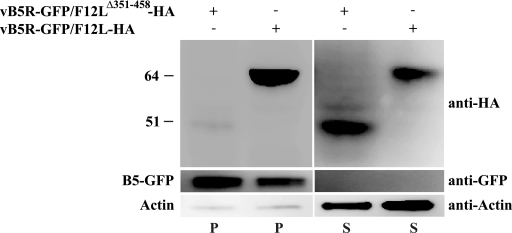
Our results have shown that F12 interacts with A36 during infection. We have also demonstrated that deletion of residues 351 to 458 of F12, the region responsible for interaction with A36, results in a defect in viral release similar to that seen following infection with a virus lacking the entire F12L ORF. We predicted that the defect seen during infection with vB5R-GFP/F12LΔ351-458-HA was due to an inability of F12Δ351-458 to associate with IEV. To test this prediction, membranes were isolated from cells infected with either vB5R-GFP/F12L-HA or vB5R-GFP/F12LΔ351-458-HA and analyzed for the presence of F12. Membranes pelleted from cells infected with vB5R-GFP/F12L-HA contain F12, as well as the integral membrane protein B5 (Fig. 9). Although B5 is still associated with membranes from cells infected with vB5R-GFP/F12LΔ351-458-HA, little to no F12Δ351-458-HA is present (Fig. 9). Importantly, F12Δ351-458-HA is present at comparable amounts to F12-HA in postnuclear supernatants (Fig. 9), demonstrating that the absence of F12Δ351-458-HA on membranes is not due to a lack of expression of the protein. These data suggest that the deletion of residues 351 to 458 results in an inability of F12 to interact with membranes.
FIG. 9.
Analysis of membranes for the presence of F12. Cells infected with the indicated viruses were harvested 24 h p.i. and lysed. Lysates were centrifuged to pellet membranes, and the resulting pellets (P) and supernatants (S) were analyzed by Western blotting for the presence of F12-HA, B5-GFP, and actin with the indicated MAbs. The positions and masses (in kilodaltons) of markers are indicated on the left.
DISCUSSION
A previous report used immunoelectron microscopy to show that F12 is only found on IEV and absent from CEV or EEV (18). F12 is not predicted to contain a transmembrane domain; therefore, its method of association with IEV was unclear. Furthermore, a recent review suggested that two putative myristoylation sites in F12 may be responsible for anchoring it to the outer membrane of IEV (14). The results from our alkaline carbonate extraction and Triton X-114 partitioning experiments suggest that F12 interacts with IEV through a peripheral association and not an integral association, such as through an unidentified transmembrane domain or protein lipidation. We hypothesized that the association of F12 with IEV may be mediated by interaction with another IEV-specific, integral membrane protein. The results of a yeast two-hybrid screen revealed an interaction between residues 351 and 458 of F12 and the cytoplasmic domain of A36. Further analysis using the yeast two-hybrid system mapped the site of interaction on A36 to amino acids 91 to 111. These results were confirmed using an in vitro, GST pull-down assay.
In order to further characterize the A36:F12 interaction and its role during virion morphogenesis, we constructed a recombinant that had the identified region on F12 removed (vB5R-GFP/F12LΔ351-458-HA). Coimmunoprecipitation confirmed that F12 and A36 interact in infected cells and that residues 351 to 458 of F12 are required for the interaction. Likewise, transfection of plasmids expressing truncated versions of A36 confirmed that residues 91 to 111 of A36 are required for interaction with F12.
Phenotypic characterization of our recombinant viruses revealed that vB5R-GFP/F12LΔ351-458-HA appeared identical to vB5R-GFP/ΔF12L. Further analysis revealed that both recombinants produced less CEV and were abrogated for actin tail formation. In addition, there was a distinct difference in the subcellular localization of F12-HA and F12Δ351-458-HA. This is most likely due to the inability of F12Δ351-458-HA to associate with A36 on the viral membrane. In corroboration of this notion, membranes isolated from cells infected with vB5R-GFP/F12LΔ351-458-HA were devoid of F12, suggesting that amino acids 351 to 458 are vital for the localization, and ultimate functioning, of F12 during virion egress.
The interaction of F12 and A36 seems logical since both proteins are found exclusively on the outermost membrane of IEV. It now appears that F12 accomplishes this localization by interacting with the IEV-exclusive, integral membrane protein A36. The mechanism by which A36 excludes itself from the inner membrane of IEV is unclear. A36 must interact with A33 for incorporation into the virion membrane (23). Unlike A36, A33 is found in both IEV membranes. This localization results in its presence on CEV and EEV. A36 must, therefore, regulate its interaction with A33 to limit its incorporation to only the outer membrane of IEV. Further work is required to elucidate the mechanism of selective incorporation of A36 and F12 into these membranes.
Four proteins have now been shown to interact with A36. The cellular proteins kinesin and Nck interact with A36 for microtubule-based movement and actin tail formation, respectively (5, 20). The cytoplasmic tail of the viral protein A33 interacts with A36, and this interaction is required for the incorporation of A36 into virion membranes (23). We now report that A36 interacts with F12 and that this interaction is required for efficient CEV/EEV formation, but a specific function for F12 has not been determined. All four of these proteins seem to bind within a narrow region of A36, residues 81 to 116, and studies have shown that the interaction of A36 is mutually exclusive to kinesin or A33 (20). Similar studies have not been carried out to determine whether A36 is able to bind Nck and any of the other three proteins simultaneously. Even if the interaction of A36 with these four proteins is mutually exclusive, this does not preclude IEV from interacting with all of them simultaneously since undoubtedly there are multiple molecules of A36 on the outer membrane of each IEV. Each of these molecules could potentially interact with a different binding partner at the same time. Much more work is required to determine the spatial and temporal dynamics, along with the potential regulation for all of the proteins that interact with A36.
During characterization of our recombinants we noticed that the majority of F12 did not localize to the site of wrapping. This is unlike A36, which has been shown to colocalize with B5 at the site of wrapping (23). This led us to hypothesize that F12 may not associate with A36 until after IEV are formed. Indeed, we were unable to detect an F12-A36 interaction in the presence of the drug rifampin, which prevents morphogenesis. This suggests that there is a regulation that prevents the interaction of F12 with A36 until after virions are formed. In addition, the lack of F12 at the site of wrapping indicates that it may not play a major role in envelopment. It is more likely that F12 is required for some aspect of IEV egress. While there has been much speculation about a role for F12, a specific function has not yet been reported.
Acknowledgments
This study was supported in part by National Institutes of Health research grant AI067391.
Footnotes
Published ahead of print on 3 December 2008.
REFERENCES
- 1.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 2561604-1607. [PubMed] [Google Scholar]
- 2.Charity, J. C., E. Katz, and B. Moss. 2007. Amino acid substitutions at multiple sites within the vaccinia virus D13 scaffold protein confer resistance to rifampicin. Virology 359227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Earley, A. K., W. M. Chan, and B. M. Ward. 2008. The vaccinia virus B5 protein requires A34 for efficient intracellular trafficking from the endoplasmic reticulum to the site of wrapping and incorporation into progeny virions. J. Virol. 822161-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkner, F. G., and B. Moss. 1990. Transient dominant selection of recombinant vaccinia viruses. J. Virol. 643108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401926-929. [DOI] [PubMed] [Google Scholar]
- 6.Hiller, G., and K. Weber. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 9.Moss, B. 2006. Poxvirus entry and membrane fusion. Virology 34448-54. [DOI] [PubMed] [Google Scholar]
- 10.Payne, L. G. 1980. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 5089-100. [DOI] [PubMed] [Google Scholar]
- 11.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schramm, B., and J. K. Locker. 2005. Cytoplasmic organization of poxvirus DNA replication. Traffic 6839-846. [DOI] [PubMed] [Google Scholar]
- 13.Smith, G. L., and M. Law. 2004. The exit of vaccinia virus from infected cells. Virus. Res. 106189-197. [DOI] [PubMed] [Google Scholar]
- 14.Smith, G. L., B. J. Murphy, and M. Law. 2003. Vaccinia virus motility. Annu. Rev. Microbiol. 57323-342. [DOI] [PubMed] [Google Scholar]
- 15.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 832915-2931. [DOI] [PubMed] [Google Scholar]
- 16.Swaffield, J. C., and S. A. Johnston. 1998. Affinity purification of proteins binding to GST fusion proteins, p. 20.2.1-20.2.10. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 3. Greene Publishing Associates/Wiley Interscience, New York, NY. [DOI] [PubMed] [Google Scholar]
- 17.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60163-178. [PubMed] [Google Scholar]
- 18.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83195-207. [DOI] [PubMed] [Google Scholar]
- 19.Ward, B. M. 2005. Visualization and characterization of the intracellular movement of vaccinia virus intracellular mature virions. J. Virol. 794755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward, B. M., and B. Moss. 2004. Vaccinia virus A36R membrane protein provides a direct link between intracellular enveloped virions and the microtubule motor kinesin. J. Virol. 782486-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 7511651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 754802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward, B. M., A. S. Weisberg, and B. Moss. 2003. Mapping and functional analysis of interaction sites within the cytoplasmic domains of the vaccinia virus A33R and A36R envelope proteins. J. Virol. 774113-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolffe, E. J., A. S. Weisberg, and B. Moss. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, W. H., D. Wilcock, and G. L. Smith. 2000. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J. Virol. 7411654-11662. [DOI] [PMC free article] [PubMed] [Google Scholar]