Abstract
Direct population sequencing and reverse hybridization (line probe assay [LiPA])-based methods are the most common methods for detecting hepatitis B virus (HBV) drug resistance mutations, although only mutations present in viral quasispecies with a prevalence of ≥20% can be detected by sequencing, and only known mutations are detected by LiPA. Massively parallel ultradeep pyrosequencing (UDPS; GS FLX platform) was used to analyze HBV quasispecies in reverse transcriptase (RT) and hepatitis B S antigen (HBsAg) from five drug-naive patients and eight drug-resistant patients. Eight primer pairs were used to obtain partially overlapping amplicons, covering the RT gene from codons 1 to 288 and the complete overlapping HBsAg sequence. A 1% mutation frequency was selected as the cutoff based on an error rate estimated on plasmid DNA. This technology enabled simultaneous analysis of between 2,852 and 18,016 clonally amplified fragments from each patient. The results indicate that UDPS has a relative sensitivity much higher than both direct sequencing and LiPA. In addition, the UDPS results are quantitative, allowing establishment of the relative frequency of both known mutations and novel substitutions. Some of the detected RT substitutions led to changes also in HBsAg. On the whole, genotype D presented a higher heterogeneity than genotype A. Considering the high quantity of information that can be provided by a single test from one patient, the short turnaround time, the information on substitution frequency, and the detection of rare variants, there are strong advantages conferred by UDPS, and the new method could play a relevant role in the clinical management of HBV infection and therapy.
The goal of treatment for chronic hepatitis B is to prevent cirrhosis, hepatic failure, and hepatocarcinoma. To date, the therapies approved for this condition are alpha 2b interferon and peginterferon, owing to their immune modulating effect and antiviral activity (25, 32), and nucleoside/nucleotide analogues, such as lamivudine (LAM), telbivudine, emtricitabine (EMC), entecavir (ETV), adefovir (ADV), and tenofovir (TDF), which have antiviral activity only.
Analogues rarely produce control of hepatitis B S antigen (HBsAg) antigenemia, and require prolonged use to control hepatitis B virus (HBV) replication in most patients (7), although recent advances suggest that combined treatment with multiple antiviral compounds may lead to a more rapid and durable HBV replication control (29, 15). In fact, prolonged therapy is associated with the emergence of resistant strains that are responsible for therapeutic failure and disease progression (5, 42). Drug resistance mutations mainly involve the reverse transcriptase (RT) region of the polymerase gene, which contains seven functional domains (A to G). The mutations conferring nucleoside/nucleotide resistance are located essentially in domains A through E (12, 21, 43).
Due to the high variability of the HBV genome, a collection of distinct, albeit closely related viral genetic variants are usually present at any given time in an infected host and are known as viral quasispecies. Antiviral treatment can result in the selection and progressive fixation of viral variants with reduced drug susceptibility, leading to virological resistance and therapy failure. Many published data have shown that the appearance of resistance mutations (genotypic resistance) may precede the increase in viral load (virological resistance) by several months (9, 18) and could be a prognostic marker for the occurrence of viral breakthrough.
Drug resistance tests have been developed in recent years, to help clinicians to choose therapeutic options attuned to the virological status of each patient.
To date, the most commonly used method for detecting drug resistance mutations is direct sequencing after PCR amplification. A major limitation of direct PCR sequencing, however, is its inability to detect variants poorly represented in the heterogeneous virus population existing in a patient's circulation. From the long-lasting experience gained with human immunodeficiency virus (HIV) therapy, it is clear that minor drug-resistant variants, although not detected by the direct sequencing approach, may be clinically relevant, as they can quickly grow out when subjected to the selective pressure exerted by the drug and become the predominant form, leading to treatment failure (16, 31). To overcome this problem, reverse hybridization line-probe assays (LiPAs) have been developed, which have the ability to detect mutations present in as few as 5% of the circulating viral population (13, 17, 34). The reverse hybridization LiPAs employ a series of short membrane-bound oligonucleotide probes to detect single mismatches in PCR-amplified HBV DNA. The major limitation of these methods is that only a limited repertoire of well-established mutations can be detected, in particular those most importantly associated with LAM and ADV resistance, but the identification of novel or recently described ones is not possible (23, 44). In addition, this technique does not allow a mutation frequency estimate, so it is not suitable for dynamic studies. Other sequence-specific genotypic resistance tests are available or are being developed, such as restriction fragment length/mass polymorphism (11, 33, 41), oligonucleotide microarray and gene chip technology (19, 36), mutation-specific real-time PCR (39), matrix-assisted laser desorption ionization-time of flight mass spectrometry (17), and pyrosequencing (14, 20). On the whole, all of the sequence-specific methods present the same drawbacks of reverse hybridization.
A very powerful tool, based on the new 454 Life Sciences platform (GS FLX, distributed by Roche), has recently become available. This technology performs massively parallel picoliter-scale amplification and pyrosequencing of individual DNA molecules (26), allowing the simultaneous analysis of thousands of clonally amplified regions of about 200 nucleotides, which increases the probability of detecting minority variants. (A description of the method can be found at http://www.454.com/index.asp.) Recently, the detection and characterization of rare drug-resistant variants in HIV-1 have been reported with the 454 Life Sciences GS20 technology (10, 28, 38).
In the present study, the ultradeep pyrosequencing (UDPS) approach, applied with the GS FLX platform, was used to detect and quantify minor variants in the RT gene of HBV present in the serum of patients. Direct sequencing and INNO-LiPA HBV DR v2 were used as standard reference tests. The corresponding mutations in the S gene open reading frame (ORF) were also determined.
(This study has been accepted for presentation at the 59th Annual Meeting of the American Association for the Study of Liver Diseases.)
MATERIALS AND METHODS
Patients.
We performed UDPS on serum samples from 13 HBV-infected patients: 9 with genotype D and 4 with genotype A, 5 drug naive and 8 under antiviral treatment but showing virological failure (4 on LAM, 1 coinfected with HIV-1 and on EMC plus TDF as part of the antiretroviral regimen, and 3 switched from LAM to ADV). These patients were selected on the basis of drug-resistance mutated codons detected by DR v2 and not by direct sequencing. HBV DNA values, measured by COBAS Taq-Man HBV test (Roche Molecular Systems, Inc., Branchburg, NJ), are reported in Table S1 in the supplemental material.
PCR amplification and UDPS.
UDPS was used to sequence overlapping segments of HBV RT after PCR amplification (ultradeep approach). Since this technique is not able to sequence target fragments longer than 250 bp, primers were designed to amplify eight partially overlapping segments covering from 1 to 288 amino acids (aa) of the RT region (including all functional domains) and the complete region of the HBsAg. The primers were designed to anneal sequences conserved among genotypes, and some were degenerate. All primers used, including 5′ extensions which provided binding sites for the pyrosequencing on the genome sequencer, are detailed in Table 1.
TABLE 1.
Sequences and positions of primers used for UDPS of HBV
| Primera | Sequence (5′→3′) | RT position |
|---|---|---|
| 1 FW | CCTGCTGGTGGCTCCAGTT | −34-53 |
| 1 RW | AGAGAAGTCCACCACGAG | 124-140 |
| 2 FW | CCTGCTCGTGTTACAGGCG | 58-72 |
| 2 RW | CCGCAGACACATCCAGCG | 242-259 |
| 3 FW | CCGTGTGTCTTGGCCAAA | 162-179 |
| 3 RW | GACAAACGGGCAACATAC | 330-347 |
| 4 FW | GCTGCTATGCCTCATCTTCT | 286-305 |
| 4 RW | GAYGATGGGATGGGAATAC | 471-489 |
| 5 FW | GCACGACTCCTGCTCAAGG | 396-414 |
| 5 RW | CCCKACGAACCACTGAACAA | 561-580 |
| 6 FW | GTATTCCCATCCCATCRTC | 471-489 |
| 6 RW | CGGTAWAAAGGGACTCAMG | 649-667 |
| 7 FW | TTGTTCAGTGGTTCGTMGGG | 561-580 |
| 7 RW | GGGTTAAATGTATACCCAVAG | 689-710 |
| 8 FW | CKTGAGTCCCTTTWTACCG | 649-667 |
| 8 RW | CKTGAGTCCCTTTWTACCG | 649-667 |
The primers included 5′ extensions (left GCCTCCCTCGCGCCATCAG and right GCCTTGCCAGCCCGCTCAG adaptors not shown in the table) which provide binding sites for pyrosequencing.
HBV DNA was extracted using the QIAamp DNA blood minikit (Qiagen, Chatsworth, CA). All samples were amplified using a proofreading enzyme (Fast Start high-fidelity enzyme; Roche, Mannheim, Germany). The conditions used for PCR were 1 cycle of 95°C for 2 min followed by 40 cycles of denaturation for 30 s at 95°C, annealing of primers for 30 s at 60°C, and extension for 45 s with a final 5-min extension at 72°C.
The PCR amplicons were purified with the QIA quick PCR purification kit (Qiagen,) and quantified by Agilent 2100 bioanalyzer (Agilent Life Sciences and Chemical Analysis).
All amplification reactions yielded DNA chains of the expected length and in amounts sufficient to support sequencing of the target. The PCR amplicons of each patient were pooled and subjected to UDPS, carried out with the GS FLX platform according to the manufacturer's instructions.
The pyrosequencing method is more error prone than the Sanger method, and therefore, a plasmid DNA was included as an error rate control, to measure the accuracy of the UDPS. The plasmid clone was obtained from a patient's sample by inserting a PCR amplicon spanning nucleotide positions 180 to 930 into a pCR4-TOPO vector (Invitrogen Corp.). The plasmid clone containing the region of interest was sequenced in parallel by UDPS and by the Sanger method.
Sanger sequencing of the clone was performed on an ABI Prism 3100, by using the BigDye terminator cycle sequencing kit, following the manufacturer's instructions (Applied Biosystems, Warrington, United Kingdom).
Any differences between the two methods were considered to be UDPS sequencing errors. Because the pyrosequencing error rate is higher in homopolymeric regions, the error rates were determined separately for homopolymeric (nucleotide repeats of three or more identical bases) and nonhomopolymeric regions. The error rate was 0.0027 (standard error, 7.4 × 10−4) within homopolymeric and 0.0012 (standard error, 7.48 × 10−5) in nonhomopolymeric regions. Based on these results, as well as on a theoretical calculation using a statistical approach (6), a conservative cutoff of 1% was adopted to estimate mutation frequency.
Analysis of HBV resistance by direct sequencing and INNO-LiPA HBV DR v2.
The presence of HBV RT mutations was also determined by direct sequencing. Two sets of primers (1 FW to 4 RW and 5 FW to 8 RW, without the adaptors) were used (covering from 1 to 288 aa). The conditions of PCR are the same as reported above. The amplified PCR products were purified by the QIA quick PCR purification kit (Qiagen). After quantification by gel electrophoresis with a low-weight molecular standard, sequencing was performed on an ABI Prism 3100, as described above.
Consensus sequences were built for RT and HBsAg of both genotypes A and D, using reference sequences derived from GenBank (genotype A, E00010 and V00866; genotype D, Y07587, AF043594, Y796031, AY721610, and AY796030). The sample sequences were compared with the genotype-matched consensus sequences by BLAST search analysis, to identify the amino acid changes in both ORFs. In the samples from the drug-experienced patients, the genotypic resistance pattern was also determined by INNO-LiPA HBV DR v2 (Innogenetics NV, Ghent, Belgium).
Treatment of sequence data and statistical analysis.
The local pairwise Smith-Waterman-Gotoh (SWG) algorithm (8, 35) was applied to sequences obtained by UDPS in order to determine changes with respect to the genotype D and A consensus sequences. A modified version of this method had already been employed by Wang et al. (38) to analyze mutations of HIV-1 sequences obtained by UDPS: Wang proposed a modified version of the Smith-Waterman algorithm, called asymmetric Smith-Waterman, where alignment scores were recalculated weighting bases by their phred-equivalent quality values for each UDPS base call. Wang compared asymmetric Smith-Waterman against standard Smith-Waterman and BLAST (2): when benchmarking the three algorithms, no significant differences were found, except for highly distant isolates (like different subtypes). Thus, we applied the standard SWG algorithm, adding space optimization as an additional parameter (with respect to open and extension gap penalties) with a grid search in [5:25] and [0.25:5], using step sizes of 5 and 0.5, respectively, maximizing the nucleotide local similarity and minimizing the number of 1-base or 2-base frameshifts. The procedure for executing the SWG algorithm and extracting mutations was implemented using the JAligner package (http://jaligner.sourceforge.net/) for java programming language (http://java.sun.com): minor refinements to the original algorithm were designed in order to correct for 3-base insertions/deletions not in frame (that were replaced in frame when possible), ambiguity translation, and 1-base or 2-base frameshift report/correction, accounting for the higher pyrosequencing error rate (6).
Variations from the reference consensus sequences, identified by the UDPS method, were considered as true mutations if previously described to be associated with drug resistance; otherwise, they were regarded as possible novel mutations or polymorphisms.
The prevalence of substitutions was calculated by means of relative frequencies in the subregions sequenced, calculating the 95% confidence interval via Agresti and Coull approximation (1).
RESULTS
A total of 128,184 sequence reads, with an average length of 189 bp, were returned from the UDPS on serum samples from 13 HBV-infected patients: the number of reads per patient ranged between 2,852 and 18,016.
In the drug-experienced patients, resistance mutations were identified by UDPS and compared with those identified by DR v2 and by direct sequencing (Table 2). None of the mutations detected at frequencies of <24% by UDPS were detected by direct sequencing, and some of these (four mutations for patient 1, four for patient 5, and one for patient 6, whose frequency ranged between 2% and 8%) were missed also by DR v2. Only in one case was a mutation (M204V in patient 4) detected by DR v2 but missed by both UDPS and direct sequencing; these latter results were confirmed by repeated testing in both systems, suggesting that the DR v2 result could be a system artifact.
TABLE 2.
Comparison of UDPS, DR v2, and direct sequencing for HBV from eight patients under treatment
| Patient | Presence or frequency of mutationa
|
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Codon 80
|
Codon 173
|
Codon 180
|
Codon 181
|
Codon 204
|
Codon N236
|
|||||||||||||||||||||||||||||||
| L80 (wt)
|
L80I/Vb
|
V173 (wt)
|
V173L
|
L180 (wt)
|
L180M
|
A181 (wt)
|
A181T/Vb
|
M204 (wt)
|
M204I/Vb
|
N236 (wt)
|
N236T
|
|||||||||||||||||||||||||
| UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | UDPS % | DR v2 | DS | |
| 1 | 96 | Y | Y | 4 | <1 | <1 | 100 | Y | Y | 92 | Y | Y | 8 | <1 | <1 | 98 | Y | Y | 2 | <1 | <1 | 95 | Y | Y | 5* | <1* | <1* | 100 | Y | Y | ||||||
| 2 | 100 | Y | Y | 100 | Y | Y | 100 | Y | Y | 97 | Y | Y | 3 | Y | <1 | 100 | Y | Y | 78 | Y | Y | 22 | Y | <1 | ||||||||||||
| 3 | 73 | Y | Y | 24/3* | Y/Y* | Y/<1* | 100 | Y | Y | 100 | Y | Y | 100 | Y | Y | 88 | Y | Y | 12 | Y | <1 | 100 | Y | Y | ||||||||||||
| 4 | 97 | Y | Y | 3* | Y* | <1* | 100 | Y | Y | 100 | Y | Y | 100 | Y | Y | 100 | Y | Y | N* | Y* | <1* | 100 | Y | Y | ||||||||||||
| 5 | 98 | Y | Y | 2 | <1 | <1 | 100 | Y | Y | 97 | Y | Y | 3 | <1 | <1 | 93 | Y | Y | 7* | <1* | <1* | 98 | Y | Y | 2* | <1* | <1* | 88 | Y | Y | 12 | Y | <1 | |||
| 6 | 100 | Y | Y | 38 | Y | Y | 62 | Y | Y | 40 | Y | Y | 60 | Y | Y | 98 | Y | Y | 2* | <1* | <1* | 39 | Y | Y | 21/40* | Y/Y* | Y/Y* | |||||||||
| 7 | 82 | Y | Y | 18* | Y* | <1* | 95 | Y | Y | 5 | Y | <1 | 23 | Y | <1 | 77 | Y | Y | 100 | Y | Y | 0 | <1 | <1 | 24/76* | Y/Y* | Y/Y* | 100 | Y | Y | ||||||
| 8 | 100 | Y | Y | 100 | Y | Y | 59 | Y | Y | 41 | Y | Y | 100 | Y | Y | 58 | Y | Y | 42* | Y* | Y* | 100 | Y | Y | ||||||||||||
The table reports all the mutations included in the DR v2. For UDPS, the results include the mutation frequency (UDPS %) in each treated patient. (Note that the frequency values have been adjusted to the nearest integral number.) Y and <1% indicate detected and not detected, respectively. DS, direct sequencing.
Where alternative mutations are found at codons 80 (I or V), 181 (T or V), and 204 (I or M), the correspondence in the columns is indicated by the asterisk.
These results indicate that all of the major drug resistance-associated mutations included in DR v2 were detected by UDPS; more importantly, the relative sensitivity of this system appears to be much higher, not only versus direct sequencing, as expected, but also versus DR v2.
In addition, a number of RT gene substitutions different from the major drug resistance-associated mutations (hence not included in DR v2) were detected at variable frequencies in both drug-treated and drug-naive patients. The complete list of the variants found in all patients is reported in Table S1 in the supplemental material. It is noteworthy that some mutations known to be associated with drug resistance (V214A and/or M204I), were detected by UDPS as minority components of viral quasispecies in two drug-naive patients (patients 11 and 12). In particular, the primary LMV-associated mutation M204I, present at a frequency of 1.5% in patient 11, corresponded to a stop codon in HBsAg.
From Table S1 in the supplemental material, it is also evident that four substitutions leading to the introduction of a stop codon in RT were detected by UDPS with frequencies ranging from 1.0% to 2.8% in five patients.
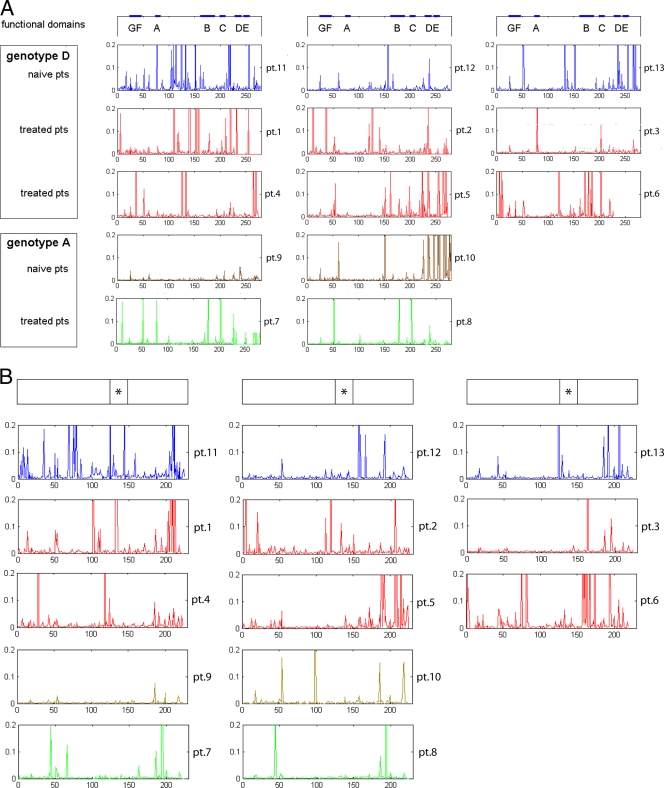
The overall distribution of all substituted codons detected by UDPS within the RT gene from each patient, according to their position in the functional and the interdomain regions, is shown in Fig. 1A. On the whole, the pattern of changes versus the genotype consensus sequence varied according to patient source and treatment schedule, including some substitutions with a frequency of >20% and several additional changes spread along the gene occurring with low frequency. Interestingly, both functional and interfunctional domains presented variant positions in both treated and drug-naive patients. The total number of changes in treated patients was always higher than in drug-naive patients, both in functional domains (48 versus 19, ratio of 2.5) and in interdomains (96 versus 63, ratio of 1.5). Generally, HBV genotype D presented a higher number of substitutions compared to genotype A in both treated and drug-naive patients. However, due to the small number of patients, it is not possible to perform statistical evaluation of the differences in the mutation patterns in relation to genotype and treatment.
FIG. 1.
Frequency of substitutions within RT (A) and HBsAg (B) detected by UDPS in 13 HBV-positive patients. Patients (pts) are reported according to genotype and treatment status. On the top of each column, the relevant protein domains are indicated with a thick line, including functional domains for RT and the “a” determinant (*) for HBsAg. The frequency scale axis has been cut at 20%, to allow better visualization of infrequent substitutions. For details, see Table S1 in the supplemental material.
For the first time, to our knowledge, the UDPS allowed the identification of novel substitutions, never described in the literature, in 71 different codons of RT (57 in genotype D only, 11 in genotype A only, and 3 in both genotypes D and A), 16 of which are present in both drug-naive and drug-treated patients (Table 3). The relative frequency of these substitutions in the quasispecies of each single patient was very variable, ranging from 1 to >99%.
TABLE 3.
Frequency of substitutions in RT detected by UDPS in the study patients not previously described in literature
| RT substitution | Treated patients (n = 8)
|
Naïve patients (n = 5)
|
||||
|---|---|---|---|---|---|---|
| na | HBV genotype | Frequency (%) | na | HBV genotype | Frequency (%) | |
| rtE8K | 1 | D | 1.40 | |||
| rtV27F | 2 | A | 1.70, 4.47 | |||
| rtV27F | 3 | D | 1.33, 2.39, 4.16 | 3 | D | 3.35, 4.49, 5.17 |
| rtH35Y | 1 | D | 2.19 | |||
| rtA38P | 1 | D | 1.48 | 1 | D | 1.87 |
| rtH53S | 1 | D | 19.04 | |||
| rtN53K | 1 | D | 2.42 | |||
| rtH54S | 1 | D | 2.33 | |||
| rtH54T | 1 | D | 2.47 | |||
| rtR55H | 1 | D | 2.47 | |||
| rtA87V | 1 | D | 1.50 | |||
| rtI91F | 1 | D | 1.61 | 1 | D | 1.13 |
| rtI91P | 1 | D | 1.13 | |||
| rtA97T | 1 | D | 1.06 | |||
| rtG107R | 2 | D | 1.17, 1.86 | 1 | D | 1.22 |
| rtN118Y | 1 | D | 1.30 | |||
| rtN118T | 1 | D | 1.38 | |||
| rtR120S | 1 | D | 8.13 | |||
| rtI121N | 1 | D | 1.10 | |||
| rtF122I | 1 | D | 2.35 | |||
| rtF122N | 1 | D | 3.20 | |||
| rtF122S | 1 | D | 1.40 | |||
| rtQ125H | 1 | D | 1.11 | |||
| rtH126Y | 1 | D | 2.90 | 2 | D | 3.17, 97.91 |
| rtD131S | 1 | D | 4.10 | |||
| rtD134G | 1 | D | 1.30 | |||
| rtD134V | 2 | D | 61.4, 54.8 | |||
| rtH135T | 1 | D | 82.47 | |||
| rtC136Y | 1 | D | 1.00 | 1 | D | 1.02 |
| rtD139K | 1 | D | 2.87 | 1 | D | 7.55 |
| rtD139Q | 1 | D | 1.10 | 1 | D | 3.15 |
| rtD139S | 1 | D | 1.09 | |||
| rtY141Q | 1 | D | 69.90 | |||
| rtV142E | 2 | D | 10.46, 18.65 | |||
| rtL145 M | 1 | D | 4.95 | 1 | D | 1.75 |
| rtK149H | 1 | D | 4.41 | |||
| rtF151Y | 1 | D | 3.94 | 1 | D | 1.65 |
| rtH160R | 1 | D | 2.19 | |||
| rtA186T | 1 | D | 1.16 | |||
| rtC188R | 1 | D | 1.16 | |||
| rtR192S | 1 | D | 1.16 | |||
| rtE218D | 1 | D | 21.9 | |||
| rtS230P | 1 | D | 2.46 | |||
| rtI233 M | 1 | D | 2.46 | |||
| rtI233T | 1 | D | 2.22 | |||
| rtL235stop | 1 | D | 2.17 | 1 | D | 1.15 |
| rtP237L | 1 | A | 1.09 | |||
| rtP237T | 1 | D | 53.08 | 2 | A, D | 46.78, 99.49 |
| rtA238H | 2 | D | 2.22, 2.5 | 1 | D | 1.76 |
| rtR242G | 1 | D | 1.23 | 1 | D | 1.17 |
| rtY245H | 1 | D | 6.60 | |||
| rtS246P | 1 | A | 1.09 | |||
| rtH248Y | 1 | D | 1.23 | |||
| rtF249L | 1 | A | 1.09 | |||
| rtM250T | 1 | D | 1.23 | |||
| rtI253T | 1 | A | 5.00 | |||
| rtY257G | 1 | D | 2.00 | |||
| rtY257W | 1 | D | 53.08 | 1 | D | 94.94 |
| rtW257L | 1 | A | 12.00 | |||
| rtT259P | 1 | A | 12.90 | |||
| rtH264Y | 1 | D | 1.23 | |||
| rtI266V | 2 | D | 46.91, 99.13 | |||
| rtQ267H | 1 | D | 1.14 | 2 | A, D | 46.78, 97.97 |
| rtI269T | 1 | D | 1.23 | |||
| rtE271H | 2 | D | 11.1, 99.13 | |||
| rtH271D | 1 | A | 46.20 | |||
| rtH271F | 1 | A | 12.90 | |||
| rtR274S | 2 | A; D | 4.76, 2.22 | |||
| rtI278A | 1 | D | 1.60 | |||
| rtV278I | 1 | A | 41.6 | |||
| rtW284R | 1 | A | 2.40 | |||
n, number of patients showing the indicated substitution.
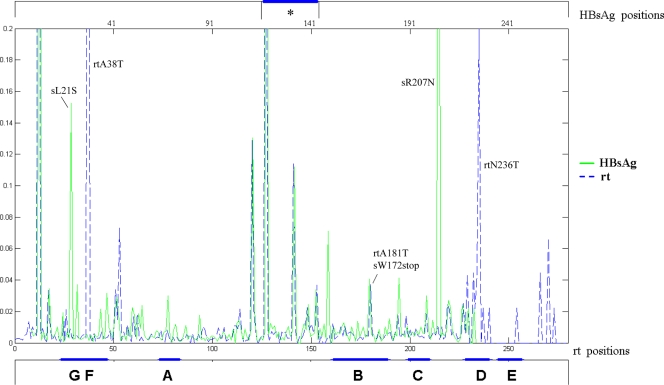
The UDPS sequence data were also used to identify the substitutions in the overlapping HBsAg ORFs. The overall distribution of HBsAg changes detected by UDPS in each patient is shown in Fig. 1B. Consistent with the RT findings, the HBsAg sequence from genotype D HBV was more variable, and the number of changes was higher in treated (n = 160) than drug-naive (n = 92) patients. However, the frequency of substituted amino acids in the corresponding positions of the two overlapping ORFs was not superimposable, implying that some nucleotide changes were synonymous in one ORF and nonsynonymous in the overlapping ORF. As an example, Fig. 2 shows the substitution frequencies in RT and HBsAg from patient 2 at the corresponding positions in the two ORFs. As can be seen, among the most striking differences, two changes in RT (rtA38T and rtN236T; frequencies of 73.38 and 21.79%, respectively), corresponded to wild-type (wt) amino acids of HBsAg; vice versa, 2 changes in HBsAg (sL21S and sR207N; frequencies of 14.80% and 56.80%, respectively), corresponded to wt amino acids of RT. It is noteworthy that all of the substitutions in the “a” determinant of HBsAg corresponded to changes in RT. In addition, the recently described mutation rtA181T (31) corresponded to a stop codon in HBsAg (sW172stop) and was present in a minority of viral quasispecies (about 3.5%); a similar situation for rtA181T was observed in patient 1 (see Table S1 in the supplemental material). The complete list of HBsAg substitutions and their relative frequencies in each patient is shown in Table S1 in the supplemental material; the correspondence between each variant position in RT and that resulting in HBsAg is reported in Table S2 in the supplemental material. Note that in one drug-naive patient (patient 11), the main immune escape mutant, sG145R, was detected at a frequency of 31.2%, corresponding to rtR153Q (see Table S1 in the supplemental material). In addition, several patients showed a number of stop codons at different positions in the S gene, leading to truncated HBsAg, whose frequency was very low (range, 1 to 3.6%) in most cases, with the exception of patient 11, in whom it was 17.1%. This patient appeared to be rather exceptional among the drug-naive patients included in the study, showing the highest number of substitutions in both RT and HBsAg, with one and eight stop codons, respectively (see Table S1 in the supplemental material).
FIG. 2.
Distribution of changed codons in the overlapping ORFs of RT and HBsAg detected by UDPS in one patient. The overall distribution of changed positions and their relative frequency are shown for patient 2, reporting the two overlapping ORFs in the same panel. The mutated codons are shown according to their localization in RT and HBsAg. The “a” determinant (*) of HBsAg is highlighted at the top, while functional and interdomain regions of RT are underlined at the bottom of the panel. The frequency scale axis has been cut at 20%, to allow better visualization of infrequent substitutions. For details, see Table S1 in the supplemental material.
DISCUSSION
To date, massively parallel UDPS has been applied to HIV drug resistance (10, 28, 38), but very few, preliminary data (including our own study) on HBV have been reported to date (M. R. Capobianchi, M. Solmone, D. Vincenti, A. Bruselles, and G. Ippolito and S. Margeridon-Thermet, N. Shulman, T. Liu, A. Ahmed, C. Wang, B. B. Simen, J. F. Simons, M. Egholm, B. Gharizadeh, and R. W. Shafer, presented at Hepatitis B and C Virus Resistance to Antiviral Therapies, Paris, France, 14 to 16 February 2008).
In this study, we describe for the first time the use of the massively parallel UDPS method, based on the next generation sequencing platform GS FLX, to analyze viral quasispecies in the RT/S region of the HBV genome.
To date, the most commonly used method for detecting drug resistance mutations is direct sequencing after PCR amplification by the Sanger method (population-based sequencing approach). This approach allows identification of all substitutions included in the amplified fragment, including primary, compensatory, and novel mutations; the contemporary determination of viral genotype; and the identification of amino acid changes in the overlapping S gene. However, direct sequence-based methods are only capable of detecting substitutions present in viral quasispecies with a prevalence of ≥20% of the total HBV population (30). On the contrary, sequence-specific methods (such as DR v2), also largely used in clinical practice, are rather sensitive and can detect minority variants represented at a frequency down to about 5%, but allow detection of only a limited set of known mutations.
The UDPS approach enables the simultaneous analysis of thousands of clonally amplified fragments from each patient, increasing the probability of highlighting minority variants. In fact, the results indicate that UDPS has a much higher relative sensitivity not only compared to direct sequencing, as expected, but also compared to DR v2. Furthermore, a large set of substitutions were detected by UDPS and the results were quantitative, allowing us to establish the relative frequency of both known and novel changes in both the RT and the overlapping S gene ORFs, in drug-experienced as well as in drug-naive patients.
Pyrosequencing (i.e., sequencing by synthesis) has been used previously to detect resistance mutations in HBV (14, 20). Compared to the classical pyrosequencing, UDPS presents a major advantage. In fact, a limitation inherent in classical pyrosequencing is that the read length is approximately 40 bp. This aspect, while allowing a large-scale screening for known point mutations, such as those at codons 180 and 204 (14, 20), renders impractical the use of pyrosequencing for whole-gene sequence analysis. On the contrary, the UDPS approach produces amplicons of about 200 bp: hence, a proper combination of primers (in this study, eight primer pairs were used) permits one to easily cover the whole RT gene, allowing identification of the entire set of mutations spread along this gene.
On the whole, UDPS results confirm the higher heterogeneity of HBV genotype D compared to genotype A (4). In addition, the patients on treatment showed a higher variability in both functional and nonfunctional domains of RT as compared to drug-naive patients, but the difference was higher for functional domains, supporting the concept that the selective pressure of the drug is stronger for domains involved in enzymatic activity. However, the older age of infection in treated patients may also play a role in the accumulation of substitutions, irrespective of selective pressure exerted by treatment. An alternative explanation is that treatment with analogues actually could be mutagenic to the virus, resulting in a higher mutation frequency in treated patients.
Interestingly, a number of changes, both novel and already described as associated with drug resistance, were found at variable frequencies in drug-naive patients. Specifically, rtM204I, corresponding to a stop codon in HBsAg, was detected at low frequency in patient 11, and V214A, which is associated with reduced sensitivity to ADV and TDF, was detected in two drug-naive patients (patients 11 and 12) at frequencies of 7.0% and 1.9%, respectively. The presence of mutations in patients never exposed to antiviral drugs is not unexpected, as it is well known that the viral replicative enzyme is error prone, and a number of casual mutations arise at each defined time point during the infection, being fixed by casual drift, or by positive selection. In fact, several studies have demonstrated that mutations associated with LAM resistance do occur naturally and can be found in low percentage in HBV carriers who have never received LAM or other antiviral therapies (24). Recent data suggested that preexistence of LAM mutations at frequencies as low as 0.1% in the viral population from patients who have not received LAM treatment may increase the risk of ETV-associated mutations (3).
In addition to the possibility to detect rare mutations before the start of therapy, because UDPS provides quantitative results, it may represent a suitable tool for the precise monitoring of viral quasispecies dynamics along treatment history, allowing the early identification of increasing frequencies of HBV mutants before the appearance of viral breakthrough. In fact, the appearance of resistance mutations (genotypic resistance) may precede the increase in viral load (virological resistance) by several months (9, 18) and could be a prognostic marker for the occurrence of viral breakthrough.
It is well known that the RT and S genes of HBV have overlapping ORFs, and thus drug resistance mutations in RT can also lead to changes in the HBsAg (27, 37). The new technology may be useful also to detect HBsAg substitutions. From our results, the frequency of changed amino acids in RT and HBsAg was not superimposable at all positions, implying that some nucleotide changes were synonymous in one ORF and nonsynonymous in the overlapping ORF; the driving force for selecting such variations is presumably acting differently on RT and HBsAg. Further studies are necessary to clarify this point, and it is foreseen that the new technology will be extremely helpful to this aim.
One particular aspect which deserves further investigation is the high number of mutations leading to stop codons in RT and in HBsAg, whose frequency was generally low (in the range of 1 to 3%) but might represent a relevant portion of the viral population in some patients (see for example, sW36stop, detected at 17.1% in patient 11). It is likely that situations similar to that of rtA181T (corresponding to sW172stop), recently described to affect the viral replication rate as a dominant-negative mutant (40), may be more diffused than expected. The new method is particularly suitable to identify mutations whose presence may have been disregarded so far, due to their low frequency. In fact, low frequencies are expected from mutated virions that are not capable of coding correct RT or HBsAg, as their function is expected to be strongly impaired and their maintenance in viral quasispecies should be dependent on the concomitant presence of wild-type virions providing the lost function(s). Similar situations may be more common than expected and may provide, for instance, a possible explanation for primary nonresponse to antiviral treatment in the absence of detectable mutations in some patients (22).
As this study included a small number of patients, HBV substitutions detected by UDPS and the different patterns observed in drug-treated versus drug-naive patients require further investigation to clarify their clinical significance. In addition, it remains to be established whether all of the variations with respect to consensus sequences represent true mutations or simple polymorphisms. Despite these limitations, our results clearly indicate that genotyping analysis based on the next generation sequencing platform is suitable for characterization of genetic diversity and detection of minor variants that may have clinical relevance for adapting patient management and adjusting treatment strategies.
Supplementary Material
Acknowledgments
This study has been partially supported by grants from the Italian Ministry of Health, “Fondi Ricerca Corrente” and “Ricerca Finalizzata (Progetto Strategico)” to INMI.
We thank Carla Nisii for revising the English in the manuscript.
Footnotes
Published ahead of print on 10 December 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agresti, A., and B. A. Coull. 1998. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52119-126. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Colonno, R. J., R. Rose, C. J. Baldick, S. Levine, K. Pokornowski, C. F. Yu, A. Walsh, J. Fang, M. Hsu, C. Mazzucco, B. Eggers, S. Zhang, M. Plym, K. Klesczewski, and D. J. Tenney. 2006. Entecavir resistance is rare in nucleoside naïve patients with hepatitis B. Hepatology 441656-1665. [DOI] [PubMed] [Google Scholar]
- 4.De Maddalena, C., C. Giambelli, E. Tanzi, D. Colzani, M. Schiavini, L. Milazzo, F. Bernini, E. Ebranati, A. Cargnel, R. Bruno, M. Galli, and G. Zehender. 2007. High level of genetic heterogeneity in S and P genes of genotype D hepatitis B virus. Virology 365113-124. [DOI] [PubMed] [Google Scholar]
- 5.Dienstag, J. L., R. D. Goldin, E. J. Heathcote, H. W. Hann, M. Woessner, S. L. Stephenson, S. Gardner, D. F. Gray, and E. R. Schiff. 2003. Histological outcome during long-term lamivudine therapy. Gastroenterology 124105-117. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson, N., L. Pachter, Y. Mitsuya, S. Y. Rhee, C. Wang, B. Gharizadeh, M. Ronaghi, R. W. Shafer, and N. Beerenwinkel. 2008. Viral population estimation using pyrosequencing. PLoS Comput. Biol. 4e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fattovich, G. 2003. Natural history of hepatitis B. J. Hepatol. 39(Suppl.)S50-S58. [DOI] [PubMed] [Google Scholar]
- 8.Gotoh, O. 1982. An improved algorithm for matching biological sequences. J. Mol. Biol. 162705-708. [DOI] [PubMed] [Google Scholar]
- 9.Hadziyannis, S. J., G. V. Papatheodoridis, E. Dimou, A. Laras, and C. Papaioannou. 2000. Efficacy of long-term lamivudine monotherapy in patients with hepatitis B e antigen-negative chronic hepatitis B. Hepatology 32847-851. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann, C., N. Minkah, J. Leipzig, G. Wang, M. Q. Arens, P. Tebas, and F. D. Bushman. 18 June 2007, posting date. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 35e91. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosseini, S. Y., F. Sabahi, S. Amini-Bavil-Olyaee, S. M. Alavian, and S. Merat. 2006. A novel accurate ACRS-PCR method with a digestion internal control for identification of wild type and YMDD mutants of hepatitis B virus strains. J. Virol. Methods 137298-303. [DOI] [PubMed] [Google Scholar]
- 12.Hussain, M., and A. S. Lok. 1999. Mutations in the hepatitis B virus polymerase gene associated with antiviral treatment for hepatitis B. J. Vir. Hepat. 6183-194. [DOI] [PubMed] [Google Scholar]
- 13.Hussain, M., S. Fung, E. Libbrecht, E. Sablon, C. Cursaro, P. Andreone, and A. S. Lok. 2006. Sensitive line probe assay that simultaneously detects mutations conveying resistance to lamivudine and adefovir. J. Clin. Microbiol. 441094-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ijaz, S., C. Arnold, S. Dervisevic, J. Mechurova, N. Tatman, R. S. Tedder, and N. V. Naoumov. 2008. Dynamics of lamivudine-resistant hepatitis B virus during adefovir monotherapy versus lamivudine plus adefovir combination therapy. J. Med. Virol. 801160-1170. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, I. M. 2008. Combination therapy for chronic hepatitis B: ready for prime time? J. Hepatol. 5687-691. [DOI] [PubMed] [Google Scholar]
- 16.Jourdain, G., N. Ngo-Giang-Huong, S. Le Coeur, C. Bowonwatanuwong, P. Kantipong, P. Leechanachai, S. Ariyadej, P. Leenasirimakul, S. Hammer, M. Lallemant, et al. 2004. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N. Engl. J. Med. 351229-240. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. S., K. H. Han, S. H. Ahn, E. O. Kim, H. Y. Chang, M. S. Moon, H. J. Chung, W. Yoo, S. O. Kim, and S. P. Hong. 2005. Evaluation of methods for monitoring drug resistance in chronic hepatitis B patients during lamivudine therapy based on mass spectrometry and reverse hybridization. Antivir. Ther. 10441-449. [PubMed] [Google Scholar]
- 18.Lampertico, P., M. Viganò, E. Manenti, M. Iavarone, G. Lunghi, and M. Colombo. 2005. Adefovir rapidly suppresses hepatitis B in HBeAg-negative patients developing genotypic resistance to lamivudine. Hepatology 41414-1419. [DOI] [PubMed] [Google Scholar]
- 19.Li, Z. G., L. Y. Chen, J. Huang, P. Qiao, J. M. Qiu, and S. Q. Wang. 2005. Quantification of the relative levels of wild-type and lamivudine-resistant mutant virus in serum of HBV-infected patients using microarray. J. Vir. Hepat. 12168-175. [DOI] [PubMed] [Google Scholar]
- 20.Lindstrom, A., J. Odeberg, and J. Albert. 2004. Pyrosequencing for detection of lamivudine-resistant hepatitis B virus. J. Clin. Microbiol. 424788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locarnini, S. 2004. Molecular virology of hepatitis B virus. Semin. Liver Dis. 24(Suppl.)3-10. [DOI] [PubMed] [Google Scholar]
- 22.Locarnini, S. 2008. Primary resistance, multidrug resistance, and cross-resistance pathways in HBV as a consequence of treatment failure. Hepatol. Int. 2147-151. doi: 10.1007/s12072-008-9048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lok, A. S., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. De Vreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 403729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lok, A. S., F. Zoulim, S. Locarnini, A. Bartholomeusz, M. G. Ghany, and J. M. Pawlotsky. 2007. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology 46254-265. [DOI] [PubMed] [Google Scholar]
- 25.Marcellin, P., G. K. Lau, F. Bonino, P. Farci, S. Hadziyannis, R. Jin, Z. M. Lu, T. Piratvisuth, G. Germanidis, C. Yurdaydin, M. Diago, S. Gurel, M. Y. Lai, P. Button, N. Pluck, et al. 2004. Peginterferon alfa-2a alone, lamivudine alone, and the two in combination in patients with HBeAg-negative chronic hepatitis B. N. Engl. J. Med. 3511206-1217. [DOI] [PubMed] [Google Scholar]
- 26.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews, G. V., A. Bartholomeusz, S. Locarnini, A. Ayres, J. Sasaduesz, E. Seaberg, D. A. Cooper, S. Lewin, G. J. Dore, and C. L. Thio. 2006. Characteristics of drug resistant HBV in an international collaborative study of HIV-HBV-infected individuals on extended lamivudine therapy. AIDS 20863-870. [DOI] [PubMed] [Google Scholar]
- 28.Mitsuya, Y., V. Varghese, C. Wang, T. F. Liu, S. P. Holmes, R. Jayakumar, B. Gharizadeh, M. Ronaghi, D. Klein, W. J. Fessel, and R. W. Shafer. 20 August 2008. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J. Virol. [Epub ahead of print.] doi: 10.1128/JVI.01827-07. [DOI] [PMC free article] [PubMed]
- 29.Nash, K. L., and G. J. Alexander. 14 May 2008, posting date. The case for combination antiviral therapy for chronic hepatitis B virus infection. Lancet Infect. Dis. 8444-448. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 30.Pallier, C., L. Castéra, A. Soulier, C. Hézode, P. Nordmann, D. Dhumeaux, and J.-M. Pawlotsky. 2006. Dynamics of hepatitis B virus resistance to lamivudine. J. Virol. 80643-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer, S., M. Kearney, F. Maldarelli, E. K. Halvas, C. J. Bixby, H. Bazmi, D. Rock, J. Falloon, R. T. Davey, Jr., R. L. Dewar, J. A. Metcalf, S. Hammer, J. W. Mellors, and J. M. Coffin. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters, M., D. M. Walling, K. Kelly, G. L. Davis, J. G. Waggoner, and J. H. Hoofnagle. 1986. Immunologic effects of interferon-alpha in man: treatment with human recombinant interferon-alpha suppresses in vitro immunoglobulin production in patients with chronic type B hepatitis. J. Immunol. 1373147-3152. [PubMed] [Google Scholar]
- 33.Sablon, E., and F. Shapiro. 2005. Advances in molecular diagnosis of HBV infection and drug resistance. Int. J. Med. Sci. 28-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sertoz, R. Y., S. Erensoy, S. Pas, U. S. Akarca, F. Ozgenc, T. Yamazhan, T. Ozacar, and H. G. Niesters. 2005. Comparison of sequence analysis and INNO-LiPA HBV DR line probe assay in patients with chronic hepatitis B. J. Chemother. 17514-520. [DOI] [PubMed] [Google Scholar]
- 35.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147195-197. [DOI] [PubMed] [Google Scholar]
- 36.Tang, X. R., J. S. Zhang, H. Zhao, Y. H. Gong, Y. Z. Wang, and J. L. Zhao. 2007. Detection of hepatitis B virus genotypes using oligonucleotide chip among hepatitis B virus carriers in Eastern China. World J. Gastroenterol. 131975-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torresi, J. 2002. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J. Clin. Virol. 2597-106. [DOI] [PubMed] [Google Scholar]
- 38.Wang, C., Y. Mitsuya, B. Gharizadeh, M. Ronaghi, and R. W. Shafer. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 171195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, R. S., H. Zhang, Y. F. Zhu, B. Han, and Z. J. Yang. 2006. Detection of YMDD mutants using universal template real-time PCR. World J. Gastroenterol. 121308-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warner, N., and S. Locarnini. 2008. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology 4888-98. [DOI] [PubMed] [Google Scholar]
- 41.Woo, H. Y., H. Park, B. I. Kim, W. K. Jeon, Y. K. Cho, and Y. J. Kim. 2007. Comparison of mass spectrometric analysis and TRUGENE HBV genotyping for monitoring lamivudine resistance in chronic hepatitis B patients. Antivir. Ther. 127-13. [PubMed] [Google Scholar]
- 42.Wright, T. L. 2004. Clinical trial results and treatment resistance with lamivudine in hepatitis B. Semin. Liver. Dis. 24(Suppl.)31-36. [DOI] [PubMed] [Google Scholar]
- 43.Zoulim, F. 2004. Mechanism of viral persistence and resistance to nucleoside and nucleotide analogs in chronic hepatitis B virus infection. Antivir. Res. 641-15. [DOI] [PubMed] [Google Scholar]
- 44.Zoulim, F. 2006. New nucleic acid diagnostic tests in viral hepatitis. Semin. Liver. Dis. 26309-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.