Abstract
The human adenovirus type 5 (Ad5) E4orf4 product has been studied extensively although in most cases as expressed from vectors in the absence of other viral products. Thus, relatively little is known about its role in the context of an adenovirus infection. Although considerable earlier work had indicated that the E4orf4 protein is not essential for replication, a recent study using dl359, an Ad5 mutant believed to produce a nonfunctional E4orf4 protein, suggested that E4orf4 is essential for virus growth in primary small-airway epithelial cells (C. O'Shea, et al., EMBO J. 24:1211-1221, 2005). Hence, to examine further the role of E4orf4 during virus infection, we generated for the first time a set of E4orf4 virus mutants in a common Ad5 genetic background. Such mutant viruses included those that express E4orf4 proteins containing various individual point mutations, those defective entirely in E4orf4 expression, and a mutant expressing wild-type E4orf4 fused to the green fluorescent protein. E4orf4 protein was found to localize primarily in nuclear structures shown to be viral replication centers, in nucleoli, and in perinuclear bodies. Importantly, E4orf4 was shown not to be essential for virus growth in either human tumor or primary cells, at least in tissue culture. Unlike E4orf4-null virus, mutant dl359 appeared to exhibit a gain-of-function phenotype that impairs virus growth. The dl359 E4orf4 protein, which contains a large in-frame internal deletion, clustered in aggregates enriched in Hsp70 and proteasome components. In addition, the late viral mRNAs produced by dl359 accumulated abnormally in a nuclear punctate pattern. Altogether, our results indicate that E4orf4 protein is not essential for virus growth in culture and that expression of the dl359 E4orf4 product interferes with viral replication, presumably through interactions with structures in the nucleus.
A considerable amount of information has accumulated over the past 15 years on the function of the human adenovirus E4orf4 protein when it is expressed in cells alone following DNA transfection or infection with adenovirus vectors (3, 6, 25). E4orf4 has been shown to induce p53-independent cell death of human cancer cells (36, 52), in many cases via a caspase-independent process (28, 29, 32, 43). Cell death has been shown to be dependent on binding of the E4orf4 protein to the Bα subunit of protein phosphatase 2A (PP2A) both in Saccharomyces cerevisiae and mammalian cells (1, 35, 47, 53). Both nuclear and cytoplasmic cell death targets, including the anaphase-promoting complex/cyclosome complex (27) as well as Src family kinases (12, 17, 28, 46), have been proposed to play a role in mitotic arrest and remodeling of the actin cytoskeleton, respectively.
Much less is known about the role of E4orf4 during adenovirus infection. Studies using viral mutants that are defective in the production of E4orf4 products (dl358 and dl359) (19) have shown that E4orf4 downregulates the response of E1A and cyclic AMP (26) as well as viral gene expression (5, 7, 34, 37), controls alternative splicing of the L1 transcript (14, 22), and activates mammalian target of rapamycin signaling in nutrient-deprived cells (42). Modulation of PP2A activity by E4orf4 toward certain substrates is believed to mediate these effects (5, 14, 22, 26, 34, 40).
Studies using several previously generated E4orf4 mutants have led to apparently contradictory results. Earlier work using mutants dl359, dl358, and pm1020 have suggested that loss of functional E4orf4 protein has little effect on viral replication in human tumor cells in culture (16, 31). In contrast, results from a more recent study using mutant dl359 have suggested that E4orf4 is essential for virus growth in primary small-airway epithelial cells (SAEC), at least in part due to its role in enhancing the synthesis of late viral proteins in cooperation with E4orf1 (42). Part of the problem in interpreting results from previous efforts is the fact that many of the various E4orf4 mutant viruses have been created in different genetic backgrounds.
To gain better insights into the role of E4orf4 during virus infection, we generated for the first time a set of E4orf4 virus mutants in a common adenovirus type 5 (Ad5) genetic background. Such mutant viruses included those that express E4orf4 proteins containing a range of individual point mutations, those defective entirely in E4orf4 expression, and a mutant expressing wild-type (wt) E4orf4 fused to the green fluorescent protein (GFP). Using these viruses we have studied viral replication and the localization of the E4orf4 proteins during productive infection in both human tumor cell lines and primary human cells. The results clearly indicated that E4orf4 protein is largely nuclear and that it is not essential to obtain normal yields of viral progeny in all tumor or primary cell types. Interestingly, the phenotype produced by dl359 appeared to result from a gain of function induced by the partial E4orf4 product encoded by this viral mutant.
MATERIALS AND METHODS
Cell lines.
Human non-small-cell lung carcinoma H1299 cells (ATCC CRL-5803) carrying a homozygous deletion of the p53 gene (39) were cultured in monolayers in Dulbecco's modified Eagle's medium (catalog no. 11995; Gibco) supplemented with 10% fetal calf serum (Gibco) and 100 U/ml of penicillin and streptomycin. The H1299-HA-E4orf4 cell line was created by transfecting H1299 cells with pcDNA3-HA-E4orf4 plasmid (where HA is hemagglutinin) and selection with 900 μg/ml G418 (Bioshop). Clones expressing low levels of E4orf4 were selected and amplified. SAEC were purchased from Cambrex and grown according to the supplier's specifications.
Antibodies.
Primary monoclonal and polyclonal antibodies used included E4orf4 polyclonal 2419 (28), Flag mouse monoclonal (Sigma), HA mouse monoclonal (Babco), actin monoclonal C4 (MP Biomedicals), fibrillarin polyclonal ab5821 (Abcam), Hsp70 rabbit polyclonal SPA-812 (Stressgen Bioreagents), 20S proteasome α1 subunit rabbit polyclonal (Calbiochem), E1B55K mouse monoclonal 2A6 (49), E4orf6 rabbit polyclonal 1807 (4), E1A mouse monoclonal M73 (20), E4orf3 rat monoclonal 6A11 (41), Ad5 capsid rabbit polyclonal L133 (24), DNA binding protein ([DBP] E2A) mouse monoclonal B6-8 (44), and antiadenovirus mouse monoclonal 059-M (BioGenex) antibodies. Secondary antibodies conjugated to horseradish peroxidase for detection in Western blotting were goat anti-mouse immunoglobulin G (IgG), goat anti-rabbit IgG, and goat anti-rat IgG (Jackson ImmunoResearch Laboratories). Secondary antibodies used for immunofluorescence were conjugated to Alexa 488 or 594 (Molecular Probes).
Expression vectors.
Cloning of cDNAs expressing HA-tagged E4orf4 (HA-E4orf4), HA-E4orf4 with the mutation L51A (HA-E4orf4-L51A), HA-E4orf4-L54A, and HA-E4orf4 with the double mutation L51A L54A (HA-E4orf4-L51/54A) has been described elsewhere (35). HA-dl359 and HA-dl358 were generated using QuickChange PCR (Stratagene) according to the manufacturer's instructions. Briefly, deletion of residues 46 to 55 (HA-dl359) or of residues 48 to 54 (HA-dl358) in E4orf4 was performed using the following forward and reverse oligonucleotides, respectively: 5′-GAA GGA GTT TAC ATA GAA CCC GAG TGG ATA TAC TAC AAC TAC-3′ and 5-GTA GTT GTA GTA TAT CCA CTC GGG TTC TAT GTA AAC TCC TTC-3′ or 5′-GTT TAC ATA GAA CCC GAA GCC G AGA GAG TGG ATA TAC TAC-3′ and 5′-GTA GTA TAT CCA CTC TCT GGC TTC GGG TTC TAT GTA AAC-3′. Deletions were made to parallel the deletions present in the dl359 and dl358 viral mutants (see above for details). All mutations were confirmed by sequencing.
Viruses.
Generation of H5pg4100, dl309 (21), dl358, dl359 (19), and pm1020 (37) has been described elsewhere as has the construction of plasmid pH5pg4100 and the transfer vector pE4-1155 (18). The Ad5 genome in plasmid pH5pg4100 is inserted into the PacI site of the bacterial cloning vector pPG-S2 (18). It lacks nucleotides (nt) 28593 to 30471 (encompassing most of E3) and contains an additional unique endonuclease restriction site at nt 30955 (BstBI) (nucleotide numbering is according to the published Ad5 sequence from the GenBank, accession no. AY339865). The transfer vector pE4-1155 contains Ad5 nt 32840 to 35934 in pPG-S2 (18). The entire sequences of pH5pg4100 (36,067 bp) and pE4-1155 (5,099 bp) are available upon request.
To generate the Ad5 with a deletion of E4orf4, GFP-E4orf4, and the mutant viruses, the desired mutation or insertions were first introduced into the E4orf4 gene in pE4-1155 by site-directed mutagenesis. For generating E4orf4 with a deletion of residues R69 to 75A (6RA), a protein with the mutations R81A and F84A (R81/F84A), and a protein carrying both of these mutations (6RA/RF), an in-frame Flag tag was first introduced at the beginning the E4orf4 gene by QuickChange PCR using the following primers: Fwd, GGT TTT GGA TAA TCT TTT GGA ATA AAA AAA AAA ACA TGG ACT ACA AAG ACG ATG ACG ATA AAA TGG TTC TTC CAG CTC TTC CCG CTC TTC CCG CTC C; and Rev, GGA GCG GGA AGA GCT GGA AGA ACC ATT TTA TCG TCA TCG TCT TTG TAG TCC ATG TTT TTT TTT TTA TTC CAA AAG ATT ATC CAA AAC C. This Flag-tagged construct was used for generating the specific mutation by QuickChange. The 6RA mutation was generated by two consecutive PCRs with the following primer sets: the pair Fwd1 (CAC AGA GCG ATC TAA GGA GGA AGA CCG GAG ACG CAG ATC) and Rev1 (GAT CTG CGT CTC CGG TCT GCC GCC TTA GAT CGC TCT GTG) and the pair Fwd2 (GAG CGA TCT AAG GCG GCA GAC GCG GCA GCG GCA TCT GTT TGT CAC GCC CGC ACC) and Rev2 (GGT GCG GGC GTG ACA AAC AGA TGC CGC TGC CGC GTC TGC CGC CTT AGA TCG CTC). The R81/F84A mutation was generated with two consecutive QuickChange PCRs with the following primers: the pair Fwd1 (CAC GCC CGC ACC TGG GCT TGC TTC AGG AAA TAT G) and Rev1 (CAT ATT TCC TGA AGC AAG CCC AGG TGC GGG CCT G) and the pair Fwd2 (CTG TTT GTC ACG CCG CCA CCT GGG CTT GCT TC) and Rev2 (GAA GCA AGC CCA GGT GGC GGC CTG ACA AAC AG). The 6RA/RF mutation was generated with the same two sequential PCRs but using the 6RA mutant as a template. For the E4orf4-null (E4orf4−) construction, the ATG start codon of E4orf4 was mutated by QuickChange PCR using the following primers: Fwd, GGA ATA AAA AAA AAA ACA AGG TTC TTC CAG CTC TTC C; and Rev, GGA AGA GCT GGA AGA ACC TTG TTT TTT TTT TTA TTC C. The construct containing the GFP cDNA in front of the E4orf4 gene was generated in two steps. First an NdeI site was inserted in the transfer vector by QuickChange PCR using the following primers: Fwd, GGT TTT GGA TAA TCT TTT GGA ATA AAA AAA AAA CAT ATG GTT TTC TTC CCA GC; and Rev, GCT GGG AAG AAA ACC ATA TGT TTT TTT TTT ATT CCA AAA GAT TAT CCA AAA CC. A second PCR product containing the GFP gene flanked by NdeI restriction sites was generated using the enhanced GFP vector (Invitrogen) as a template with the following primers: Fwd, GGC ATA TGG TGA GCA AGG GCG AGC; and Rev, GGC ATA TGA CCA GCG ATA CCA GCA CCA AGA GCC TTG TAC AGC TCG TCC ATG CCG AG. This PCR product was cloned through the NdeI site in the modified transfer vector, resulting in a GFP tag upstream of the E4orf4 gene separated by the following linker: ALGAGIAG. Following mutation in the transfer vector, the E4 region was swapped in the bacmid containing the wt adenoviral genome as described previously (18). Recombinant plasmids were partially sequenced to confirm the mutations in the E4orf4 gene. Other E4orf4 virus mutants not discussed in this paper (K88A, 4YF, D71A, and F84A) were also generated in a similar fashion. A summary of the predicted E4orf4 products of these mutants is shown in Fig. 1. For the generation of mutant viruses, the viral genomes were released from the recombinant plasmids by PacI digestion and transfected in H1299 cells or in the H1299-HA-E4orf4 stable cell line. After 5 days, cells were harvested, and viruses were released by four freeze-thaw cycles. The viruses were propagated in H1299 monolayer cells (except for dl359, which was propagated in H1299-HA-E4orf4 cells), and titers were determined by the fluorescence-forming unit (FFU) method (18). Briefly, serial dilutions of the viruses were used to infect H1299 cells, which were fixed at 20 h postinfection (hpi). Infected cells were then identified by immunostaining with an antibody against the early E2A protein (antibody B6-8). Viral DNA was isolated as described previously (50) from viral particles purified by cesium chloride equilibrium density centrifugation (18, 50) and analyzed by HindIII restriction endonuclease digestion. In addition, the viral DNA was partially sequenced to verify the presence of the E4orf4 mutations/insertion.
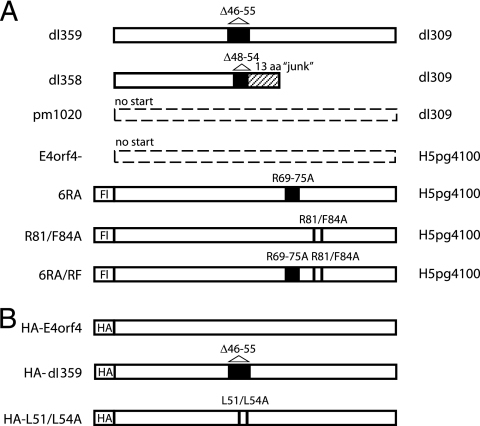
FIG. 1.
E4orf4 mutants. Schematic representation of the predicted products of the E4orf4 mutant viruses (A) and cDNAs (B). Black boxes and bars indicate deleted or mutated amino acids while the hashed box represents an out-of-frame sequence. The newly created viruses were made in the H5pg4100 background, unlike the previously published mutant viruses which were made in the dl309 background. The background of the viruses is indicated on the right. Fl and HA indicate that the open reading frames were tagged with a Flag tag or an HA tag, respectively. aa, amino acids.
Transfection, infections, immunoprecipitations, time courses, and growth curves.
Cells were transfected in six-well dishes with 1 μg of DNA using the liposome reagent DMRIE-C (1,2-dimyristyloxypropyl-3-dimethyl-hydroxy ethyl ammonium bromide and cholesterol) (Gibco-BRL) or infected with viruses diluted in infection medium (0.2 mM CaCl2, 0.2 mM MgCl2, and 2% serum in phosphate-buffered saline plus 100 U/ml of penicillin and streptomycin) for 90 min, after which the infection medium was removed and normal growth medium was added. For time course studies, cells were infected at a multiplicity of infection (MOI) of 5 FFU/cell and harvested by scraping at different times postinfection. The cells were washed in phosphate-buffered saline and collected by centrifugation, and the pellets were frozen until completion of the experiment. For time course analysis, pellets were lysed in radio immunoprecipitation assay buffer (10 mM sodium phosphate, pH 7.2, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol) plus inhibitors (4 mM NaF, 2 mM sodium polyphosphate, 500 μM sodium vanadate, 200 μg/ml phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, and 5 μg/ml leupeptin), subjected to a freeze-thaw step in liquid nitrogen, and sonicated three times for 20 s. Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes. For immunoprecipitations, cells were infected at an MOI of 20 FFU/cell, and at 20 hpi, they were lysed in 1% SDS, 50 mM Tris, pH 7.4, and inhibitors (4 mM NaF, 2 mM sodium polyphosphate, 500 μM sodium vanadate, 200 μg/ml phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, and 5 μg/ml leupeptin). The extracted samples were then diluted 1 in 5 in a second buffer containing 1.25% Triton X-100, 50 mM Tris, pH 7.4, 190 mM NaCl and the same inhibitors and sonicated. Equal protein amounts (1 mg) were immunoprecipitated with 2419 (E4orf4) antibodies before the addition of protein G-Sepharose (Upstate). Following extensive washing in the final lysis buffer plus phenylmethylsulfonyl fluoride, the beads were eluted in Laemmli buffer and run on SDS-polyacrylamide gels. For the viral growth curves, cells were infected at an MOI of 5 FFU/cell and harvested at 24, 48, and 72 hpi. Viruses were released from the cells by three freeze-thaw cycles, and titers were determined by a fluorescent antibody assay (FFU assay). Total progeny virions in each sample were calculated, and values were plotted on a graph.
Immunofluorescence and microscopy.
Infected or transfected cells grown on coverslips were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked in 1% goat serum before incubation with antibodies; before mounting, cell nuclei were also stained with 4′,6′-diamidino-2-phenylindole (DAPI). For in vivo localization studies, live cells expressing GFP-E4orf4 were observed using a Zeiss LSM 510 Axiovert 100 M confocal microscope equipped with a Plan-Achromat 63× (1.4 numerical aperture) oil differential interference contrast objective. For regular confocal microscopy, samples were scanned at a speed of 7 using the linear mode and the mean method as well as a pinhole of 106 (optical slice, <2.2 μm) and a zoom setting of 2. Eight scans were compiled and averaged using Zeiss LSM 510 software to give the final image, which was captured as an 8-bit TIF file. The detector gain was set to between 800 and 1,000 while the amplifier onset and amplifier gain were kept constant. The palette tool was also used in order to ensure that the signals recorded did not reach pixel saturation. For live-cell imaging, pictures were taken in a similar manner except that four scans instead of eight were compiled per picture in order not to overbleach the samples while the detector gain was set at around 800 at the beginning of the procedure and left untouched during the whole duration of the procedure. Pictures were taken every 15 min between 8 and 21 hpi. Cells were kept in a 37°C chamber supplemented with 5% CO2. Regular fluorescence microscopy was also performed using a Zeiss Axiovision 3.1 microscope equipped with an Axiocam HR (Zeiss, Thornwood, NY) digital camera.
RNA fluorescence in situ hybridization.
H1299 cells were infected with the wt dl309 or mutant dl359 viruses at an MOI of 5 PFU for 24 h. Cells were first extracted with 0.5% Triton X-100 in buffer containing 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8 and 2 mM vanadyl riboside complex before being fixed in 4% paraformaldehyde. The tripartite leader probe (TGT GAC TGG TTA GAC GCC TTT CTC GAG AGG TTT TCC GAT CCG GTC GAT GCG GAC TCG CTC AGG TCC CTC GGT GGT T) or the poly(dT) probe was prepared as described by Singer (http://singerlab.org/protocols/). Briefly, the amino-allyl T-amino-acid modified nucleotides were incorporated in the oligonucleotides during their synthesis and conjugated to the Cy3 label. The labeled oligonucleotideswere hybridized to the cells in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5% dextran sulfate, 12.5 mM NaPO4, and 2 mM vanadyl riboside complexovernight at 30°C. Following washes in 50% formamide plus 2× SSC and with 2× SSC alone, the cells were blocked for immunofluorescence and then subjected to a standard immunofluorescence assay with the antibody against adenovirus.
RESULTS
Generation of E4orf4 virus point mutants.
To analyze the function and localization of Ad5 E4orf4 during productive infection, a series of virus mutants (Fig. 1 shows predicted E4orf4 products) was generated in a common Ad5 genetic background using a direct cloning approach (18). In addition to an E4orf4-null virus (E4orf4−) in which the initiator ATG codon was mutated and a variant (GFP-E4orf4) that produces an E4orf4 protein that is tagged at the amino terminus with GFP, several mutants containing alterations affecting individual amino acids were generated. These included E4orf4 mutations that had been shown in DNA transfection experiments to result in proteins defective for binding to PP2A and/or cell killing (the R81/F84A, 6RA, and 6RA/RF proteins, the last two of which also affect the nuclear/nucleolar localization signal [38]). The viruses were grown and amplified, and titers were determined in H1299 or H1299 cells stably expressing small amounts of E4orf4 (see Materials and Methods for details). They were then purified, viral DNA was isolated, and several adenovirus genes were sequenced to ensure that the viruses contained the desired mutations in the E4orf4 gene but not in other genes important for the virus life cycle (data not shown).
GFP-E4orf4 associates with several nuclear structures during infection.
The E4orf4 protein, when overexpressed from plasmid cDNAs following transfection, has previously been shown by our group and others to be predominantly localized in the nucleus of cells, with small amounts also present in the cytoplasm (28, 38, 45, 52). A nuclear/nucleolar localization signal rich in arginine residues seems to mediate this localization and to be important for E4orf4-induced cell death (38). Using the virus variant that expresses GFP-E4orf4, we studied the cellular localization of the wt E4orf4 protein produced at normal levels under the viral E4 promoter during productive infection. H1299 cells were infected at an MOI of 5, and the localization of GFP-E4orf4 was monitored between 8 and 24 hpi using live-cell imaging and confocal microscopy. Figure 2A shows representative images from these immunofluorescence studies. GFP-E4orf4 was first detected throughout the nucleus of infected cells at about 8 hpi. Frequently, distinct, small, and very intense dots were observed early after infection (Fig. 2A, frame a). As the infection progressed, GFP-E4orf4 protein in many cells became enriched in distinct nuclear bodies that looked like replication centers (Fig. 2A, frame b), and in some cells it accumulated in what appeared to be nucleoli (Fig. 2A, frame c) and in perinuclear bodies resembling aggresomes (Fig. 2A, frame d). At times following 18 hpi cells started swelling, rounding up, blebbing, and finally detaching from the plate. In many cells throughout the infection, small amounts of GFP-E4orf4 protein could also be detected in cytoplasmic structures as well as at the plasma membrane (data not shown). For live-cell imaging illustrating many of these observations, see Movie S1 in the supplemental material.
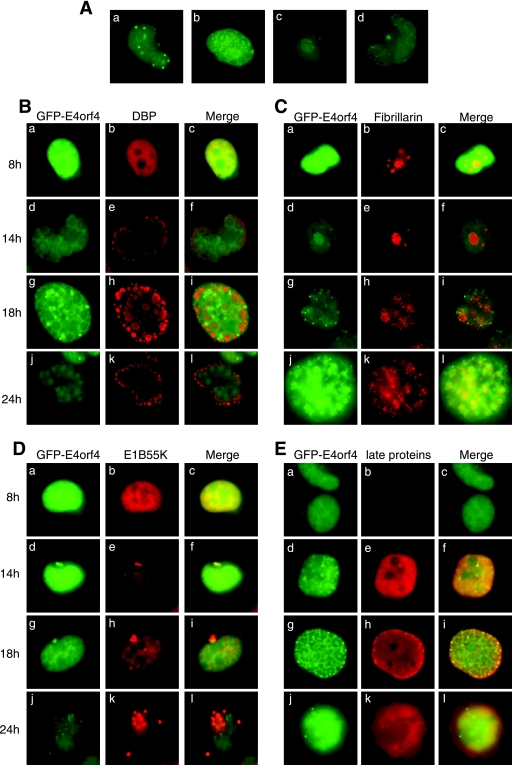
FIG. 2.
Localization of GFP-E4orf4 protein during infection. (A) H1299 cells were infected with the GFP-E4orf4 virus at an MOI of 5 FFU/cell, and pictures of live cells were taken during the 8- to 24-hpi period and represent the various structures that the GFP-E4orf4 protein localizes. (B to E) H1299 cells were infected with the GFP-E4orf4 virus at an MOI of 5 FFU/cell and at the indicated time on the left were fixed with 4% paraformaldehyde. Cells were stained with antibodies against DBP (B), fibrillarin (C), E1B55K (D), or the viral late proteins (E), as indicated on the figure.
To confirm the identity of the nuclear bodies containing GFP-E4orf4 protein, colocalization studies were performed with proteins known to associate with adenovirus replication centers, nucleoli, and aggresomes, namely, the adenovirus 72-kDa DBP (Fig. 2B), fibrillarin (Fig. 2C), and E1B55K that was shown previously to be present in aggresomes (2, 31) (Fig. 2D), respectively, as well as with late viral proteins (Fig. 2E). Figure 2B shows that although GFP-E4orf4 protein is present throughout the nucleus between 8 and 18 hpi, it does not specifically colocalize with DBP within viral replication centers; rather, it has an opposite pattern, with strong staining in the middle and outside of the replication rings delineated by DBP staining but not in the ring itself. Figure 2C shows that GFP-E4orf4 protein is enriched in nucleoli at 8 to 14 hpi as it colocalized with fibrillarin, but by 18 hpi the nucleoli started to be disrupted, resulting in a change in the E4orf4 protein localization. Figure 2D shows clear colocalization with E1B55K at earlier times in what appear to be aggresomes, but unlike with E1B55K, which continues to accumulate to large amounts in aggresomes until 24 hpi, only small amounts of E4orf4 protein accumulate there at later times. Figure 2E shows that by 14 hpi both E4orf4 and late proteins were present in the nucleus, and at 18 hpi both colocalized in small dots toward the periphery of the nucleus. At present, the significance of these structures in late viral events is not known (but see below). Coimmunofluorescence of GFP-E4orf4 with proteins known to be present in bodies that exhibit these morphological characteristics, including PML (PML bodies), coilin (Cajal bodies), and SC35 (splicing speckles), failed to show any colocalization with E4orf4 protein (data not shown), suggesting that the nuclear dots seen with GFP-E4orf4 do not correspond to these structures. Similar localization results were obtained in A549 cells (data not shown). The significance of this finding remains unknown.
Attempts were made to conduct comparable studies using wt virus and anti-E4orf4 antibodies (28); however, the avidity of these antibodies was not such that meaningful results could be obtained. Thus, it cannot be ruled out that the association with some of these structures may, at least in part, be due to interactions with the GFP portion of the tagged E4orf4 product. Nevertheless, the present studies have strongly suggested that during the infectious cycle, E4orf4 protein is present in nucleoli (presumably due to the presence of a nucleoli targeting sequence) (38) and aggresomes and localizes at the periphery of viral replication centers. Potential implications of these observations will be discussed below.
E4orf4 is not essential for virus growth in human cancer or primary SAEC.
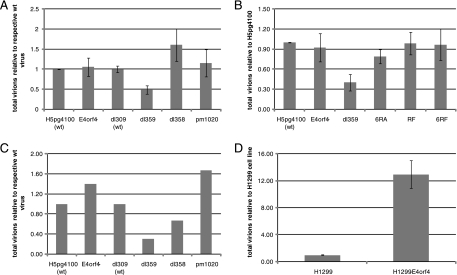
Recently in studies involving mutant dl359, E4orf4 was reported to be essential for virus growth in SAEC, causing a significant reduction in late viral protein levels and progeny virions (42). These results differed from previous studies conducted in human tumor cell lines infected with other E4orf4 mutants that indicated little effect on virus production in the absence of functional E4orf4 protein (19, 37). We therefore examined the growth of the newly created E4orf4 mutant viruses in both H1299 human tumor cells and in primary SAEC. H1299 cells were infected at an MOI of 5 with wt adenovirus H5pg4100 or with an E4orf4-null virus. Cells were also infected with dl309 (wt), dl359, which is the E4orf4 deletion virus reported to impair virus growth (42), and with dl358 (19) and pm1020 (37), which are E4orf4 deletion and E4orf4-null viruses, respectively, made in the same genetic background (dl309) as dl359 and used in previous studies. Cells were harvested and lysed at 48 hpi. The supernatant containing the progeny virions was used to reinfect a second set of cell cultures, which were fixed at 20 hpi and used to quantify virus growth by fluorescence microscopy. As seen in Fig. 3A, while a significant reduction in virus growth was detected for dl359, no major difference was observed with the other E4orf4 deletion and E4orf4-null viruses (E4orf4−, dl358, and pm1020) compared to the wt virus controls (H5pg4100 and dl309). Furthermore, when virus growth was investigated in the same cells using E4orf4 virus mutants containing point mutations previously reported to have defects in PP2A Bα binding and E4orf4-mediated killing, in localization, or in Src binding and extranuclear cell death (the 6RA, R81/F84A, and 6RA/RF mutations), no significant reduction in virus growth was observed (Fig. 3B) compared to the wt viruses (H5pg4100 and dl309), with the exception of dl359. Similar results were also obtained using A549 cells (data not shown). In addition, to examine the growth of these viruses in normal human primary cells, infections of SAEC were carried out, and Fig. 3C shows results from a representative experiment indicating that, again, only dl359 showed any consistent impairment for production of progeny virions. Hence, E4orf4 did not appear to be essential for adenovirus growth, at least under tissue culture conditions, whereas dl359 exhibited an apparent gain of function resulting in inhibition of virus production. To test this hypothesis, control H1299 cells carrying an empty vector or H1299-HA-E4orf4, a stable cell line expressing small amounts of HA-tagged wt E4orf4, were infected with dl359 at an MOI of 5 to determine if the wt E4orf4 protein could rescue dl359 growth (Fig. 3D). The viral progeny present at 48 hpi were measured, and, perhaps surprisingly, mutant dl359 replicated considerably better in H1299-HA-E4orf4 cells, suggesting that the presence of wt E4orf4 protein was able to restore growth of dl359 to nearly normal levels and overcome the gain-of-function defect induced by the dl359 E4orf4 product (see below for further discussion).
FIG. 3.
Effect of E4orf4 mutations on the virus growth. H1299 cells (A and B) or primary SAEC (C) were infected with the indicated viruses at an MOI of 5 FFU/cell and harvested at 48 h. The collected cells were broken by freeze-thaw cycles, and the progeny virions were titrated by immunofluorescence, as described in Materials and Methods. (D) H1299 cells stably expressing either an empty vector (H1299-EV) or the wt HA-tagged E4orf4 proteins (H1299-E4orf4) were infected with the dl359 virus at an MOI of 5 and harvested at 48 h. The collected cells were broken by freeze-thaw cycles, and the progeny virions were titrated by immunofluorescence, as described in Materials and Methods.
Production of viral proteins by E4orf4 mutants.
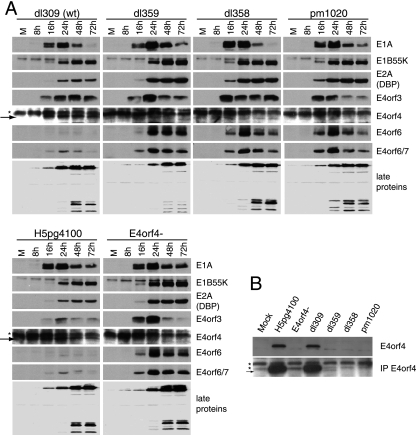
To examine the synthesis of E4orf4 and other viral proteins by the mutant viruses, H1299 cells were infected at an MOI of 5 with either of the two wt viruses (H5pg4100 and dl309) or with E4orf4−, dl359, dl358, or pm1020. Cells were harvested at 8, 16, 24, 48, and 72 hpi and lysed, and equal protein levels were subjected to SDS-PAGE, followed by Western blotting. Detection of the early viral proteins E1A, E1B55K, E2A (DBP), E4orf3, E4orf4, E4orf6, and E4orf6/7 and the late viral proteins (structural proteins) was performed using appropriate antibodies (see Materials and Methods). Figure 4A shows that while both wt viruses H5pg4100 and dl309 (wt) expressed E4orf4 protein, no expression of E4orf4 could be detected under these conditions for any of the E4orf4-null or E4orf4 deletion viruses E4orf4−, dl359, dl358, or pm1020. Based on the known sequence for dl359, it was predicted that a smaller version of the E4orf4 protein should be produced, and thus another approach was used to identify this potential product. In this case cell extracts were prepared in a more stringent buffer and were first immunoprecipitated with anti-E4orf4 antibodies and then subjected to Western blotting analysis, again using anti-E4orf4 antibodies. Figure 4B shows that under these conditions, a species was detected with dl359 that migrated slightly faster than wt E4orf4 protein (in H5pg4100 and dl309) and that was absent with the other E4orf4 mutants (E4orf4−, dl358, and pm1020). Thus, dl359 could express only very small amounts of a mutant E4orf4 protein; this species might be unstable, or it may be poorly recognized by anti-E4orf4 antibodies. Mutant dl358 is also predicted to encode a greatly truncated E4orf4 species; however, repeated attempts to identify such a product have been unsuccessful, suggesting that it may be difficult to detect for one of the reasons listed above.
FIG. 4.
Expression profile of virus proteins during infection with various E4orf4 mutants. (A) H1299 cells were infected with the indicated viruses at an MOI of 5 FFU/cell. Cells were harvested at several times, as indicated, and cell extracts were analyzed by SDS-PAGE followed by Western blotting using appropriate antibodies, as indicated to the right. The stars indicate background bands, and the arrows point at the lower bands that represent the mutant form of E4orf4 in dl359. (B) H1299 cells were infected with the indicated virus at an MOI of 20 FFU/cell for 20 h. Proteins were extracted using a more stringent buffer, as described in Materials and Methods, and equal amounts of protein were separated on SDS gels (top panel) or were first immunoprecipitated (IP) with antibodies to E4orf4 (2419) (bottom panel) and were immunoblotted with antibodies against E4orf4. Nonspecific bands are marked with asterisks, and the smaller dl358 fragment is marked with an arrow.
In addition, Fig. 4A shows that most of the other early viral proteins (E1A products, E1B55K, DBP, E4orf3, E4orf6, and E4orf6/7) were slightly or significantly overexpressed with all mutant viruses relative to the wt viruses. This phenomenon has been observed by several groups of investigators previously and is believed to result, especially for E2 and E4 products, from a relief of negative transcriptional regulation by the E4orf4 protein (5, 37, 57). Figure 4A also shows that, while late viral protein levels were comparable with wt viruses for most of the mutants, they were clearly reduced with dl359. Similar results were also obtained in A549 cells (data not shown). Hence, although dl359 exhibited a pattern of early protein expression similar to the other E4orf4 mutants, it was clearly defective in late protein production, as reported previously (42).
Analysis of the properties of the dl359 E4orf4 product.
To gain a better understanding of the unique properties of the dl359 virus, we first looked at the genome integrity of all these E4orf4 mutant viruses by purifying virus particles, isolating the viral DNA, and sequencing the E4orf4 gene as well as a few other genes including E1A, E4orf6, and E1B55K. Sequencing results confirmed that there were no unintended mutations in any of these genes in the E4orf4-null and E4orf4 deletion viruses tested, including dl359, that there was no methionine start codon in the E4orf4 gene of the E4orf4− and pm1020 viruses, and that there was a stop codon at the beginning of the E4orf4 coding sequence of the E4orf4− virus as designed (Fig. 1; also data not shown). Furthermore, sequencing results also showed that residues 48 to 54 of E4orf4 were deleted in dl358, resulting in a 20-bp out-of-frame deletion, while residues 46 to 55 were deleted in dl359, resulting in a 30-bp in-frame deletion, (data not shown). Consequently, dl358 should encode the first 47 residues of E4orf4 in addition to 14 random amino acids; however, as mentioned above, we have never detected this species. Likewise, dl359 should encode the first 46 amino-terminal residues of E4orf4 fused to residues 55 to 114 of the protein and thus lack an internal E-A-R-G-R-L-D-A-L-R 10-amino-acid sequence (Fig. 1). As shown above in Fig. 4B, we believe that this species is present in dl359-infected cells.
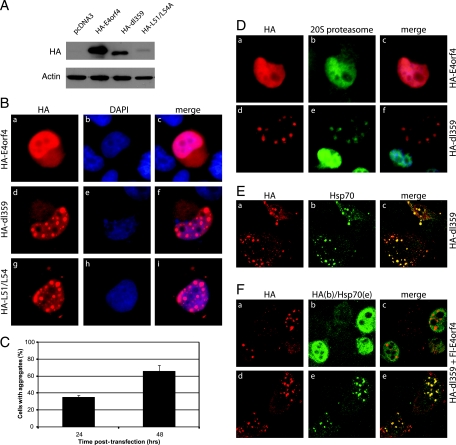
Because we were able to detect only low levels of this altered dl359 product using anti-E4orf4 antibodies, the predicted dl359 product tagged by HA was generated and used in studies that also included plasmid cDNAs expressing wt HA-E4orf4 and HA-E4orf4-L51/54A, an E4orf4 mutant containing two point mutations in the region deleted in dl359 (35). H1299 cells were transfected with these cDNAs, and protein expression as well as localization were investigated 48 h posttransfection. Figure 5A shows that, by Western blotting using anti-HA antibodies, the dl359 E4orf4 product was detected at somewhat reduced levels relative to wt HA-E4orf4 and that HA-E4orf4-L51/54A was present at greatly reduced levels. Similar results were also obtained using anti-E4orf4 antibodies (data not shown). These results clearly suggest that deletion or mutation of the sequence between amino acids 49 and 58 in E4orf4 may render the product at least partially unstable. Figure 5B shows results of initial localization studies employing immunofluorescence using anti-HA antibodies. Clearly, both HA-dl359 and HA-E4orf4-L51/54A localized differently from wt in that, instead of being evenly distributed throughout the nucleus, they were present in a punctate nuclear pattern. At later times of expression, dot-like structures also appeared in the cytoplasm. The size of the structures containing these proteins was seen to increase with time following infection (data not shown) as did the number of cells exhibiting this characteristic (Fig. 5C). It should be pointed out that parallel studies on the localization of E1B55K and the E4orf6 protein were undertaken and that no differences were seen with these proteins in wt- or dl359-infected cells (data not shown). The reduced stability of the dl359 product suggested that it may be misfolded and targeted for degradation by the 20S proteasome. Figure 5D shows that, indeed, this mutant product but not the wt E4orf4 protein colocalized with proteasomes. Since misfolded proteins tend to aggregate, often the heat shock protein Hsp70 will be present in these aggregates (10, 55, 56). Figure 5E shows that Hsp70 does indeed colocalize with the HA-dl359 aggregates. The E4orf4 mutation in the dl359 virus has a gain of function that results in reduced growth of the virus and that was relieved when the virus was grown in a cell line stably expressing wt E4orf4 (Fig. 3C). We therefore asked whether expression of wt E4orf4 eliminated or reduced the formation of the HA-dl359 aggregates. Figure 5F shows that even with strong expression of Flag-tagged E4orf4, the aggregates were still present (Fig. 5F, frames a to c) and still colocalized with Hsp70 (frames d and e). After large field of cells was counted, neither the percentage of cells containing aggregates nor the size of the aggregates was affected by the expression of wt E4orf4 (data not shown). These results suggested that the compensatory effect for viral growth of wt E4orf4 protein in dl359-infected cells did not involve increased degradation or redistribution of the mutant dl359 E4orf4 product.
FIG. 5.
Effect of the dl359 mutation on the expression and localization of the E4orf4 protein. (A) Plasmid DNAs encoding HA-tagged wt E4orf4, a mutant containing the dl359 deletion, or a mutant form containing L51A and L54A alterations were transfected in H1299 cells. Cells were harvested at 48 h, and cell extracts were analyzed by SDS-PAGE followed by Western blotting using appropriate antibodies against the HA tag and actin. (B) H1299 cells were transfected with HA-E4orf4, HA-dl359, or HA-E4orf4-L51/54A (HA-L51/54A), and the subcellular localizations of the E4orf4 products were analyzed at 48 h posttransfection by immunofluorescence using HA antibody. Frames are stained as indicated at the top of the panel. (C) The percentages of cells containing aggregates from the study on HA-dl359 shown in panel B (frames d to f) at 24 and 48 h were counted. (D) H1299 cells were transfected with HA-wt-E4orf4 (a to c) or HA-dl359 (d to f), and the localization of E4orf4 products was determined by immunofluorescence (a and d) and compared to that of proteasomes (b and e). (E) H1299 cells were transfected with plasmid DNA expressing HA-dl359 and its localization (a) was compared to that of the Hsp70 protein (b) by immunofluorescence. (F) H1299 cells were cotransfected with plasmid cDNAs expressing HA-dl359 and Flag-wt-E4orf4 for 48 h. Cells were costained using antibodies against HA (red) and Flag (green) (a to c) or with HA (red) and Hsp70 (green) (d to f).
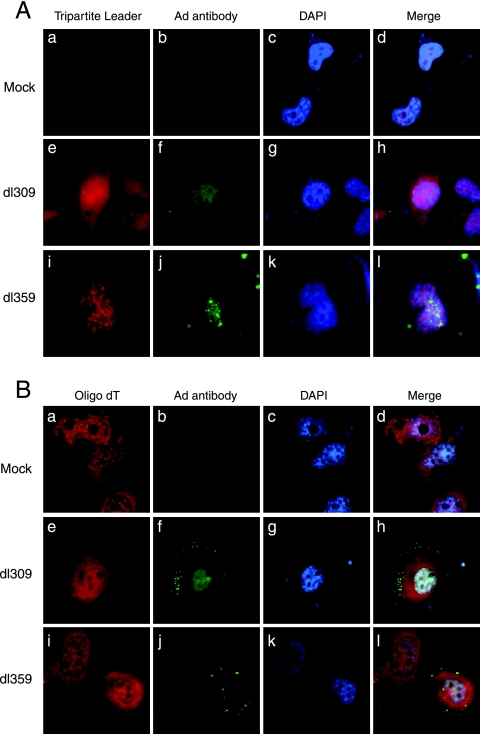
It seemed possible that the growth defect exhibited by dl359 could result from either reduced translation of late viral mRNAs or defects in splicing or transport of these messages. To examine the latter possibility, localization of late viral messages was investigated at 24 hpi in H1299 cells infected with dl359 or with wt dl309 at an MOI of 5. Late viral messages were visualized using a probe designed to bind to the spliced tripartite leader sequence of these mRNAs while adenovirus infection was monitored using antibodies against adenovirus. Figure 6A shows that no mRNAs were evident in mock-infected cells (frames a to d), but, strikingly, the spliced late viral messages in dl359-infected cells (frames i to l) were enriched in a nuclear punctate pattern similar to that observed for HA-dl359 (Fig. 5B, frames d to f). In contrast, they were present in a diffuse, homogeneous, nuclear, and cytoplasmic pattern with wt dl309 (frames e to h). Figure 6B shows the pattern obtained using an oligo(dT) probe of total polyadenylated cell mRNA in mock-, dl309-, and dl359-infected samples. Thus, the results in Fig. 6 suggest that dl359 growth might be impaired due to the specific accumulation of late viral messages in aggregates in the nucleus of cells. It is tempting to suggest that these aggregates may correspond to the structures shown in Fig. 5 to contain dl359 E4orf4 protein; however, thus far it has been impossible to detect E4orf4 during infection using the antibodies available.
FIG. 6.
Effect of the dl359 mutation on the localization of late viral and total cell mRNAs. H1299 cells were either mock infected or infected with wt dl309 or the mutant dl359, as indicated on the left. At 24 hpi fixed cells were hybridized with either a fluorescent oligonucleotide complementary to the spliced tripartite leader (frames a, e, and i in A) or a fluorescent poly(dT) oligo (frames a, e, and i in B). Following hybridization, the late viral proteins were immunostained with an antibody recognizing the capsid proteins (frames b, f, and j in both panels). DAPI was used to stain the nucleus, as indicated on the figure. Images in the first three columns are combined at right (merge).
DISCUSSION
E4orf4 associates with nuclear bodies during infection but is not essential for virus growth.
Previous studies have demonstrated that E4orf4 expressed by DNA transfection is predominantly enriched in the nucleus of cells (38, 45) and that this enrichment is mediated via the E4ARM, an arginine-rich sequence that serves as a nuclear/nucleolar localization signal (38). Results presented here show for the first time that GFP-E4orf4 is also predominantly localized in the nucleus of live cells during a lytic infection and that, in addition, it is specifically enriched in nucleoli, viral replication centers, and perinuclear bodies. In addition, E4orf4 is also enriched in small round structures in the nucleus of cells that perfectly colocalize with late viral proteins.
Many cellular and viral proteins have been shown to localize to adenovirus replication centers and to participate in DNA replication (30); however, the results presented here suggest that E4orf4 does not participate directly in DNA replication since it does not colocalize with the DNA replication protein DBP. Rather, the results suggest that it could be involved in viral transcriptional and/or posttranscriptional processes as it exhibits a localization pattern reminiscent of viral RNA and splicing factors (8). Indeed, these observations agree with previous results obtained in in vitro assays reporting that E4orf4 is not directly involved in viral DNA replication (37) and that it participates in the switch from proximal to distal splicing of the viral L1 transcript during the viral life cycle by dephosphorylating SR splicing factors with the help of PP2A (22) and by associating with the splicing factors ASF/SF2 and SRp30c (14).
When overexpressed from DNA plasmid vectors, E4orf4 had previously been reported to associate with nucleoli (38). The fact that GFP-E4orf4 was also found to accumulate in nucleoli of live cells during infection suggests a putative role in this compartment. Nonetheless, the potential nucleolar functions of E4orf4 remain obscure as E4orf4 does not appear to be the adenovirus protein responsible for redistribution of key nucleolar proteins like fibrillarin outside nucleoli during infection (43; also data not shown). It is possible that E4orf4 may be involved in modulating RNA functions as other proteins associating with nucleoli and containing nuclear localization signal-nucleolar localization signal motifs similar to that of E4orf4 perform such roles (15, 33, 48, 51).
Finally, GFP-E4orf4 protein was present in perinuclear bodies throughout infection. Perinuclear bodies, also called aggresomes, are regions adjacent to the nucleus surrounding centrosomes and microtubule organizing centers. This region has generally been shown to be involved in cell cycle regulation, cytoskeletal rearrangements, and endosome transport as well as protein degradation (13). The adenovirus E1B55K protein sequesters p53 in aggresomes during infection (58), while E1B55K, E4orf3, and E4orf6 proteins have previously been reported to localize to aggresomes and to be involved in the degradation of double-strand break repair system proteins (2, 31). In addition, E4orf4 expressed by DNA transfection has also been recently shown to localize to this area and to induce the assembly of an actin-myosin ring and de novo polymerization of actin as well as recruitment of endosomes in an Src-dependent fashion (46). Whether E4orf4 is involved in these processes during the course of an infection remains to be determined.
Taken together, the results presented here suggest a role for E4orf4 in certain nuclear components during infection; however, association with these nuclear structures is clearly not essential for virus growth, as, with the exception of dl359, all E4orf4 mutant viruses tested in H1299 or A549 human cancer cells and in primary SAEC exhibited normal viral growth and produced normal late viral protein levels. The fact that E4orf4 is not essential for virus growth is in contradiction with a recent report (42) but is in agreement with other results from past studies (19, 37).
dl359 exhibits a gain of function.
The severe defect in the production of late viral proteins and in virus growth observed with dl359 is reminiscent of other severe defects observed with viruses harboring mutations in essential viral genes (19). The unique subcellular localization (aggregates) exhibited by HA-dl359, its colocalization with the chaperone protein Hsp70, and the fact that late viral messages produced by dl359 were abnormally enriched in nuclear aggregates suggested that dl359 exhibits a gain-of-function mutation that, in contrast to the complete absence of E4orf4, is detrimental to virus growth. Similar toxic gain of functions have been reported with polyglutamine (11, 23, 54) and non-polyglutamine proteins (16) in which aggregation-prone or misfolded proteins accumulate at PML bodies, recruiting in the process Hsp70 or Hsp40 chaperone family members, proteasome components, and transcription factors such as CREB binding protein, p53, and Sp1. Even though the mechanisms of action of these effects are not clearly understood at present, various models propose that the unusual accumulation of these proteins at PML bodies results in the inhibition of PML body functions such as transcriptional repression or protein degradation, ultimately leading to cellular toxicity and cell death (16, 23, 54). Likewise, accumulation of the E4orf4 mutant protein encoded by dl359 in nuclear aggregates that localize adjacent to PML bodies (data not shown) may result in the accumulation and sequestration of nuclear proteins such as transcription factors important for virus growth. Subversion of the host cellular transcription machinery by adenovirus and, in particular, of the transcription factor CREB binding protein has been well documented (9). Hence, failure to recruit important transcription factors to viral replication centers may negatively impact virus growth. Perhaps more importantly, accumulation of late viral messages in similar nuclear aggregates could have a direct impact on late viral production and virus growth by preventing cytoplasmic translation of these messages to occur. These possibilities need to be investigated in further studies.
An additional or alternative hypothesis suggested by our data is that the aberrant dl359 E4orf4 product directly or indirectly interferes with selective transport of viral mRNA transport induced by the E4orf6 protein and E1B55K. The levels of these two viral proteins were as high or higher than those in wt-infected cells, and their localization appeared to be normal (data not shown). Clearly, this effect alone could account for the poor replication and low levels of late viral product in dl359-infected cells, and studies are under way to investigate this possibility.
The reason that dl359 growth can be rescued by overexpressing the wt E4orf4 protein remains unclear. As mentioned above, it is possible that the mutant E4orf4 protein produced by dl359 binds and sequesters cellular or viral factors essential for virus growth. It is also possible that overexpressed wt E4orf4 protein competes for binding to these factors and somehow alleviates or negates virus growth defects. The fact that several binding partners have been observed for E4orf4 in the past and that E4orf4 is a “sticky” protein that coimmunoprecipitates with a great number of cellular proteins (53; also our unpublished observations) could support this hypothesis. Further experiments addressing this issue are clearly indicated.
Supplementary Material
Acknowledgments
We thank Tom Shenk for providing us with the dl309, dl359, and dl358 viruses; Gary Ketner for the pm1020 virus; Josee Lavoie for her anti-E4orf4 2419 antibodies; and Jacynthe Laliberté for technical assistance with the confocal microscopy.
This work was supported by grants from the Canadian Cancer Society though the National Cancer Institute of Canada (P.E.B.) and the Canadian Institutes of Health Research. M.-J.M. had a studentship from the Fédération de recherche en santé au Québec.
Footnotes
Published ahead of print on 10 December 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Afifi, R., R. Sharf, R. Shtrichman, and T. Kleinberger. 2001. Selection of apoptosis-deficient adenovirus E4orf4 mutants in Saccharomyces cerevisiae. J. Virol. 754444-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 7911382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Israel, H., and T. Kleinberger. 2002. Adenovirus and cell cycle control. Front Biosci. 7d1369-dl1395. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, D., M. R. Morrison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 731245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondesson, M., K. Ohman, M. Manervik, S. Fan, and G. Akusjarvi. 1996. Adenovirus E4 open reading frame 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J. Virol. 703844-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branton, P. E., and D. E. Roopchand. 2001. The role of adenovirus E4orf4 protein in viral replication and cell killing. Oncogene 207855-7865. [DOI] [PubMed] [Google Scholar]
- 7.Bridge, E., S. Medghalchi, S. Ubol, M. Leesong, and G. Ketner. 1993. Adenovirus early region 4 and viral DNA synthesis. Virology 193794-801. [DOI] [PubMed] [Google Scholar]
- 8.Bridge, E., D. X. Xia, M. Carmo-Fonseca, B. Cardinali, A. I. Lamond, and U. Pettersson. 1995. Dynamic organization of splicing factors in adenovirus-infected cells. J. Virol. 69281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockmann, D., and H. Esche. 2003. The multifunctional role of E1A in the transcriptional regulation of CREB/CBP-dependent target genes. Curr. Top. Microbiol. Immunol. 27297-129. [DOI] [PubMed] [Google Scholar]
- 10.Chai, Y., S. L. Koppenhafer, N. M. Bonini, and H. L. Paulson. 1999. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J. Neurosci. 1910338-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai, Y., S. L. Koppenhafer, S. J. Shoesmith, M. K. Perez, and H. L. Paulson. 1999. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 8673-682. [DOI] [PubMed] [Google Scholar]
- 12.Champagne, C., M. C. Landry, M. C. Gingras, and J. N. Lavoie. 2004. Activation of adenovirus type 2 early region 4 ORF4 cytoplasmic death function by direct binding to Src kinase domain. J. Biol. Chem. 27925905-25915. [DOI] [PubMed] [Google Scholar]
- 13.Doxsey, S., D. McCollum, and W. Theurkauf. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21411-434. [DOI] [PubMed] [Google Scholar]
- 14.Estmer Nilsson, C., S. Petersen-Mahrt, C. Durot, R. Shtrichman, A. R. Krainer, T. Kleinberger, and G. Akusjarvi. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 861495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu, L., Y. S. Gao, and E. Sztul. 2005. Transcriptional repression and cell death induced by nuclear aggregates of non-polyglutamine protein. Neurobiol. Dis. 20656-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingras, M. C., C. Champagne, M. Roy, and J. N. Lavoie. 2002. Cytoplasmic death signal triggered by SRC-mediated phosphorylation of the adenovirus E4orf4 protein. Mol. Cell. Biol. 2241-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groitl, P., and T. Dobner. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol. Med. 13029-39. [DOI] [PubMed] [Google Scholar]
- 19.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow, E., B. R. Franza, Jr., and C. Schley. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17683-689. [DOI] [PubMed] [Google Scholar]
- 22.Kanopka, A., O. Muhlemann, S. Petersen-Mahrt, C. Estmer, C. Ohrmalm, and G. Akusjarvi. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393185-187. [DOI] [PubMed] [Google Scholar]
- 23.Kaytor, M. D., L. A. Duvick, P. J. Skinner, M. D. Koob, L. P. Ranum, and H. T. Orr. 1999. Nuclear localization of the spinocerebellar ataxia type 7 protein, ataxin-7. Hum Mol. Genet. 81657-1664. [DOI] [PubMed] [Google Scholar]
- 24.Kindsmuller, K., P. Groitl, B. Hartl, P. Blanchette, J. Hauber, and T. Dobner. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. USA 1046684-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinberger, T. 2000. Induction of apoptosis by adenovirus E4orf4 protein. Apoptosis 5211-215. [DOI] [PubMed] [Google Scholar]
- 26.Kleinberger, T., and T. Shenk. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J. Virol. 677556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornitzer, D., R. Sharf, and T. Kleinberger. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J. Cell Biol. 154331-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavoie, J. N., C. Champagne, M. C. Gingras, and A. Robert. 2000. Adenovirus E4 open reading frame 4-induced apoptosis involves dysregulation of Src family kinases. J. Cell Biol. 1501037-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavoie, J. N., M. Nguyen, R. C. Marcellus, P. E. Branton, and G. C. Shore. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J. Cell Biol. 140637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, H., J. H. Naismith, and R. T. Hay. 2003. Adenovirus DNA replication. Curr. Top. Microbiol. Immunol. 272131-164. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 7914004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livne, A., R. Shtrichman, and T. Kleinberger. 2001. Caspase activation by adenovirus E4orf4 protein is cell line specific and is mediated by the death receptor pathway. J. Virol. 75789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338254-257. [DOI] [PubMed] [Google Scholar]
- 34.Mannervik, M., S. Fan, A. C. Strom, K. Helin, and G. Akusjarvi. 1999. Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256313-321. [DOI] [PubMed] [Google Scholar]
- 35.Marcellus, R. C., H. Chan, D. Paquette, S. Thirlwell, D. Boivin, and P. E. Branton. 2000. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Bα subunit of protein phosphatase 2A. J. Virol. 747869-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcellus, R. C., J. N. Lavoie, D. Boivin, G. C. Shore, G. Ketner, and P. E. Branton. 1998. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J. Virol. 727144-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medghalchi, S., R. Padmanabhan, and G. Ketner. 1997. Early region 4 modulates adenovirus DNA replication by two genetically separable mechanisms. Virology 2368-17. [DOI] [PubMed] [Google Scholar]
- 38.Miron, M. J., I. E. Gallouzi, J. N. Lavoie, and P. E. Branton. 2004. Nuclear localization of the adenovirus E4orf4 protein is mediated through an arginine-rich motif and correlates with cell death. Oncogene 237458-7468. [DOI] [PubMed] [Google Scholar]
- 39.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, and et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7171-180. [PubMed] [Google Scholar]
- 40.Muller, U., T. Kleinberger, and T. Shenk. 1992. Adenovirus E4orf4 protein reduces phosphorylation of c-Fos and E1A proteins while simultaneously reducing the level of AP-1. J. Virol. 665867-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 731591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Shea, C., K. Klupsch, S. Choi, B. Bagus, C. Soria, J. Shen, F. McCormick, and D. Stokoe. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J. 241211-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puvion-Dutilleul, F., and M. E. Christensen. 1993. Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur. J. Cell Biol. 61168-176. [PubMed] [Google Scholar]
- 44.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128480-484. [DOI] [PubMed] [Google Scholar]
- 45.Robert, A., M. J. Miron, C. Champagne, M. C. Gingras, P. E. Branton, and J. N. Lavoie. 2002. Distinct cell death pathways triggered by the adenovirus early region 4 ORF 4 protein. J. Cell Biol. 158519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robert, A., N. Smadja-Lamere, M. C. Landry, C. Champagne, R. Petrie, N. Lamarche-Vane, H. Hosoya, and J. N. Lavoie. 2006. Adenovirus E4orf4 hijacks Rho GTPase-dependent actin dynamics to kill cells: a role for endosome-associated actin assembly. Mol. Biol. Cell 173329-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roopchand, D. E., J. M. Lee, S. Shahinian, D. Paquette, H. Bussey, and P. E. Branton. 2001. Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A. Oncogene 205279-5290. [DOI] [PubMed] [Google Scholar]
- 48.Rosen, C. A., J. G. Sodroski, and W. A. Haseltine. 1985. Location of cis-acting regulatory sequences in the human T-cell leukemia virus type I long terminal repeat. Proc. Natl. Acad. Sci. USA 826502-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120510-517. [DOI] [PubMed] [Google Scholar]
- 50.Schmid, S. I., and P. Hearing. 1999. Adenovirus DNA packaging. Construction and analysis of viral mutants, p. 47-59. In W. S. Wold (ed.), Adenovirus methods and protocols, vol. 21. Humana Press Inc., Totowa, NJ. [Google Scholar]
- 51.Seiki, M., J. Inoue, M. Hidaka, and M. Yoshida. 1988. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 857124-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shtrichman, R., and T. Kleinberger. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J. Virol. 722975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shtrichman, R., R. Sharf, and T. Kleinberger. 2000. Adenovirus E4orf4 protein interacts with both Bα and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Bα. Oncogene 193757-3765. [DOI] [PubMed] [Google Scholar]
- 54.Skinner, P. J., B. T. Koshy, C. J. Cummings, I. A. Klement, K. Helin, A. Servadio, H. Y. Zoghbi, and H. T. Orr. 1997. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature 389971-974. [DOI] [PubMed] [Google Scholar]
- 55.Stenoien, D. L., C. J. Cummings, H. P. Adams, M. G. Mancini, K. Patel, G. N. DeMartino, M. Marcelli, N. L. Weigel, and M. A. Mancini. 1999. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum. Mol. Genet. 8731-741. [DOI] [PubMed] [Google Scholar]
- 56.Suhr, S. T., M. C. Senut, J. P. Whitelegge, K. F. Faull, D. B. Cuizon, and F. H. Gage. 2001. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J. Cell Biol. 153283-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whalen, S. G., R. C. Marcellus, A. Whalen, N. G. Ahn, R. P. Ricciardi, and P. E. Branton. 1997. Phosphorylation within the transactivation domain of adenovirus E1A protein by mitogen-activated protein kinase regulates expression of early region 4. J. Virol. 713545-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, L. Y., and D. Liao. 2003. Sequestration of p53 in the cytoplasm by adenovirus type 12 E1B 55-kilodalton oncoprotein is required for inhibition of p53-mediated apoptosis. J. Virol. 7713171-13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.