Abstract
The control of human immunodeficiency virus type 1 (HIV-1) associated with particular HLA class I alleles suggests that some CD8+ T-cell responses may be more effective than others at containing HIV-1. Unfortunately, substantial diversities in the breadth, magnitude, and function of these responses have impaired our ability to identify responses most critical to this control. It has been proposed that CD8 responses targeting conserved regions of the virus may be particularly effective, since the development of cytotoxic T-lymphocyte (CTL) escape mutations in these regions may significantly impair viral replication. To address this hypothesis at the population level, we derived near-full-length viral genomes from 98 chronically infected individuals and identified a total of 76 HLA class I-associated mutations across the genome, reflective of CD8 responses capable of selecting for sequence evolution. The majority of HLA-associated mutations were found in p24 Gag, Pol, and Nef. Reversion of HLA-associated mutations in the absence of the selecting HLA allele was also commonly observed, suggesting an impact of most CTL escape mutations on viral replication. Although no correlations were observed between the number or location of HLA-associated mutations and protective HLA alleles, limiting the analysis to mutations selected by acute-phase immunodominant responses revealed a strong positive correlation between mutations at conserved residues and protective HLA alleles. These data suggest that control of HIV-1 may be associated with acute-phase CD8 responses capable of selecting for viral escape mutations in highly conserved regions of the virus, supporting the inclusion of these regions in the design of an effective vaccine.
Despite substantial advances in antiretroviral therapies, development of an effective human immunodeficiency virus type 1 (HIV-1) vaccine remains a critical goal (6, 39, 82). Unfortunately, current vaccine efforts have failed to reduce infection rates in humans (9, 75) and have only achieved modest decreases in viral loads in the simian immunodeficiency virus (SIV)/SHIV macaque model (21, 44, 81). A majority of these vaccine approaches have focused on inducing T-cell responses, utilizing large regions of the virus in an attempt to induce a broad array of immune responses (6, 34, 44, 81). While it is well established that CD8+ T-cell responses play a critical role in the containment of HIV-1 (45, 49, 67), supported in part by the strong association of particular HLA class I alleles with control of HIV (20, 33, 42, 61), it remains unclear which particular CD8+ T-cell responses are best able to control the virus and thus should be preferentially targeted by a vaccine. Studies comparing the magnitude, breadth, and function of CD8+ T-cell responses in subjects exhibiting either enhanced or poor control of HIV-1 have yielded few clues as to the specific factors associated with an effective CD8+ T-cell response (2, 28, 64, 67). Various differences in the functional capacity of T-cell responses have been observed in long-term nonprogressors (1, 26, 64), although it is possible that these differences may be reflective of an intact immune response, as opposed to having had directly enhanced immune control. As such, efforts are needed to identify factors or phenotypes associated with protective CD8+ T-cell responses in order to enable vaccines to induce the most effective responses.
Recent studies have begun to suggest that the specificity of the CD8+ T-cell response, or the targeting of specific regions of the virus, may be associated with control of HIV-1. Preferential targeting of Gag, a structurally conserved viral protein responsible for multiple functions, has been associated with lower viral loads (25, 43, 56, 60, 77, 85). Furthermore, Kiepiela et al. (43) recently illustrated in a large cohort of 578 clade C-infected subjects that Gag-specific responses were associated with lowered viremia, in contrast to Env-specific responses, which were associated with higher viremia. These data are in line with previous observations that many of the major histocompatibility complex (MHC) class I alleles most strongly associated with control of HIV-1 and SIV, namely, HLA-B57, HLA-B27, and Mamu-A*01, restrict immunodominant CD8+ T-cell responses against the Gag protein (8, 10, 24, 63, 68, 83). However, other alleles associated with slower disease progression, such as HLA-B51 in humans and Mamu-B08 and B-17 in the rhesus macaque, do not immunodominantly target Gag, suggesting that targeting of some other regions of the virus may also be capable of eliciting control (8, 52-54). In addition, recent studies investigating the pattern of HIV-1-specific CD8+ T-cell responses during acute infection reveal that only a small subset of CD8+ T-cell responses restricted by any given HLA allele arise during acute infection and that there exist clear immunodominance patterns to these responses (8, 77, 85). Since control of HIV-1 is likely to be established or lost during the first few weeks of infection, these data suggest that potentially only a few key CD8+ T-cell responses may be needed to adequately establish early control of HIV-1.
One of the major factors limiting the effectiveness of CD8+ T-cell responses is the propensity for HIV-1 to evade these responses through sequence evolution or viral escape (3, 13, 66). Even single point mutations within a targeted CD8 epitope can effectively abrogate recognition by either the HLA allele or the T-cell receptor. However, recent studies have begun to highlight that many sequence polymorphisms will revert to more common consensus residues upon transmission of HIV-1 to a new host, including many cytotoxic T-lymphocyte (CTL) escape mutations (4, 30, 33, 48, 50). Notably, the more rapidly reverting mutations have been observed to preferentially occur at conserved residues, indicating that structurally conserved regions of the virus may be particularly refractory to sequence changes (50). In support of these data, many CTL escape mutations have now been observed to directly impair viral replication (15, 23, 55, 74), in particular those known to either revert or require the presence of secondary compensatory mutations (15, 23, 73, 74). Taken together, these data suggest that, whereas CTL escape mutations provide a benefit to the virus to enable the evasion of host immune pressures, some of these mutations may come at a substantial cost to viral replication. These data may also imply that the association between Gag-specific responses and control of HIV-1 may be due to the targeting of highly conserved regions of the virus that are difficult to evade through sequence evolution.
The propensity by which HIV-1 escapes CD8+ T-cell responses, and the reproducibility by which mutations arise at precise residues in targeted CD8 epitopes (3, 48), also enables the utilization of sequence data to predict which responses may be most capable of exerting immune selection pressure on the virus. Studies in HIV-1, SIV, and hepatitis C virus (16, 58, 65, 78) are now rapidly identifying immune-driven CTL escape mutations across these highly variable pathogens at the population level by correlating sequence polymorphisms in these viruses with the expression of particular HLA alleles. We provide here an analysis of HLA-associated mutations across the entire HIV-1 genome using a set of sequences derived from clade B chronically infected individuals. Through full-length viral genome coverage, these data provide an unbiased analysis of the location of these mutations and suggest that the control of HIV-1 by particular HLA alleles correlates with their ability to preferentially restrict early CD8+ T-cell responses capable of selecting for viral escape mutations at highly conserved residues of the virus. These data provide support for the inclusion of specific highly conserved regions of HIV-1 into vaccine antigens.
MATERIALS AND METHODS
Subjects.
Ninety-eight chronic untreated HIV-1 subtype B-infected subjects were enrolled in Boston through the Massachusetts General Hospital, the Lemuel-Shattuck Hospital, and the Fenway Community Health Center. The study was approved by the Massachusetts General Hospital Review Board, and all subjects gave written informed consent.
HLA typing.
Molecular HLA typing was performed by the Tissue Typing Laboratory at the Churchill Hospital in Oxford, the Massachusetts General Hospital Tissue Typing Laboratory, and the Centre for Clinical Immunology and Biomedical Statistics at the Royal Perth Hospital and Murdock University in Perth, Australia.
Viral sequencing.
Genomic DNA was extracted from peripheral blood mononuclear cell samples (5 million cells) by using a QIAamp DNA blood minikit (Qiagen catalog no. 51104). Nested PCR protocols with limiting dilution adapted from Salminen et al. (71, 72) were used to amplify HIV-1 genomes lacking 5′ and 3′ long terminal repeat regions using EXL DNA polymerase (Stratagene catalog no. 600344). The sequences of the primary forward and reverse PCR primers, respectively, were 5′-AAATCTCTAGCAGTGGCGCCCGAACAG-3′ and 5′-TGAGGGATCTCTAGTTACCAGAGTC-3′, while the nested forward and reverse primers were 5′-GCGGAGGCTAGAAGGAGAGAGATGG-3′ and 5′-GCACTCAAGGCAAGCTTTATTGAGGCTTA-3′. The PCR cycling conditions were as follows: 92°C for 2 min; 10 cycles of 10 s at 92°C, 30 s at 60°C, and 10 min at 68°C; 20 cycles of 10 s at 92°C, 30 s at 55°C, and 10 min at 68°C; and a final extension of 10 min at 68°C. Five independent PCR products of each sample were pooled and purified by using a QIAquick PCR purification kit (Qiagen catalog no. 28104) and directly population sequenced at the Massachusetts General Hospital DNA Sequencing Core facility using 70 clade B consensus sequencing primers as previously described (7).
Phylogenetic analysis of HLA-associated sequence polymorphisms.
To identify associations between HLA alleles and HIV polymorphisms, we used a previously described phylogenetic correction method (12, 19), with a slight modification to account for HLA linkage disequilibrium. Briefly, a maximum-likelihood tree was generated for each gene. To compute the pairwise correlation between an HLA allele and an observed amino acid, two models of evolution were compared by using a likelihood ratio test. In the null model, the amino acid was allowed to evolve independently down the tree (“independent evolution model”). In the alternative model, the presence (or absence) of the HLA allele in a given patient was allowed to influence the final transition at the leaf node (“conditional evolution model”). To account for HLA linkage disequilibrium, we used a decision tree built using forward selection. First, for every amino acid at each codon, the allele with the strongest association was added to the list of identified associations. Then, individuals expressing this allele were removed from the data set, and the analysis was repeated. This forward-selection procedure was iterated until no HLA allele yielded an association with an uncorrected P value of <0.05, thus tending to eliminate spurious associations due to HLA linkage disequilibrium. Rare HLA-amino acid pairs were not considered; specifically, we required that the observed or expected count in each bin of the two-by-two contingency table was at least three. To account for multiple comparisons, P values were converted to q-values (76) by using a permutation test as previously described (19). Associations with q ≤ 0.2, corresponding to a 20% false discovery rate (11), are reported. In the present study, correlations in the presence of an HLA allele are called “escape associations,” and correlations in the absence of an HLA allele are called “reversion associations”.
HLA RH and immunodominance.
The relative hazards (RHs) for disease progression for each HLA allele were previously determined (21, 33, 62; data not shown). Here we used the RH values for progression to AIDS disease definition of 1987 as a marker of “protective” versus “hazardous” HLA alleles. Based on a previous study by Altfeld et al. (8), acute-phase immunodominant responses were defined as any HIV-1-specific CD8+ T-cell response that was detected in at least one HIV-1-infected subject within the first 2 months following presentation with acute HIV-1 infection. We then ranked the epitopes restricted by each HLA allele in the order of their frequency of recognition, such that the epitope most frequently recognized by individuals expressing the corresponding allele received a ranking of 1, and the second most frequently recognized epitope received a ranking of 2, and so on.
Conservation and covariation analysis.
Conservation scores for each residue were calculated by using PFAAT (17) for clade B sequence alignments derived from the LANL HIV Sequence Database (http://www.hiv.lanl.gov), which removed highly similar sequences such as multiple clones from a single isolate and multiple sequences from a single patient. The conservation score S is defined as S = 1 − H, where H represents the normalized Shannon entropy of a particular column in the alignment. We applied a previously derived P value and algorithm (79) for pairwise residue covariation analysis on all clade B sequences, limited to one sequence per patient, for all HIV coding genes. P values are adjusted by Bonferroni correction. Sequences were aligned to HXB2, and positions where either the residue conservation score was <0.6 or a gap represented the most abundant variant were removed from the analysis. VisANT was used for network visualization (37, 38).
Statistical analysis.
Mann-Whitney, Spearman correlated coefficient tests, and Fisher exact tests were conducted using Prism 4.0 (GraphPad, San Diego, CA). Cook's distances (D) (22) were calculated using MatLab 7.0 (MathWorks, Natick, MA). The data points are consider outliers (35) if D ≥ 4/n, where n is number of data points.
Predicted CD8 epitopes.
Predicted CD8 epitopes were determined based on scanning the cohort consensus sequence using the epitope prediction algorithm developed by Heckerman et al. (36).
Nucleotide sequence accession numbers.
All sequence data in the present study were deposited in GenBank under accession numbers FJ469682 to FJ469772 and DQ886031 to DQ886038 (27).
RESULTS
Identification of HLA-associated sequence polymorphisms across the HIV-1 genome.
Viral escape from CD8+ T-cell responses frequently occurs through evolution of specific residues within or flanking a given CD8 epitope (3, 48). This phenomenon, typically driven by the selection of mutations at the most variable residue, enables CD8 escape mutations, or HLA-associated mutations, to be rapidly identified at the population level (16, 58, 65, 78). In order to identify frequent HLA-associated mutations across the HIV-1 genome, we sequenced the entire coding region (from Gag to Nef; nucleotides 790 to 9414 on HXB2) of viruses derived from 98 chronic untreated HIV-1 subtype B-infected subjects with an average sequence length of 8,794 nucleotide bases. A total of 18 HLA-A, 26 HLA-B, and 14 HLA-C alleles were represented in this cohort (see Table S1 in the supplemental material). HLA-associated sequence polymorphisms across the viral genome were then identified by using a phylogenetics-based likelihood-ratio approach (PhyloD) (12, 19). The PhyloD approach corrects for confounding founder effects by considering phylogeny as a source of hierarchical structure in a generative model, which can provide much higher specificity to the assessment of HLA-associated polymorphisms. The PhyloD approach captures positive selection and purifying negative selection processes within four states: polymorphisms away from a particular residue (“escape”) or toward a specific nonconsensus residue (“attraction”) in the presence of a specific HLA allele and polymorphisms away from nonconsensus residues (“repulsion”) or toward a particular residue (“reversion”) in the absence of a specific HLA. For the current analyses, these four states were consolidated into either “escaping” (escape or attraction in the presence of HLA) or “reverting” (reversion or repulsion in the absence of HLA).
The PhyloD approach identified a total of 76 HLA-associated polymorphisms located at 47 distinct amino acid residues across HIV-1 (see Table S2 in the supplemental material). These were further broken down into 41 escaping and 35 reverting associations, with 29 positions overlapping between them. These HLA-associated mutations were then categorized into four classes based on their location relative to epitopes: (i) located within previously defined CD8 epitopes (46), (ii) flanking described CD8 epitopes (within 3 amino acid residues), or within (iii) or flanking (iv) predicted CD8 epitopes. Using these criteria there were 17, 1, 18, and 5 escaping associations found in each class, respectively, and 13, 1, 15, and 6 reverting associations for each class (Table 1). As expected, we observed HLA-associated escape mutations in common CD8 epitopes such as B57-TW10, B27-KK10, and A3-RK9, where CTL escape has previously been described in longitudinal analyses (see Fig. S1 in the supplemental material) (3, 4, 41, 48). Moreover, we also identified HLA-associated reversions in the B57-TW10 and A3-RK9 epitopes as previously described (48), verifying the use of the PhyloD approach for identifying both of these types of associations at the population level. Notably, the numbers of HLA-associated polymorphism detected are significantly different from the frequencies of specific HLA alleles (P < 0.0001 [Mann-Whitney test]), illustrating lack of any bias introduced by high frequency alleles in this analysis. Therefore, due to the modest size of this cohort (n = 98), these HLA associations likely represent some of the strongest associations, reflective of those CD8+ T-cell responses capable of exerting the greatest selective pressure.
TABLE 1.
Distribution of HLA-associated escape and reversing mutations observed across the HIV-1 genome relative to the location of the CD8+ T-cell restricted epitope
| Protein | No. of epitopes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Escape
|
Reversion
|
|||||||
| Described epitope | Flank described epitope | Predicted epitope | Flank predicted epitope | Described epitope | Flank described epitope | Predicted epitope | Flank predicted epitope | |
| Gag | 7 | 0 | 3 | 1 | 5 | 0 | 2 | 1 |
| Pol | 3 | 1 | 10 | 3 | 2 | 1 | 7 | 4 |
| Rev | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Vif | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Vpr | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Vpu | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Env | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 |
| Nef | 6 | 0 | 2 | 0 | 5 | 0 | 3 | 0 |
| Total | 17 | 1 | 18 | 5 | 13 | 1 | 15 | 6 |
HLA-associated polymorphisms are concentrated in Gag, Pol, and Nef.
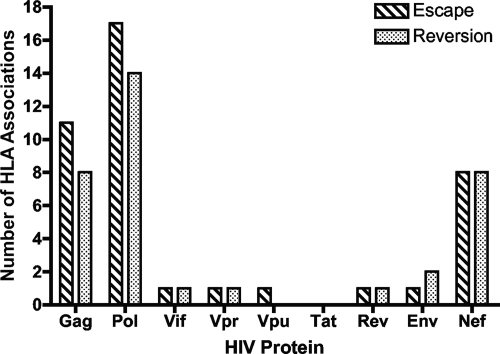
We first examined whether HLA-associated mutations might be clustering in specific regions of HIV-1, comparing the frequency and location of escaping and reverting HLA-associated polymorphisms across the HIV-1 genome. As illustrated in Fig. 1, the majority of HLA-associated sequence changes were located in Gag (n = 19), Pol (n = 31), and Nef (n = 16), where previous studies have illustrated that HIV-specific CD8+ T-cell responses are most frequently detected (2, 28). In contrast, very few HLA-associations were detectable in Env (n = 3) and the accessory and regulatory proteins. Furthermore, 32 of 76 of the polymorphisms, both escaping and reverting mutations, were located within or flanking described epitopes in Gag and Nef (Table 1). Therefore, the HLA-associated mutations reflective of immune-driven viral evolution support the dominant targeting of the viral proteins Gag and Nef by CD8+ T-cell responses. In contrast, the majority (24 of 31 [77%]) of the associated polymorphisms occurring in Pol were located within or flanking predicted epitopes that have not been previously described. In comparing these predicted epitopes with CD8+ T-cell responses detected using genome-wide overlapping peptides in cells from clade B chronically infected subjects (28), we found that for 10 of 27 (37%) overlapping peptides containing predicted epitopes, at least 10% of the individuals expressing the specific HLA allele exhibited a response, suggesting that a substantial number of CD8 epitopes in this protein have not yet been mapped.
FIG. 1.
HLA class I-associated mutations are distributed across HIV-1. Near-full-length viral genome sequencing was conducted on 98 chronically infected subjects. HLA-associated escape and reverting mutations were then identified for all positions across the HIV-1 genome and plotted by their location in HIV-1, illustrating their predominance in Gag, Pol, and Nef.
HLA-associated mutations arise at both conserved and variable residues across HIV-1.
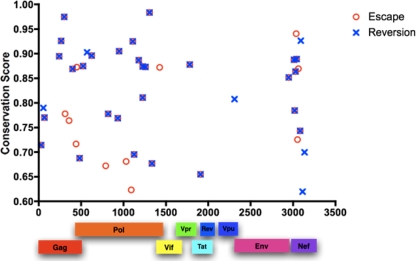
In Fig. 1, the relative proportions of escaping (n = 41) and reverting (n = 35) sequence polymorphisms were similar within any given protein, suggesting an overall balance between positive (escaping) and purifying (reverting) evolutionary selective pressures. To more closely examine the relationship between escaping and reverting HLA-associated mutations, the location of these associations across the viral genome was plotted against the conservation score (1-entropy) of the residue at which they arose. Figure 2 illustrates that HLA-associated mutations were equally present at both highly conserved and more variable residues. Moreover, in the majority of residues where forward escape mutations were identified a matching HLA-associated reversion was also detected (30 of 41 cases [71%]). Our previous study examining viral evolution in longitudinally monitored subjects illustrated that reverting mutations preferentially arose within structurally conserved residues, with mutations in Gag and Pol proteins reverting much more rapidly than in other proteins (50). Again, we observed that escaping residues without an associated reversion were significantly less conserved than escaping residues with an associated reversion (Fig. 2; mean conservation score of 0.825 versus 0.741, respectively [P = 0.018]). Taken together, these data support that many CTL escape mutations are reverting upon transmission to a new host and that the inherent conservation of a given residue can strongly influence the propensity for reversion to occur, likely reflecting different impacts of mutations on viral fitness.
FIG. 2.
Location and residue conservation of HLA-associated escape and reverting mutations. The location of all HLA-driven escaping (red circle) and reverting (blue cross) mutations in the HIV-1 proteome were plotted against the conservation scores of those residues at which they arose. Crosses overlaying circles represent positions where both escape and reversion were observed for the same HLA-associated mutation. HLA-associated escape and reverting mutations arose at both conserved and variable residues, and the majority of escaping residues were accompanied by a corresponding reversion.
HLA-associated polymorphisms are predominantly driven by immunodominant CD8+ T-cell responses.
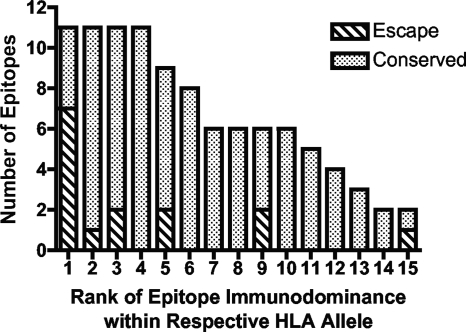
Recent studies have begun to examine the immunodominance patterns of CD8+ T-cell responses, illustrating that only a small subset of defined CD8+ T-cell responses are actually present during acute infection and therefore may have a disproportional effect on early control of HIV-1 (8, 77). Since this subset of CD8+ T-cell responses represents the first line of responses that HIV-1 encounters, we examined whether HLA-associated mutations reflective of viral escape were preferentially occurring within epitopes targeted by the most frequent acute-phase responses. We counted the number of the respective HLA-associated mutations in defined CD8 epitopes targeted during the early phase of infection. These epitopes were plotted in ranked order as described in Materials and Methods. Of the 15 described CD8 epitopes recognized during acute-phase infection that contain HLA-associated escape mutations, close to half of these (7 of 15 [47%]) were targeted by the most frequently mounted CD8+ T-cell response of any given HLA allele (Fig. 3). These data suggest that the most immunodominant acute-phase CD8+ T-cell responses are preferentially driving viral escape or that, conversely, HIV-1 is actively evading responses that dominate the acute phase of infection. These data are in line with previous reports indicating preferential viral escape from the earliest CD8+ T-cell responses in HIV-1 and SIV infection (40, 62).
FIG. 3.
HLA-associated mutations are predominantly selected by acute-phase immunodominant CD8+ T-cell responses. The total number of CD8 epitopes targeted during the acute phase of HIV-1 infection (9) were plotted in ranked order of their frequency of recognition as described in Materials and Methods. Dotted bars represent epitopes that are immunodominant but do not contain HLA-associated escape mutations identified in the present study; hatched bars represent those epitopes containing HLA-associated escape mutations defined in the present study, illustrating that HLA-associated mutations are predominantly driven by the most immunodominant CD8 responses within those tested HLA alleles.
Protective HLA alleles restrict acute-phase CD8+ T-cell responses that are associated with viral escape mutations at highly conserved residues.
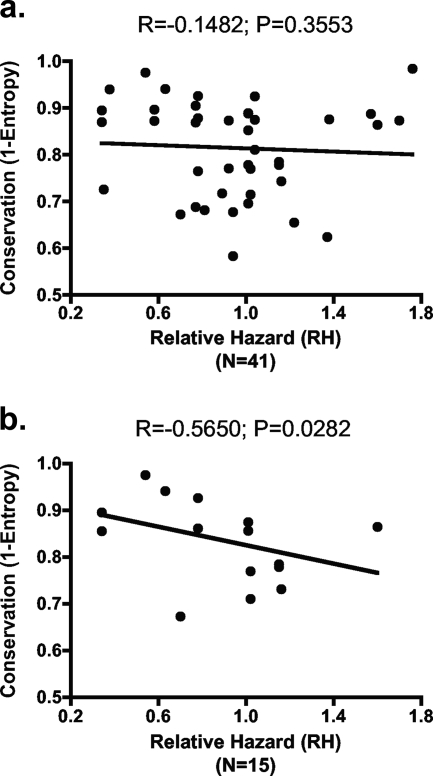
Numerous studies now illustrate that targeting of the Gag protein by HIV-specific CD8+ T-cell responses, as measured by a gamma interferon enzyme-linked immunospot assay, correlates with the control of HIV-1 (25, 56, 60, 77, 85). These data are supported by recent studies illustrating that certain CTL escape mutations in Gag impair viral replication (15, 23, 55, 74), suggesting that HIV-1 may be constrained in its ability to effectively escape from cellular immune pressures against this highly conserved region of the virus. To determine whether immune control of HIV-1 may more broadly correlate with the ability of CD8+ T-cell responses to exhibit immune selection pressure at conserved residues, we examined the correlation between residue conservation at immune-driven escaping positions with the RH (80) of each restricting HLA allele for disease progression (20, 32, 61). No correlation was observed when the residue conservation of all 41 immune-driven escape mutations was compared to the RH of the mutations' restricting HLA allele (Fig. 4a, R = −0.1482, P = 0.3553 [Spearman rank]). This analysis was the same regardless of whether all associations, or only those associated with defined CD8 epitopes, were included in the analysis. However, given the potential for acute-phase CD8+ T-cell responses to disproportionately influence early outcome of HIV-1 infection, the analysis was then limited only to those HLA-associated mutations localized within epitopes targeted by acute-phase CD8+ T-cell responses (8, 77) (see Table S2 in the supplemental material) to test the hypothesis of whether escape mutations selected by CD8 responses restricted by protective alleles are more conserved than those selected by hazardous HLA alleles. Here, we observed a significant inverse correlation between the RH of the restricting HLA allele and the conservation of the CD8+ T-cell selected escaping residues (R = −0.5650, P = 0.0282 [Spearman rank test]). In this analysis, there were two clear potential outliers (T290 in the B51-TI8 epitope in Pol and Y81 in the B35-VY8 epitope in Nef) based on Cook's distance (22) as described in Materials and Methods. When these outliers were removed from the analysis, this inverse correlation remained significant and was further strengthened (R = −0.7746, P = 0.0019 [Spearman rank test]) (data not shown). Therefore, while Altfeld et al. (8) have previously shown that epitope sequence heterogenicity does not influence immunodominance patterns, suggesting that conserved epitopes are not preferentially targeted during the acute phase, we examined here residue conservation at the exact residue where the HLA-associated escape mutation occurs as a measurement of the potential for an escape mutation to impact viral replication. Taken together, these data suggest that protective HLA alleles are associated with acute-phase CD8+ T-cell responses that select for viral escape mutations at more conserved residues, whereas the early responses restricted by hazardous HLA alleles select for mutations at more variable residues. These results provide broader support for the observation that targeting of the highly conserved p24 region of Gag by CD8+ T-cell responses correlates with control of HIV-1 (29, 43) and moreover suggest that the capacity of early responses to select for viral escape mutations may represent an important component of this phenomenon.
FIG. 4.
Protective HLA alleles restrict acute-phase CD8+ T-cell responses that select for viral escape mutations at highly conserved residues. The conservation scores of residues at which HLA-associated escape mutations arose were plotted against the RH (81) to disease progression of the HLA allele restricting the response. (a) Plotting of all HLA-associated escape mutations observed within the HIV genome revealed no correlation between RH of the HLA allele and the conservation of the escaping residue. (b) HLA-associated escape mutations observed within CD8 epitopes targeted during acute HIV-1 infection revealed a strong correlation between protective HLA alleles (low RH) and HLA-associated escape mutations arising at conserved residues.
Residue conservation and compensatory mutations influence the reversion of HLA-associated mutations.
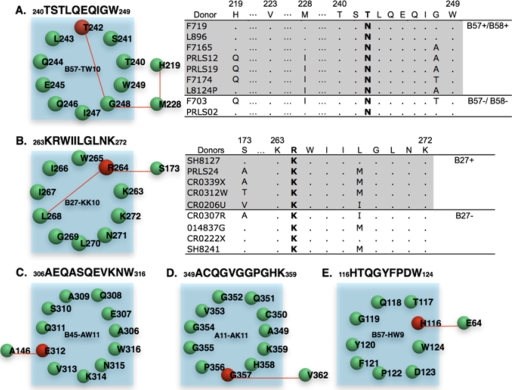
We have previously observed that during acute HIV-1 infection transmitted mutations revert more quickly when arising in highly conserved residues (50). In this regard, it is notable that of the 11 HLA-associated escape mutations in Fig. 2 that did not exhibit a matching HLA-associated reversion, 5 were located at variable residues with entropy values of <0.75. It is possible that these mutations may have had only a minimal impact on viral replication and, therefore, were less apt to revert upon transmission. It is also possible that compensatory mutations could have arisen during viral escape which functioned to temper the impact of these escape mutations on viral replication (15, 23, 31, 74). To examine whether compensatory mutations might be influencing the nonreverting HLA-associated mutations, we performed a covariation analysis between all pairs of residues within each individual HIV-1 protein to identify pairs of mutations having a high probability of occurring in conjunction with one another. This analysis was conducted on a large data set of 284 Gag, 118 Pol, and 600 Nef sequences derived from the Los Alamos National Laboratory (LANL) HIV database (www.hiv.lanl.gov). As validation of this approach, we detected previously described compensatory mutations for the T242N escape mutation in the B57-TW10 epitope and the R264K escape mutation in the B27-KK10 epitope (Fig. 5) (15, 74). The analysis identified three of the four known covarying residues for B57-TW10 (H219Q, M228I, and G248A), where H219Q and M228I mutations were previously shown to partially restore the viral replication defect of T242N (Fig. 5A). It also successfully identified both the prerequisite (L268M) and the compensatory (S173A) mutations for the R264K escape mutation in B27-KK10 (Fig. 5B). In applying this covariation analysis to the 98 HIV-1 sequences in our data set, we observed that six of nine sequences exhibiting the T242N mutation contained one or more of the predicted compensatory residues, in one case from an HLA-B57-negative subject, suggesting the stability of some of these mutations upon transmission. Of the nine sequences containing the R264K mutation, seven contained one or more of the predicted covarying residues.
FIG. 5.
Identification of covarying residues linked to HLA-associated escape mutations. Covarying residue pairs were determined as described in Materials and Methods. Residues located within each epitope are present within each circle, with HLA-associated escape residues colored red. Covarying residues are then connected by lines and located outside the circle. Each residue is labeled by its consensus residue and HXB2 position. The epitope sequences are noted in the following format: SXXXXE, where S is the starting position, E is the end position, and XXXX is the epitope sequence. (A and B) Covariation networks for B57-TW10 and B27-KK10 epitopes, respectively. Sequences from subjects in our cohort with T242N or R264K escape mutations are given. The presence or absence of B57/B58 or B27 allele in these patients is noted. (C to F) Covariation networks for CD8 epitopes B45-AW11, A11-AK11, and B57-HW9, respectively.
In extending this analysis to the 11 HLA-associated escape mutations in Fig. 2 for which no reversions were detected, we observed strong covarying residues for the three intraepitope associations at residues Gag E312, Gag G354, and Nef H116 (Fig. 5C to E) (Table 2). Notably, these three HLA-associated mutations were also among the six most conserved residues without detectable reversions, a finding suggestive of a possible contribution of compensatory mutations to the lack of reversions detected at these more conserved residues. Taken together, these data suggest an important contribution of both the conservation of escaping residues and covarying mutations, which potentially compensate for the impact of specific mutations on viral fitness and their propensity to revert upon transmission.
TABLE 2.
Covarying residue positions for HLA-associated escaping residues without detectable reversiona
| Protein | HLA | Position | Optimal epitope | Conservation | Covarying position | Covarying position conservation |
P
|
No. of patientsd | |
|---|---|---|---|---|---|---|---|---|---|
| LANLb | 98Chronicc | ||||||||
| Gag | B45 | E312D | AEQASQEVKNW | 0.7781 | A146P | 0.7449 | 8.31E-05 | 1.04E-02 | 11 |
| Gag | A11 | G357S | ACQGVGGPGHK | 0.7643 | V362I | 0.8242 | 7.22E-12 | 1.34E-05 | 7 |
| Gag | A74 | H441 | LGKIWPSHK | 0.7167 | ND | NA | NA | NA | NA |
| Gag | B15 | L449 | SHKGRPGNF | 0.8726 | ND | NA | NA | NA | NA |
| Pol | B51 | T290 | TAFTIPSI | 0.6723 | ND | NA | NA | NA | NA |
| Pol | C12 | A531 | KIATESIVIW | 0.681 | ND | NA | NA | NA | NA |
| Pol | C04 | I590 | EPIVGAETFY | 0.6235 | ND | NA | NA | NA | NA |
| Pol | C14 | K926 | KELQKQITK | 0.8723 | ND | NA | NA | NA | NA |
| Nef | C08 | H89 | AALDLSHFL | 0.9407 | ND | NA | NA | NA | NA |
| Nef | B57 | K105 | KRQDILDLW | 0.7257 | ND | NA | NA | NA | NA |
| Nef | B57 | H116N | HTQGYFPDW | 0.8697 | E64Q | 0.8876 | 2.82E-04 | 2.51E-03 | 5 |
Mutations are labeled in the form XNNNY, where X is the consensus residue, Y is the mutant residue, and NNN is the HXB2 position. ND, not detected; NA, not available.
LANL P values were determined by using sequences from the LANL database.
98Chronic P values were determined by using HIV sequences from our cohort of 98 subjects.
That is, the number of patients from our cohort that simultaneously contain both mutations.
DISCUSSION
Large population-based studies of HLA-associated polymorphisms serve to rapidly identify the location and frequency of CTL escape mutations which function to evade host CD8+ T-cell responses. Although previous population studies of HLA class I-associated mutations in HIV-1 have demonstrated immune selection pressures against specific HIV-1 residues (16, 29, 58, 65, 69), we present here an unbiased assessment of HLA-mediated sequence polymorphisms across the full subtype B HIV genome. Our data reveal that these polymorphisms are most frequently observed in Gag, Pol, and Nef, which are known to be dominantly targeted by CD8+ T-cell responses (2, 28). Escape mutations were also observed to occur at both highly conserved and variable residues, with matching reverting HLA-associated mutations also detectable for most mutations. Notably, escape mutations associated with acute-phase CD8+ T-cell responses restricted by protective HLA alleles were preferentially located at more conserved residues than those restricted by hazardous HLA alleles. Taken together, these data suggest that acute-phase CD8+ T-cell responses capable of selecting for viral escape mutations within highly conserved regions of the virus may be critical to the control of HIV-1. These data lend support to recent suggestions that eliciting CTL responses targeting highly conserved regions of HIV-1 such as Gag may be critical to an effective HIV-1 vaccine.
Longitudinal studies of CTL escape have observed that CD8+ T-cell responses represent a major driving force of sequence evolution in HIV, SIV, and hepatitis C virus (3, 16, 47, 58, 62). Viral escape from CD8+ T-cell responses has especially been described during the acute phases of both HIV-1 and SIV infection, illustrating rapid evasion of specific host immune responses during the first few weeks or months after infection (3, 5, 13, 62, 66). Recent studies examining the immunodominance of CD8+ T-cell responses in HIV-1 infection are beginning to reveal predictable patterns to these responses, with some CD8 epitopes preferentially recognized during the acute phase of infection (8, 77). Moreover, responses restricted by some HLA alleles appear to dominate over responses restricted by other HLA alleles (8). Such studies have important implications for vaccine design since control of HIV-1 is predominantly established during the first few weeks or months after infection, with steady-state viral set points strongly predictive of disease progression (57). Therefore, some CD8+ T-cell responses are undoubtedly more critical to this early control than others. In the present study, not only were HLA-associated escape mutations preferentially located within the viral proteins most frequently targeted by CD8+ T-cell responses (Gag, Pol, and Nef) but they were also located within the most immunodominant CD8 epitopes targeted during acute infection. These data strongly support that HIV-1 is preferentially evading the strongest and earliest CD8+ T-cell responses that represent the front line responses.
It has been observed that CTL escape mutations will revert to consensus residues when transmitted to a new host expressing different HLA alleles, since these mutations are no longer required to evade specific CD8+ T-cell responses (4, 30, 33, 48, 50). During the acute phase of infection, such reversions may account for upward of 60% of mutations and notably arise most rapidly when located at more conserved residues (50). These data, and more recent studies directly illustrating the impact of CTL escape mutations on viral replication, provide strong support that a majority of adaptive mutations are capable of impairing the replicative capacity of HIV-1 (15, 23, 50, 73, 74). Our observation that the majority of detected escape mutations are accompanied by a corresponding reversion provides further support for these data at a genome-wide level. Interestingly, however, we observed that a subset of CTL escape mutations were not associated with detectable reversions, with several of the more conserved residues associated with compensatory mutations that may have tempered the effects of the escape mutation on viral replication capacity. These data support the important role that compensatory mutations play in the impact of CTL escape mutations on viral fitness (15, 23, 73) and highlight that due to structural constraints some CTL escape mutations may not be able to acquire the necessary compensatory mutations and thereby may disproportionately impact viral replication capacity. It will be important, therefore, to more broadly examine the frequency and complexity by which compensatory mutations can arise, since as in the case of the B57-TW10 epitope the network of compensatory mutations may be highly complex (15, 74).
The ability to more rapidly generate HIV-1 sequence data and correlate sequence changes with the expression of specific host HLA alleles has enabled a growing number of studies identifying HLA-associated mutations in HIV-1 (14, 16, 51, 58, 65). These studies have begun to provide substantial insight not only into the frequency and complexity of immune-driven mutations but also into the mechanisms associated with “protective” HLA alleles. Frater et al. (29) recently illustrated that HLA alleles associated with slower disease progression were most often associated with CD8+ T-cell responses that drive viral escape in Gag, Pol, and Nef, supporting that “protective” HLA alleles may restrict uniquely strong CD8+ T-cell responses. Our study builds upon this work by extending these analyses to the entire HIV-1 genome, revealing the importance of distinguishing between acute-phase immunodominant responses and chronic responses, and specifically addressing the issue of viral sequence constraints by taking into account the entropy of escaping residues. Through these combined analyses we were able to illustrate that early immunodominant CD8 responses restricted by “protective” HLA alleles primarily select for viral escape mutations in highly conserved regions of HIV-1, suggesting that the protection conferred by specific HLA alleles on HIV-1 disease control may depend upon the costs to replicative capacities required to successfully escape from early immunodominant responses. HIV-1 mutants with diminished replication fitness have been known to provide clinical benefit in practice. For example, many clinicians have chosen to continue to use 3TC even after the 3TC resistance mutation M184V has emerged to take advantage of the viral replicative defect caused by this mutation (18).
Many of the protective MHC class I alleles, such as HLA-B27, HLA-B57, and Mamu-A*01, preferentially restrict CD8+ T-cell responses targeting the highly conserved Gag protein (15, 59, 74, 77). The importance of targeting CD8 epitopes in Gag not only may be due to the sequence conservation of Gag but also may be due to the early presentation of CD8 epitopes in Gag derived from the incoming capsid particle prior to de novo protein synthesis of other viral proteins (70). It is noteworthy, however, that other MHC class I alleles such as HLA-B51, Mamu-B08, and Mamu-B17 associated with the control of HIV-1 and SIV do not immunodominantly target CD8 epitopes in Gag but rather target other viral proteins such as Pol, Vif, and Nef (8, 52-54). Indeed, many of the immunodominant CD8 responses restricted by “protective” HLA alleles were found to drive viral escape in non-Gag proteins, although notably still at highly conserved residues. Therefore, in the development of an HIV-1 vaccine it may be important to consider other highly conserved regions of HIV-1 outside of Gag in order to provide ample targets for the diversity of HLA alleles in the population.
The moderate size of the current data set limited the present study to HLA-associated mutations restricted by common HLA alleles, as well as those HLA alleles for which immunodominance patterns are known, since the original study by Altfeld et al. (8) was not an exhaustive analysis for all known HLA alleles. Similarly, it is likely that we did not have the power to detect CTL escape mutations associated with subdominant CD8+ T-cell responses. In addition, since our study does not focus on end-stage individuals, late-arising CTL escape mutations will not be represented in our study, as well as early escape mutations targeted by acute-phase CD8+ responses. Therefore, while the correlation between the most immunodominant CD8+ responses and likelihood for viral escape was very strong in our study, this correlation might diminish as subjects are monitored later into infection. Further broadening of the data set to a larger cohort will be required to extend our analyses to examine the relative contribution of these additional responses to early immune control of HIV-1. More importantly, the approach of utilizing viral sequence polymorphisms as a measurement of the impact of particular CD8+ T responses on HIV-1 fitness obviously does not take into account CD8 epitopes that do not escape. Here, larger datasets will enable determining whether in fact some epitopes are completely refractory to escape or escape only late in infection. Finally, it will be important to extend these analyses to other clades of HIV-1 to determine whether similar correlates of immune control exist in the setting of different viral strains and different frequencies of HLA alleles.
In conclusion, these data provide an initial assessment of HLA-associated sequence polymorphisms across the entire HIV-1 proteome and suggest an important influence of early immunodominant CD8+ T-cell responses not only on viral evolution but also potentially on the outcome of HIV-1 infection. More importantly, we show that the HLA alleles that correlate with disease control are capable of selecting for viral sequence polymorphisms in highly conserved regions of HIV-1. Taken together, these findings suggest that vaccines designed to elicit CD8+ T-cell responses may need to focus responses against highly conserved regions of the virus that would exact a substantial impact on viral fitness.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health grant R01-AI054178 (T.M.A.) and as part of the Collaboration for AIDS Vaccine Discovery with support from the Bill & Melinda Gates Foundation.
Footnotes
Published ahead of print on 26 November 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Addo, M. M., R. Draenert, A. Rathod, C. L. Verrill, B. T. Davis, R. T. Gandhi, G. K. Robbins, N. O. Basgoz, D. R. Stone, D. E. Cohen, M. N. Johnston, T. Flynn, A. G. Wurcel, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2007. Fully differentiated HIV-1 specific CD8+ T effector cells are more frequently detectable in controlled than in progressive HIV-1 infection. PLoS ONE 2e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, K. M. O'Sullivan, I. Desouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 7913239-13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 787069-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407386-390. [DOI] [PubMed] [Google Scholar]
- 6.Altfeld, M., and T. M. Allen. 2006. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 27504-510. [DOI] [PubMed] [Google Scholar]
- 7.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420434-439. [DOI] [PubMed] [Google Scholar]
- 8.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous. 2007. HIV vaccine failure prompts Merck to halt trial. Nature 449390. [DOI] [PubMed] [Google Scholar]
- 10.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 777367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57289. [Google Scholar]
- 12.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 3151583-1586. [DOI] [PubMed] [Google Scholar]
- 13.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 14.Boutwell, C. L., and M. Essex. 2007. Identification of HLA class I-associated amino acid polymorphisms in the HIV-1C proteome. AIDS Res. Hum. Retrovir. 23165-174. [DOI] [PubMed] [Google Scholar]
- 15.Brockman, M. A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C. A. Derdeyn, S. Allen, E. Hunter, J. Mulenga, P. A. Goepfert, B. D. Walker, and T. M. Allen. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 8112608-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brumme, Z. L., C. J. Brumme, D. Heckerman, B. T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, T. Mo, R. S. Hogg, J. S. Montaner, N. Frahm, C. Brander, B. D. Walker, and P. R. Harrigan. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caffrey, D. R., P. H. Dana, V. Mathur, M. Ocano, E. J. Hong, Y. E. Wang, S. Somaroo, B. E. Caffrey, S. Potluri, and E. S. Huang. 2007. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC Bioinform. 8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell, T. B., N. S. Shulman, S. C. Johnson, A. R. Zolopa, R. K. Young, L. Bushman, C. V. Fletcher, E. R. Lanier, T. C. Merigan, and D. R. Kuritzkes. 2005. Antiviral activity of lamivudine in salvage therapy for multidrug-resistant HIV-1 infection. Clin. Infect. Dis. 41236-242. [DOI] [PubMed] [Google Scholar]
- 19.Carlson, J., C. Kadie, S. Mallal, and D. Heckerman. 2007. Leveraging hierarchical population structure in discrete association studies. PLoS ONE 2e591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 2831748-1752. [DOI] [PubMed] [Google Scholar]
- 21.Casimiro, D. R., F. Wang, W. A. Schleif, X. Liang, Z. Q. Zhang, T. W. Tobery, M. E. Davies, A. B. McDermott, D. H. O'Connor, A. Fridman, A. Bagchi, L. G. Tussey, A. J. Bett, A. C. Finnefrock, T. M. Fu, A. Tang, K. A. Wilson, M. Chen, H. C. Perry, G. J. Heidecker, D. C. Freed, A. Carella, K. S. Punt, K. J. Sykes, L. Huang, V. I. Ausensi, M. Bachinsky, U. Sadasivan-Nair, D. I. Watkins, E. A. Emini, and J. W. Shiver. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J. Virol. 7915547-15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cook, R. D. 2000. Detection of influential observation in linear regression. Technometrics 4265-68. [Google Scholar]
- 23.Crawford, H., J. G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, J. I. Mullins, R. Kaslow, P. Goepfert, S. Allen, E. Hunter, J. Mulenga, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 818346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Sorrentino, A. H., K. Marinic, P. Motta, A. Sorrentino, R. Lopez, and E. Illiovich. 2000. HLA class I alleles associated with susceptibility or resistance to human immunodeficiency virus type 1 infection among a population in Chaco Province, Argentina. J. Infect. Dis. 1821523-1526. [DOI] [PubMed] [Google Scholar]
- 25.Edwards, B. H., A. Bansal, S. Sabbaj, J. Bakari, M. J. Mulligan, and P. A. Goepfert. 2002. Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 762298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emu, B., E. Sinclair, H. Hatano, A. Ferre, B. Shacklett, J. N. Martin, J. M. McCune, and S. G. Deeks. 2008. HLA class I-restricted T cell responses may contribute to the control of HIV infection, but such responses are not always necessary for long-term virus control. J. Virol. 825398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frahm, N., D. E. Kaufmann, K. Yusim, M. Muldoon, C. Kesmir, C. H. Linde, W. Fischer, T. M. Allen, B. Li, B. H. McMahon, K. L. Faircloth, H. S. Hewitt, E. W. Mackey, T. Miura, A. Khatri, S. Wolinsky, A. McMichael, R. K. Funkhouser, B. D. Walker, C. Brander, and B. T. Korber. 2007. Increased sequence diversity coverage improves detection of HIV-specific T-cell responses. J. Immunol. 1796638-6650. [DOI] [PubMed] [Google Scholar]
- 28.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. St. John, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 782187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frater, A. J., H. Brown, A. Oxenius, H. F. Gunthard, B. Hirschel, N. Robinson, A. J. Leslie, R. Payne, H. Crawford, A. Prendergast, C. Brander, P. Kiepiela, B. D. Walker, P. J. Goulder, A. McLean, and R. E. Phillips. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 816742-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10275-281. [DOI] [PubMed] [Google Scholar]
- 31.Friedrich, T. C., A. B. McDermott, M. R. Reynolds, S. Piaskowski, S. Fuenger, I. P. De Souza, R. Rudersdorf, C. Cullen, L. J. Yant, L. Vojnov, J. Stephany, S. Martin, D. H. O'Connor, N. Wilson, and D. I. Watkins. 2004. Consequences of cytotoxic T-lymphocyte escape: common escape mutations in simian immunodeficiency virus are poorly recognized in naive hosts. J. Virol. 7810064-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao, X., A. Bashirova, A. K. Iversen, J. Phair, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, M. Altfeld, S. J. O'Brien, and M. Carrington. 2005. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 111290-1292. [DOI] [PubMed] [Google Scholar]
- 33.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412334-338. [DOI] [PubMed] [Google Scholar]
- 34.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4630-640. [DOI] [PubMed] [Google Scholar]
- 35.Hadi, A. S., and J. S. Simonoff. 1993. Procedures for the identification of multiple outliers in linear models. J. Am. Stat. Assoc. 881264-1272. [Google Scholar]
- 36.Heckerman, D., C. Kadie, and J. Listgarten. 2007. Leveraging information across HLA alleles/supertypes improves epitope prediction. J. Comput. Biol. 14736-746. [DOI] [PubMed] [Google Scholar]
- 37.Hu, Z., J. Mellor, J. Wu, M. Kanehisa, J. M. Stuart, and C. DeLisi. 2007. Towards zoomable multidimensional maps of the cell. Nat. Biotechnol. 25547-554. [DOI] [PubMed] [Google Scholar]
- 38.Hu, Z., D. M. Ng, T. Yamada, C. Chen, S. Kawashima, J. Mellor, B. Linghu, M. Kanehisa, J. M. Stuart, and C. DeLisi. 2007. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 35W625-W632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston, M. I., and A. S. Fauci. 2007. An HIV vaccine-evolving concepts. N. Engl. J. Med. 3562073-2081. [DOI] [PubMed] [Google Scholar]
- 40.Jones, N. A., X. Wei, D. R. Flower, M. Wong, F. Michor, M. S. Saag, B. H. Hahn, M. A. Nowak, G. M. Shaw, and P. Borrow. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 2001243-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432769-775. [DOI] [PubMed] [Google Scholar]
- 43.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 44.Koff, W. C., P. R. Johnson, D. I. Watkins, D. R. Burton, J. D. Lifson, K. J. Hasenkrug, A. B. McDermott, A. Schultz, T. J. Zamb, R. Boyle, and R. C. Desrosiers. 2006. HIV vaccine design: insights from live attenuated SIV vaccines. Nat. Immunol. 719-23. [DOI] [PubMed] [Google Scholar]
- 45.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuiken, C., B. Korber, and R. W. Shafer. 2003. HIV sequence databases. AIDS Rev. 552-61. [PMC free article] [PubMed] [Google Scholar]
- 47.Kuntzen, T., J. Timm, A. Berical, L. L. Lewis-Ximenez, A. Jones, B. Nolan, J. Schulze Zur Wiesch, B. Li, A. Schneidewind, A. Y. Kim, R. T. Chung, G. M. Lauer, and T. M. Allen. 2007. Viral sequence evolution in acute hepatitis C virus infection. J. Virol. 8111658-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10282-289. [DOI] [PubMed] [Google Scholar]
- 49.Letvin, N. L., J. E. Schmitz, H. L. Jordan, A. Seth, V. M. Hirsch, K. A. Reimann, and M. J. Kuroda. 1999. Cytotoxic T lymphocytes specific for the simian immunodeficiency virus. Immunol. Rev. 170127-134. [DOI] [PubMed] [Google Scholar]
- 50.Li, B., A. D. Gladden, M. Altfeld, J. M. Kaldor, D. A. Cooper, A. D. Kelleher, and T. M. Allen. 2007. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J. Virol. 81193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, Y., J. McNevin, H. Zhao, D. M. Tebit, R. M. Troyer, M. McSweyn, A. K. Ghosh, D. Shriner, E. J. Arts, M. J. McElrath, and J. I. Mullins. 2007. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J. Virol. 8112179-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loffredo, J. T., A. T. Bean, D. R. Beal, E. J. Leon, G. E. May, S. M. Piaskowski, J. R. Furlott, J. Reed, S. K. Musani, E. G. Rakasz, T. C. Friedrich, N. A. Wilson, D. B. Allison, and D. I. Watkins. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 821723-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loffredo, J. T., T. C. Friedrich, E. J. Leon, J. J. Stephany, D. S. Rodrigues, S. P. Spencer, A. T. Bean, D. R. Beal, B. J. Burwitz, R. A. Rudersdorf, L. T. Wallace, S. M. Piaskowski, G. E. May, J. Sidney, E. Gostick, N. A. Wilson, D. A. Price, E. G. Kallas, H. Piontkivska, A. L. Hughes, A. Sette, and D. I. Watkins. 2007. CD8 T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS ONE 2e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maness, N. J., L. J. Yant, C. Chung, J. T. Loffredo, T. C. Friedrich, S. M. Piaskowski, J. Furlott, G. E. May, T. Soma, E. J. León, N. A. Wilson, H. Piontkivska, A. L. Hughes, J. Sidney, A. Sette, and D. I. Watkins. 2008. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive SIV-infected rhesus macaques. J. Virol. 825245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 803617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masemola, A., T. Mashishi, G. Khoury, P. Mohube, P. Mokgotho, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 783233-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mellors, J. W., J. B. Margolick, J. P. Phair, C. R. Rinaldo, R. Detels, L. P. Jacobson, and A. Munoz. 2007. Prognostic value of HIV-1 RNA, CD4 cell count, and CD4 Cell count slope for progression to AIDS and death in untreated HIV-1 infection. JAMA 2972349-2350. [DOI] [PubMed] [Google Scholar]
- 58.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2961439-1443. [DOI] [PubMed] [Google Scholar]
- 59.Navis, M., I. Schellens, D. van Baarle, J. Borghans, P. van Swieten, F. Miedema, N. Kootstra, and H. Schuitemaker. 2007. Viral replication capacity as a correlate of HLA B57/B5801-associated nonprogressive HIV-1 infection. J. Immunol. 1793133-3143. [DOI] [PubMed] [Google Scholar]
- 60.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7379-381. [DOI] [PubMed] [Google Scholar]
- 62.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8493-499. [DOI] [PubMed] [Google Scholar]
- 63.O'Connor, D. H., B. R. Mothe, J. T. Weinfurter, S. Fuenger, W. M. Rehrauer, P. Jing, R. R. Rudersdorf, M. E. Liebl, K. Krebs, J. Vasquez, E. Dodds, J. Loffredo, S. Martin, A. B. McDermott, T. M. Allen, C. Wang, G. G. Doxiadis, D. C. Montefiori, A. Hughes, D. R. Burton, D. B. Allison, S. M. Wolinsky, R. Bontrop, L. J. Picker, and D. I. Watkins. 2003. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 779029-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pereyra, F., M. M. Addo, D. E. Kaufmann, Y. Liu, T. Miura, A. Rathod, B. Baker, A. Trocha, R. Rosenberg, E. Mackey, P. Ueda, Z. Lu, D. Cohen, T. Wrin, C. J. Petropoulos, E. S. Rosenberg, and B. D. Walker. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197563-571. [DOI] [PubMed] [Google Scholar]
- 65.Poon, A. F., S. L. Kosakovsky Pond, P. Bennett, D. D. Richman, A. J. Leigh Brown, and S. D. Frost. 2007. Adaptation to human populations is revealed by within-host polymorphisms in HIV-1 and hepatitis C virus. PLoS Pathog. 3e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 941890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reimann, K. A., K. Tenner-Racz, P. Racz, D. C. Montefiori, Y. Yasutomi, W. Lin, B. J. Ransil, and N. L. Letvin. 1994. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J. Virol. 682362-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rohowsky-Kochan, C., J. Skurnick, D. Molinaro, and D. Louria. 1998. HLA antigens associated with susceptibility/resistance to HIV-1 infection. Hum. Immunol. 59802-815. [DOI] [PubMed] [Google Scholar]
- 69.Rousseau, C. M., M. G. Daniels, J. M. Carlson, C. Kadie, H. Crawford, A. Prendergast, P. Matthews, R. Payne, M. Rolland, D. N. Raugi, B. S. Maust, G. H. Learn, D. C. Nickle, H. Coovadia, T. Ndung'u, N. Frahm, C. Brander, B. D. Walker, P. J. R. Goulder, T. Bhattacharya, D. E. Heckerman, B. T. Korber, and J. I. Mullins. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 826364-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sacha, J. B., C. Chung, E. G. Rakasz, S. P. Spencer, A. K. Jonas, A. T. Bean, W. Lee, B. J. Burwitz, J. J. Stephany, J. T. Loffredo, D. B. Allison, S. Adnan, A. Hoji, N. A. Wilson, T. C. Friedrich, J. D. Lifson, O. O. Yang, and D. I. Watkins. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 1782746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salminen, M. O., B. Johansson, A. Sonnerborg, S. Ayehunie, D. Gotte, P. Leinikki, D. S. Burke, and F. E. McCutchan. 1996. Full-length sequence of an ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res. Hum. Retrovir. 121329-1339. [DOI] [PubMed] [Google Scholar]
- 72.Salminen, M. O., C. Koch, E. Sanders-Buell, P. K. Ehrenberg, N. L. Michael, J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology 21380-86. [DOI] [PubMed] [Google Scholar]
- 73.Schneidewind, A., M. A. Brockman, J. Sidney, Y. E. Wang, H. Chen, T. J. Suscovich, B. Li, R. I. Adam, R. L. Allgaier, B. R. Mothé, T. Kuntzen, C. Oniangue-Ndza, A. Trocha, X. G. Yu, C. Brander, A. Sette, B. D. Walker, and T. M. Allen. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 825594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. LaBute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 8112382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sekaly, R. P. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 2057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 1009440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Streeck, H., M. Lichterfeld, G. Alter, A. Meier, N. Teigen, B. Yassine-Diab, H. K. Sidhu, S. Little, A. Kelleher, J. P. Routy, E. S. Rosenberg, R. P. Sekaly, B. D. Walker, and M. Altfeld. 2007. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 817725-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Timm, J., B. Li, M. G. Daniels, T. Bhattacharya, L. L. Reyor, R. Allgaier, T. Kuntzen, W. Fischer, B. E. Nolan, J. Duncan, J. Schulze zur Wiesch, A. Y. Kim, N. Frahm, C. Brander, R. T. Chung, G. M. Lauer, B. T. Korber, and T. M. Allen. 2007. Human leukocyte antigen-associated sequence polymorphisms in hepatitis C virus reveal reproducible immune responses and constraints on viral evolution. Hepatology 46339-349. [DOI] [PubMed] [Google Scholar]
- 79.Wang, Y. E., and C. DeLisi. 2006. Inferring protein-protein interactions in viral proteins by co-evolution of conserved side chains. Genome Inform. 1723-35. [PubMed] [Google Scholar]
- 80.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, L. Y. Geer, W. Helmberg, Y. Kapustin, D. L. Kenton, O. Khovayko, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, K. D. Pruitt, G. D. Schuler, L. M. Schriml, E. Sequeira, S. T. Sherry, K. Sirotkin, A. Souvorov, G. Starchenko, T. O. Suzek, R. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2006. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 34D173-D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson, N. A., J. Reed, G. S. Napoe, S. Piaskowski, A. Szymanski, J. Furlott, E. J. Gonzalez, L. J. Yant, N. J. Maness, G. E. May, T. Soma, M. R. Reynolds, E. Rakasz, R. Rudersdorf, A. B. McDermott, D. H. O'Connor, T. C. Friedrich, D. B. Allison, A. Patki, L. J. Picker, D. R. Burton, J. Lin, L. Huang, D. Patel, G. Heindecker, J. Fan, M. Citron, M. Horton, F. Wang, X. Liang, J. W. Shiver, D. R. Casimiro, and D. I. Watkins. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 805875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang, O. O. 2008. Aiming for successful vaccine-induced HIV-1-specific cytotoxic T lymphocytes. AIDS 22325-331. [DOI] [PubMed] [Google Scholar]
- 83.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 805074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu, X. G., M. M. Addo, E. S. Rosenberg, W. R. Rodriguez, P. K. Lee, C. A. Fitzpatrick, M. N. Johnston, D. Strick, P. J. Goulder, B. D. Walker, and M. Altfeld. 2002. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J. Virol. 768690-8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zuñiga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 803122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.