Abstract
Replication-defective adenovirus (Ad) vectors can vary considerably in genome length, but whether this affects virion stability has not been investigated. Helper-dependent Ad vectors with a genome size of ∼30 kb were 100-fold more sensitive to heat inactivation than their parental helper virus (>36 kb), and increasing the genome size of the vector significantly improved heat stability. A similar relationship between genome size and stability existed for Ad with early region 1 deleted. Loss of infectivity was due to release of vertex proteins, followed by disintegration of the capsid. Thus, not only does the viral DNA encode all of the heritable information essential for virus replication, it also plays a critical role in maintaining capsid strength and integrity.
Adenovirus (Ad) vectors are the most commonly used vehicle for delivery of foreign genes into mammalian cells for gene therapy, as recombinant vaccines, or as general-purpose expression vectors in experimental studies (2). First-generation Ad (fgAd) vectors with both early region 1 (E1) and E3 deleted have a minimal genome size of 30.2 kb (84% of the wild-type genome size) and a maximum cloning capacity of almost 8 kb (3). For small transgenes, the genome size may increase by only a few kilobases; for example, an fgAd that encodes a green fluorescent protein expression cassette is only 32.2 kb (14). For helper-dependent Ad (hdAd) vectors, the genome is typically constructed to be close to 30 kb in length, in order to provide maximum genetic stability (24) and also to aid in separation of the hdAd virion from residual helper virus during virus purification on a cesium chloride gradient (23). In this study, we show that reduction in the size of the packaged Ad genome significantly reduces virion stability. We subjected a number of hdAd vectors and the helper viruses used for their propagation to heating at 47°C for 0, 15, or 30 min and examined the effect on vector titer. All of the helper viruses were relatively resistant to inactivation by heating, whereas all of the hdAds showed significantly reduced infectivity (Fig. 1A). After 30 min of incubation at 47°C, the helper viruses tested showed a 20 to 70% drop in titer, whereas the hdAds generated with these helper viruses dropped in titer by 100- to 1,000-fold. Previous studies showed that the capsid protein constituents of virions containing unusually small genomes (∼9 to 12 kb) are altered compared to wild-type Ad (AdWT), which could contribute to virion instability (15, 28). We analyzed the protein constituents of purified virions of a 30-kb hdAd compared to its parental helper virus and AdWT. As shown in Fig. 1B, all three of the viruses had identical protein contents, including pIX, which has been implicated previously in stabilizing the Ad virion against heat denaturation (9, 22). Thus, although hdAd vectors have capsid protein constituents identical to those of AdWT, they exhibit significant thermal lability.
FIG. 1.
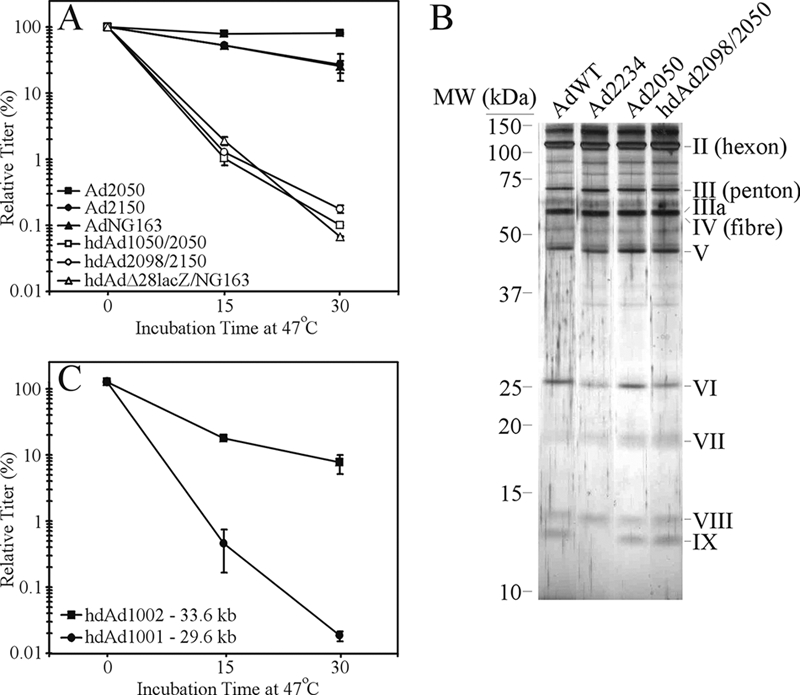
hdAd vectors exhibit reduced heat stability. (A) Three different hdAd vectors amplified with three different helper viruses were analyzed for their heat stability at 47°C. In parallel, the helper viruses used to generate these hdAds were evaluated. The genome sizes of these vectors are as follows: Ad2050, 35.8 kb; Ad2150, 35.9 kb; AdNG163, 37.2 kb; hdAd1050, 30.0 kb; hdAd2098, 30.2 kb; and hdAdΔ28lacZ, 28.9 kb. These data are representative of two experiments. Error bars indicate standard deviations. (B) Protein constituents of hdAd capsids are identical to those of AdWT. Aliquots of AdWT, Ad2234 (E1+ pIX−), Ad2050, and hdAd2098/2050 were separated by 12% SDS-PAGE and the resulting gel silver stained to visualize the capsid proteins. (C) The DNA genome size of the hdAd affects heat stability. Duplicate aliquots of hdAd1001 (29.6 kb) and hdAd1002 (33.6 kb) were incubated at 47°C for 0, 15, or 30 min, and the titer of each vector was determined at the end of the assay. These data are representative of two experiments. All of the hdAds and helper viruses have been described previously (5, 21, 23, 24) and were propagated using standard methods (21, 23).
All of the hdAds used for Fig. 1A have a genome size of less than ∼30 kb, and although such genomes are genetically stable (i.e., do not rearrange their DNA [24]), the observed reduced heat stability suggests that further biochemical constraints upon the Ad capsid exist whereby the DNA-capsid interactions help to stabilize the virion. To test the involvement of genome size in conferring virion stability, we examined the stability of two hdAd vectors that varied in the size of their stuffer DNA, resulting in genome sizes of 29.6 kb (hdAd1001) and 33.6 kb (hdAd1002). Increasing the genome size of the hdAd resulted in a significant improvement in vector stability (Fig. 1C). Thus, improved capsid stability is observed in hdAds as the genome size approaches the size of AdWT.
To determine if traditional E1-deleted viruses also exhibit a similar dependence of virion stability on genome size, we analyzed the stability of a series of E1-deleted vectors ranging from 30.2 kb to 36 kb. As with the hdAd, we observed a strong correlation between the size of the E1-deleted virus DNA and its stability (Fig. 2A). AdWT with a genome size of 36 kb exhibited almost complete stability over the 30-min time course at 47°C, as did other vectors larger than 33.3 kb. In contrast, the heat stability of vectors smaller than 32.2 kb was dramatically influenced by genome size. A 10- to 200-fold drop in vector titer, dependent on the size of the genome, was observed. For the vector with the smallest genome, AdJR34, the time required to reduce the titer by half was only 4.2 min. Loss of infectivity was accompanied by an increase in permeability of the capsid, suggesting a loss of capsid integrity (Fig. 2B). Of note, neither AdJR34 nor AdWT showed a loss in infectivity when heated at 37°C for 6 h (data not shown), indicating that the inherent instability of Ad vectors containing small genomes may be manifest only under certain conditions. Taken together, our data indicate that both hdAd and E1-deleted Ad show a similar dependence of virion stability on the size of the genome contained within the capsid.
FIG. 2.
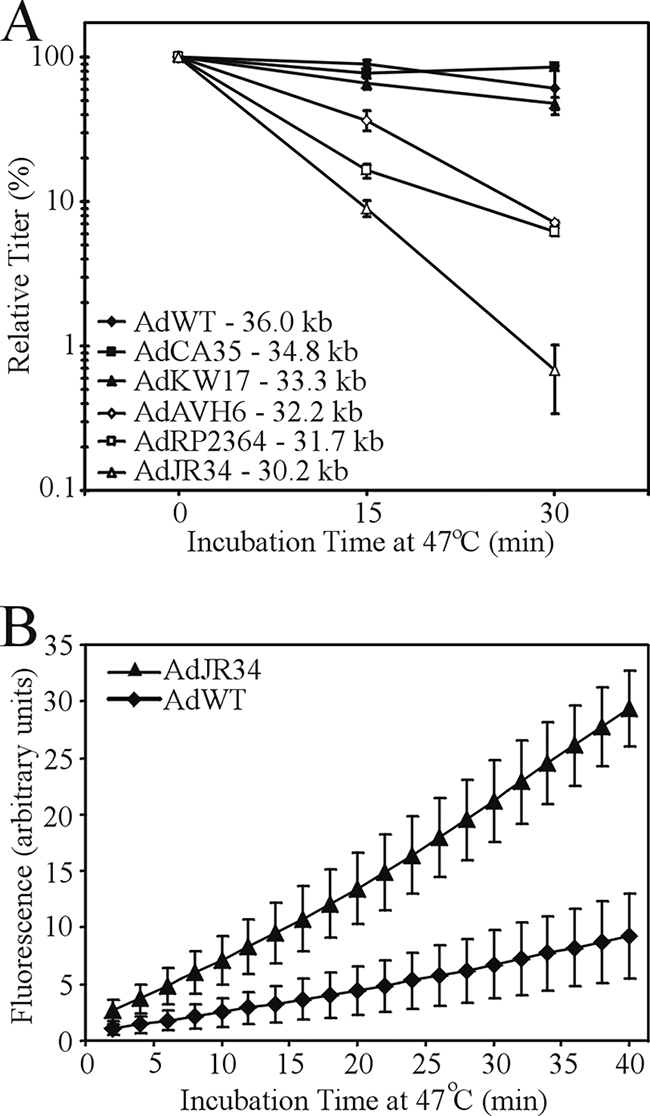
The heat stability of E1-deleted Ad is affected by DNA genome size. (A) Duplicate aliquots of various E1/E3-deleted Ad vectors ranging in size from 30.2 kb to 34.8 kb were incubated at 47°C for 0, 15, or 30 min, and the titer of each vector was determined at the end of the assay. In parallel, AdWT was also subjected to the same analysis. These data are representative of three experiments. Error bars indicate standard deviations. (B) AdJR34 (30.2 kb) or AdWT (36 kb) was mixed with Picogreen double-stranded DNA fluorescent dye and subjected to heating at 47°C, and fluorescence was measured at 2-min intervals using a Stratagene Mx3000P quantitative PCR machine. These data are based on three experiments. The E1/E3-deleted Ad vectors used in this study are as follows: AdRP2364 (31.7 kb) encodes the cDNA for DsRed (Discosoma sp. red fluorescent protein) under regulation by the human cytomegalovirus immediate-early promoter/enhancer and bovine growth hormone polyadenylation sequence, AdKW17 (33.3 kb) encodes the murine secreted alkaline phosphatase cDNA (mSEAP) (16) under regulation by the mouse ubiquitin C promoter and bovine growth hormone polyadenylation sequence, and AdJR34 (30.2 kb) is deleted of E1 and E3 but contains no transgene and contains a FLAG epitope-tagged core protein VII. AdAVH6 (32.2 kb) and AdCA35 (34.8 kb) have been described previously (1, 14, 27). The AdWT and the E1-deleted vectors were propagated and purified using standard methods (25).
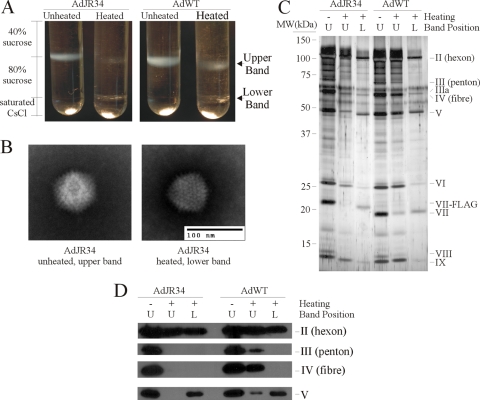
We next examined the changes in capsid morphology and protein content of heated virions containing small or large genomes. Virions were heated at 47°C for 30 min and then separated on a 40%/80% sucrose step gradient, and the identities of the resulting virus bands were examined by electron microscopy (EM) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Unheated AdJR34 (30.2 kb) and AdWT (36 kb) formed a single band at the interface of the 40% and 80% sucrose (Fig. 3A). Heating of AdWT at 47°C for 30 min resulted in a reduction in the intensity of the band present at the 40%/80% interface (upper band) and the appearance of a new band at the bottom of the 80% sucrose layer (lower band). Heating of AdJR34 resulted in a more pronounced decrease in the upper band and the appearance of a similar lower band. EM images of samples from each band showed that all contained intact capsid with the classic Ad morphology (although the fiber protein was not readily discernible in our images) (Fig. 3B and data not shown), with the exception of the upper band from heated AdJR34, for which we did not see any capsid structures. This observation is consistent our data suggesting that heating of AdJR34 results in increased accessibility of the viral DNA to fluorescent dye (Fig. 2B), due to a loss of capsid architecture.
FIG. 3.
Heating of Ad vectors causes a change in migration in a sucrose gradient and alters capsid morphology. (A) AdJR34 or AdWT was incubated at 0°C or 47°C for 30 min, layered on a 40%/80% sucrose step gradient, and centrifuged at 35,000 rpm for 2 h. (B) Representative electron micrographs of AdJR34 either unheated (left panel) or heated (right panel). (C) Samples from the various virus bands identified in panel A were separated by SDS-PAGE and silver stained to visualize the proteins. (D) Samples from the various bands identified in panel A were separated by SDS-PAGE and analyzed by immunoblotting for various capsid proteins using the following antibodies, as previously described (27): all Ad capsid proteins (ab6982, 1/10,000; Abcam), Ad fiber (MS-1027-P0, 1/1000; Neomarkers), or core V (1/100, a kind gift of S. J. Flint [Princeton]).
The data from our EM analysis were corroborated by protein analysis of the various bands. The upper bands from the unheated samples of both AdJR34 and AdWT contained all of the Ad capsid proteins (Fig. 3C) and represent intact virus. For heated AdJR34, the lower band contained a protein pattern that was consistent with capsids devoid of penton and fiber (Fig. 3C and D). The upper band of heated AdJR34 was devoid of many capsid proteins, including penton, fiber, protein VIII, and core proteins V and VII, indicating that material in this band lacks the Ad nucleoprotein core and likely represents dissociated capsid protein (that these proteins migrate to a similar position as intact capsid in our gradient appears to be strictly coincidental). These data are consistent with the relative paucity of infectious virus in the heated AdJR34 sample (Fig. 2) and the lack of visible capsids by EM analysis. For heated AdWT, the lower band contained a protein pattern consistent with capsid devoid of the vertex proteins, similar to AdJR34, whereas the upper band contained all capsid proteins. However, based on the ratio of hexon to the other capsid proteins, the upper band likely contains a mix of fully intact virus and partially dissociated virions. That a significant portion of heated AdWT appears to retain all protein from mature capsid (Fig. 3) and is infectious (Fig. 2) is in clear contrast to our observations for AdJR34.
Based on the morphological analysis of virion structure and the biochemical analysis of capsid composition, we conclude that a reduction in the size of the Ad genome results in a significant destabilization of the Ad capsid with heating. Within the virion, the viral DNA does not make direct contact with the major capsid proteins (12) but associates with three highly basic proteins, VII, V, and μ (7, 17, 26). Protein VII is a protamine-like protein and is primarily responsible for wrapping and condensing the viral DNA (19). The protein VII-DNA nucleoprotein complex is organized into a central dense core from which extend 12 large spherical units, termed adenosomes, that are directed toward each vertex (6, 20). Protein V is believed to form a shell around the protein VII-DNA complex (6, 11) and tethers the protein VII-wrapped DNA to the inner capsid (8, 10) through its direct association with protein VI (8, 18), which is directly connected to each peripentonal hexon (29). Thus, the only positions of contact between the viral DNA and the capsid are at the vertices, bridged through protein V. Interestingly, deletion of protein V results in virus that is heat labile but can be compensated for through secondary mutations in protein μ (30); thus, the bridging function of protein V is crucial to the stability of the Ad capsid.
Our studies indicate that reducing the size of the Ad genome destabilizes the vertex regions of the capsid. Destabilization of this region also occurs as a very early step during natural Ad infection: integrin binding to penton base stimulates virus endocytosis (31) and may induce a conformation change in the peripentonal capsid region (13), liberating protein VI, which then acts to lyse the endosomal membrane (32). Given the importance of the timely removal of the vertex proteins for efficient infection, it is likely a tightly orchestrated event and requires proper positioning of all viral proteins involved. Tight packaging of wild-type-length DNA into the capsid may force the adenosome DNA in the vertex regions into the proper position/orientation to achieve the required linkage between the DNA and the peripentonal hexons, bridged by proteins V and VI. Misalignment or prevention of the interaction between this series of proteins is a conceivable consequence of reducing the genome size, leading to destabilization of the capsid. Interestingly, compared to an fgAd of nearly identical genome length, hdAd showed an ∼5-fold-reduced stability (Fig. 1 and 2), suggesting that specific DNA sequences within the Ad genome may contribute to virion stability. However, all of the E1/E3-deleted Ads used in our studies contain identical Ad sequences yet display dramatically different thermal stabilities. Thus, the overall genome length is likely the primary determinant in DNA-mediated capsid stabilization.
We and others have shown that Ad has a fairly strict genome size requirement for optimal DNA packaging. Genomes below 75% or above 105% tend to undergo rearrangement to increase or decrease the size of the genome to closer-to-wild-type length (4, 24). In this study, we elucidated one of the mechanisms by which this occurs: packaging of viral DNA that is too small results in destabilized virions, which would provide a growth disadvantage. Thus, evolutionary pressures over millions of years may have optimized both the length of the viral DNA and the size of the capsid to maximize the stability and fitness of Ad.
Acknowledgments
We thank Frank L. Graham and Michael Kennedy for critical evaluation of the manuscript and John Bell, Doug Gray, Jane Flint, and Phil Ng for reagents. We also thank Tim Karnauchow for assistance with generating electron microscopy images.
Research in the Parks laboratory is supported by grants from the Canadian Institutes of Health Research (CIHR) and the Jesse Davidson Foundation for Gene and Cell Therapy and by a CIHR/Muscular Dystrophy Canada/Amyotrophic Lateral Sclerosis Society of Canada Partnership Grant.
Footnotes
Published ahead of print on 26 November 2008.
REFERENCES
- 1.Addison, C. L., M. Hitt, D. Kunsken, and F. L. Graham. 1997. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol. 781653-1661. [DOI] [PubMed] [Google Scholar]
- 2.Amalfitano, A., and R. J. Parks. 2002. Separating fact from fiction: assessing the potential of modified adenovirus vectors for use in human gene therapy. Curr. Gene Ther. 2111-133. [DOI] [PubMed] [Google Scholar]
- 3.Bett, A. J., W. Haddara, L. Prevec, and F. L. Graham. 1994. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl. Acad. Sci. USA 918802-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bett, A. J., L. Prevec, and F. L. Graham. 1993. Packaging capacity and stability of human adenovirus type 5 vectors. J. Virol. 675911-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramson, J. L., N. Grinshtein, R. A. Meulenbroek, J. Lunde, D. Kottachchi, I. A. Lorimer, B. J. Jasmin, and R. J. Parks. 2004. Helper-dependent adenoviral vectors containing modified fibre for improved transduction of developing and mature muscle cells. Hum. Gene Ther. 15179-188. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. T., M. Westphal, B. T. Burlingham, U. Winterhoff, and W. Doerfler. 1975. Structure and composition of the adenovirus type 2 core. J. Virol. 16366-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, P. K., M. E. Vayda, and S. J. Flint. 1986. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J. Mol. Biol. 18823-37. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, P. K., M. E. Vayda, and S. J. Flint. 1985. Interactions among the three adenovirus core proteins. J. Virol. 55379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colby, W. W., and T. Shenk. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J. Virol. 39977-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everitt, E., L. Lutter, and L. Philipson. 1975. Structural proteins of adenoviruses. XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation and chemical cross-linking. Virology 67197-208. [DOI] [PubMed] [Google Scholar]
- 11.Everitt, E., B. Sundquist, U. Pettersson, and L. Philipson. 1973. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology 52130-147. [DOI] [PubMed] [Google Scholar]
- 12.Fabry, C. M., M. Rosa-Calatrava, J. F. Conway, C. Zubieta, S. Cusack, R. W. Ruigrok, and G. Schoehn. 2005. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 241645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75477-486. [DOI] [PubMed] [Google Scholar]
- 14.Hubberstey, A. V., M. Pavliv, and R. J. Parks. 2002. Cancer therapy utilizing an adenoviral vector expressing only E1A. Cancer Gene Ther. 9321-329. [DOI] [PubMed] [Google Scholar]
- 15.Lieber, A., C. Y. He, I. Kirillova, and M. A. Kay. 1996. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J. Virol. 708944-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maelandsmo, G. M., P. J. Ross, M. Pavliv, R. A. Meulenbroek, C. Evelegh, D. A. Muruve, F. L. Graham, and R. J. Parks. 2005. Use of a murine secreted alkaline phosphatase as a non-immunogenic reporter gene in mice. J. Gene Med. 7307-315. [DOI] [PubMed] [Google Scholar]
- 17.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology 36126-136. [DOI] [PubMed] [Google Scholar]
- 18.Matthews, D. A., and W. C. Russell. 1998. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 791671-1675. [DOI] [PubMed] [Google Scholar]
- 19.Mirza, M. A., and J. Weber. 1982. Structure of adenovirus chromatin. Biochim. Biophys. Acta 69676-86. [DOI] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., J. W. Boring, and J. C. Brown. 1984. Ion etching of human adenovirus 2: structure of the core. J. Virol. 5152-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer, D., and P. Ng. 2003. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 8846-852. [DOI] [PubMed] [Google Scholar]
- 22.Parks, R. J. 2005. Adenovirus protein IX: a new look at an old protein. Mol. Ther. 1119-25. [DOI] [PubMed] [Google Scholar]
- 23.Parks, R. J., L. Chen, M. Anton, U. Sankar, M. A. Rudnicki, and F. L. Graham. 1996. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA 9313565-13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks, R. J., and F. L. Graham. 1997. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 713293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross, P. J., and R. J. Parks. 2006. Construction of first-generation adenoviral vectors, p. 149-165. In T. Freidman and J. Rossi (ed.), Gene therapy vectors: a techniques manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Russell, W. C., W. G. Laver, and P. J. Sanderson. 1968. Internal components of adenovirus. Nature 2191127-1130. [DOI] [PubMed] [Google Scholar]
- 27.Sargent, K., R. A. Meulenbroek, and R. J. Parks. 2004. Activation of adenoviral gene expression by protein IX is not required for efficient virus replication. J. Virol. 785032-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shayakhmetov, D. M., Z. Y. Li, A. Gaggar, H. Gharwan, V. Ternovoi, V. Sandig, and A. Lieber. 2004. Genome size and structure determine efficiency of postinternalization steps and gene transfer of capsid-modified adenovirus vectors in a cell-type-specific manner. J. Virol. 7810009-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart, P. L., S. D. Fuller, and R. M. Burnett. 1993. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 122589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ugai, H., A. V. Borovjagin, L. P. Le, M. Wang, and D. T. Curiel. 2007. Thermostability/infectivity defect caused by deletion of the core protein V gene in human adenovirus type 5 is rescued by thermo-selectable mutations in the core protein X precursor. J. Mol. Biol. 3661142-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73309-319. [DOI] [PubMed] [Google Scholar]
- 32.Wiethoff, C. M., H. Wodrich, L. Gerace, and G. R. Nemerow. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 791992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]