Abstract
Lamivudine therapy selects for the M184V mutation. Although this mutation reduces the replicative capacity of human immunodeficiency virus in vitro, its impact on viral fitness in vivo has not been well defined. We used quantitative allele-specific PCR to precisely calculate the fitness differences between the mutated M184V virus and one that had reverted to the wild type in a cohort of patients by selectively interrupting reverse transcriptase inhibitor therapy, and we found that the M184V variants were consistently 4 to 8% less fit than the wild type in the absence of drug. After a lag phase of variable duration, wild-type variants emerged due to continued evolution of pol and back mutation rather than through emergence of an archived wild-type variant.
Despite recent advances, the management of multidrug-resistant (MDR) human immunodeficiency virus type 1 (HIV-1) remains a major clinical problem. One strategy to avoid immune deterioration while minimizing toxicity in viremic patients infected with MDR HIV-1 is to prevent the emergence of wild-type (WT) HIV-1 by continuing selected drugs in a failing antiretroviral regimen. Studies show that reverse transcriptase (RT) inhibitors (RTIs) continue to exert antiviral activity in the presence of resistance mutations(8, 21). In particular, continuation of lamivudine (3TC) or emtricitabine in the presence of the M184V mutation may provide clinical benefit (3, 4). We previously showed that M184V is lost at a median of 20 weeks following interruption of 3TC together with other RTIs while the use of protease (PR) inhibitors (PIs) was continued in viremic subjects with MDR HIV-1(8). In this study, we performed a detailed analysis of the decay of the M184V-carrying mutants in these individuals.
(This work was presented in part at the 13th [abstract 628] and 14th [Abstract 588] Conferences on Retroviruses and Opportunistic Infections, held in 2006 in Denver, CO, and in 2007 in Los Angeles, CA, respectively.)
Subjects were antiretroviral treatment-experienced HIV-1-infected patients enrolled in an ongoing prospective cohort study (8). This particular substudy focused on five adherent, highly treatment-experienced, viremic subjects with HIV-1 resistant to antiretroviral drugs from at least two classes who interrupted 3TC together with other RTIs but remained on PIs. A sixth participant (subject 3158) enrolled in the parent study while receiving 3TC, stavudine (d4T), and nelfinavir (NFV) and selectively interrupted NFV. At week 52, this patient discontinued 3TC and d4T therapy and subsequently remained off all antiretroviral therapy. Subjects were followed weekly for the first 4 weeks, every 2 weeks for the next 8 weeks, and every 4 weeks thereafter for at least 48 weeks or until treatment was modified. Participants provided written, informed consent for participation in these studies, which were approved by the University of California, San Francisco, Committee on Human Research, and the Partners HealthCare Systems Institutional Review Board.
At the time of the partial treatment interruption, the median plasma HIV-1 RNA level was 3.65 log copies/ml, and the median CD4+ count was 336 cells/mm3. Further details of this cohort have been reported previously (8).
Population sequencing of plasma viruses and phenotypic antiretroviral susceptibility tests (GeneSeq and Phenosense HIV; Monogram Biosciences, South San Francisco, CA) obtained before the RTI interruption and at multiple time points thereafter showed a slow decay of nucleoside RTI (NRTI) resistance. Mutations decayed gradually at a rate roughly proportional to their associated fitness costs, as estimated in vitro in the absence of drug (7) (Table 1). The thymidine analogue mutations waned more slowly than the M184V mutations. Changes in phenotypic susceptibility to 3TC and other NRTIs paralleled the genotypic changes (data not shown). As expected, all subjects maintained viruses with high-level PI resistance throughout the study period (data not shown).
TABLE 1.
Mutations in HIV-1 RT at codons associated with drug resistance in subjects interrupting RTI therapy, as assessed by population sequencing of plasma viruses
| Subject or isolate | Treatment interrupted | Treatment continued | No. of weeks post-PTIa | Codonb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41 | 62 | 67 | 69 | 70 | 103 | 118 | 181 | 184 | 190 | 210 | 215 | 219 | ||||
| Clade B consensus | M | A | D | T | K | K | V | Y | M | G | L | T | K | |||
| 3005 | ZDV/3TC | IDV/RTV | 0 | L | - | N | D | - | - | - | - | V | - | W | Y | - |
| 34 | M/L | - | N | D | - | - | V/I | - | M/V | - | L/W | Y | - | |||
| 36 | M | - | N | D | - | - | V/I | - | M/V | - | L/W | Y | - | |||
| 48 | - | - | N | D | - | - | V/I | - | - | - | L/W | D/N/Y | - | |||
| 3040 | d4T/3TC | LPV/RTV | 0 | - | - | N | T/A | R | - | - | Y/C | V | - | - | T/S | Q |
| 12 | - | - | N | T | R | - | - | - | V | - | - | - | Q | |||
| 16 | - | - | N | - | R | - | - | - | V | - | - | - | Q | |||
| 22 | - | - | N | - | R | - | - | - | M/V | - | - | - | Q | |||
| 24 | - | - | N | - | R | - | - | - | M/V | - | - | - | Q | |||
| 36 | - | - | N | - | R | - | - | - | - | - | - | - | Q | |||
| 3057 | d4T/3TC | IDV | 0 | L | V | - | T/A | K/R | - | - | - | V | - | - | - | K/Q |
| 7 | L | V | - | - | - | - | - | - | V | - | - | - | ||||
| 36 | L | V | - | - | - | - | - | - | - | - | - | - | - | |||
| 48 | L | V | - | - | - | - | - | - | - | - | - | - | - | |||
| 3151 | DDI/d4T/3TC/EFV | APV/RTV | 0 | L | - | N | D | R | - | I | C | V | S | W | F | Q |
| 16 | L | - | N | D | R | - | I | C | V | S | L/W | F | Q | |||
| 3167 | ZDV/3TC | NFV | 0 | - | - | - | - | - | - | - | - | V | - | - | - | - |
| 4 | - | - | - | - | - | - | - | - | V | - | - | - | - | |||
| 8 | - | - | - | - | - | - | - | - | V | - | - | - | - | |||
| 12 | - | - | - | - | - | - | - | - | M/V | - | - | - | - | |||
| 3158 | d4T/3TC | None | 0 | - | - | N | - | R | - | - | - | V | - | - | - | Q |
| 5 | - | - | N | - | R | - | - | - | V | - | - | - | Q | |||
| 12 | - | - | N | - | R | - | - | - | M/V | - | - | - | Q | |||
| 16 | - | - | N | - | R | - | - | - | M/V | - | - | - | Q | |||
PTI, partial treatment interruption.
Amino acid residues are indicated by single-letter abbreviations. The HIV-1 clade B consensus sequence is shown for comparison. Dashes indicate identity with the consensus sequence. ZDV, zidovudine; ddI, didanosine; EFV, efavirenz; IDV, indinavir; RTV, ritonavir; SQV, saquinavir; APV, amprenavir.
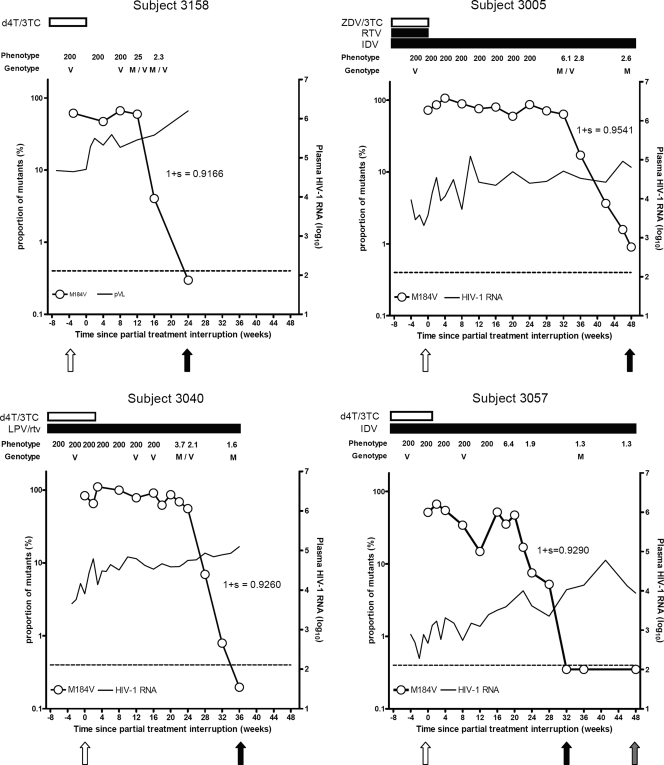
An allele-specific PCR (AS-PCR) assay that detects M184V variants present in at least 0.4% of the quasispecies population (18) was used to quantify more precisely the proportion of viruses carrying the M184V mutation over time and to estimate the fitness of M184V-containing viruses in vivo. By AS-PCR, M184V mutants were present as the predominant viral species at baseline in all subjects. (Fig. 1) In subjects 3167 and 3151, there was no significant change in the proportion of M184V-containing virus through 12 and 16 weeks of follow-up, respectively (data not shown), consistent with the population sequencing results. In subjects 3005, 3040, 3057, and 3158, M184V variants exhibited a biphasic decay: after an initial lag phase of variable duration, M184V variants decreased exponentially (Fig. 1). The lag phase lasted 20 weeks in subject 3057, 24 weeks in subject 3040, and 32 weeks in subject 3005 but was much shorter (12 weeks) in subject 3158, who was receiving only d4T and 3TC and therefore interrupted all antiretroviral drugs in his regimen. Despite the variable lag times, once decay began, the duration and slopes of the exponential decay phase were similar in all subjects, lasting approximately 12 weeks, although minor M184V variants remained detectable for up to 48 weeks after 3TC interruption in subject 3005.
FIG. 1.
Dynamics of M184V variants after the interruption of RTI therapy. Only subjects achieving complete M184V mutant decay are shown. For the AS-PCR assay, viral RNA was extracted from 500 μl of plasma. Open circles, proportion of M184V mutants as determined by AS-PCR testing (in logarithmic scale); continuous line, HIV-1 RNA levels (in log10 copies/ml); horizontal dashed line, sensitivity threshold to detect M184V variants (0.4%); horizontal bars in the superior part of each graph, duration of antiretroviral treatment. Phenotype refers to the relative change in 3TC susceptibility. Genotype refers to the codon 184 allele detected by population-based sequencing: methionine (M), valine (V), or a mixture of variants with methionine and variants with valine in codon 184 (M/V). Arrows show the time points when T0 (white), T1 (black), and T2 (gray) clonal sequences were obtained. The fitness (1+S) values of M184V viruses relative to 184M variants are shown next to the exponential M184V virus decay phase. ZVD, zidovudine; RTV, ritonavir; IDV, indinavir; LPV/rtv, lopinavir boosted with ritonavir.
Relative fitness was calculated in the four subjects with M184V reversion using the averaging method, which incorporated multiple AS-PCR measurements during the exponential M184V mutant decay phase, the HIV-1 RNA level at each time point, and the death rate of infected cells (1, 15) (we used a death rate [δ] of 0.5, as this value approximates the mean of many independent estimates [2]). The relative fitness disadvantage of the M184V mutation ranged from 4.6% to 8.3% in these MDR viruses (Fig. 1).
To further characterize the viral variants emerging after NRTI discontinuation, clonal HIV-1 pol sequences were obtained from subjects 3040, 3057, 3005, and 3158 at the time of RTI interruption (T0) and at the first time point at which the M184V mutation became undetectable by AS-PCR (T1). An additional time point (T2) was available for subject 3057 at 16 weeks after T1. A mean of 24 clones (range, 21 to 29) (Table 2) was analyzed at each time point. All sequences obtained at T0 carried the M184V mutation, whereas all sequences obtained at T1 and T2 were WT at this codon (Table 2). In addition to the changes in the frequency of thymidine analogue mutations observed by population sequencing, clonal analysis revealed a decrease in the frequency of K70R variants in subjects 3040 and 3158 and evidence of back mutation at codon 215 from the mutant TAC (Tyr) to the partial revertants GAC (Asp) and AAC (Asn) in subject 3005.
TABLE 2.
Clonal analysisa
| Subject or isolateb | Week no. | Total no. of clones | No. of clones | Codonc
|
Sequence diversity (d ± SE)d
|
Ka/Ks ratiod,e
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41 | 62 | 67 | 69 | 70 | 184 | 210 | 215 | 219 | PR+RT | PR | RT | PR+RT | PR | RT | ||||
| Clade B consensus | M | A | D | T | K | M | L | T | K | |||||||||
| 3005 | 0 | 24 | 24 | L | - | N | D | - | V | W | Y | - | 0.0052 ± 0.0014 | 0.0010 ± 0.0007 | 0.0067 ± 0.0017 | 0.20 | 0.00 | 0.22 |
| 48 | 29 | 16 | - | - | N | D | - | - | - | Y | - | 0.0111 ± 0.0016* | 0.0045 ± 0.0011* | 0.0135 ± 0.0021* | 0.19 | 0.22 | 0.18 | |
| 4 | - | - | N | D | - | - | - | N | - | |||||||||
| 4 | - | - | N | D | - | - | W | N | - | |||||||||
| 3 | - | - | N | D | - | - | W | D | - | |||||||||
| 1 | L | - | N | D | - | - | R | D | - | |||||||||
| 1 | - | - | N | D | - | - | - | D | - | |||||||||
| Overall | 0.0112 ± 0.0017 | 0.0047 ± 0.0017 | 0.0118 ± 0.0028 | 0.29 | 0.44 | 0.25 | ||||||||||||
| 3040 | 0 | 22 | 22 | - | - | N | - | R | V | - | - | Q | 0.0065 ± 0.0016 | 0.0073 ± 0.0032 | 0.0063 ± 0.0016 | 0.08 | 0.01 | 0.11 |
| 36 | 27 | 15 | - | - | N | - | R | - | - | - | Q | 0.0056 ± 0.0016 | 0.0078 ± 0.0034 | 0.0048 ± 0.0015 | 0.40 | 0.78 | 0.21 | |
| 12 | - | - | N | - | - | - | - | - | Q | |||||||||
| Overall | 0.0149 ± 0.0024 | 0.0172 ± 0.0054 | 0.0128 ± 0.0028 | 0.26 | 0.27 | 0.16 | ||||||||||||
| 3057 | 0 | 22 | 22 | L | V | - | - | - | V | - | - | - | 0.0061 ± 0.0014 | 0.0061 ± 0.0027 | 0.0061 ± 0.0015 | 0.50 | NA | 0.40 |
| 32 | 21 | 20 | L | V | - | - | - | - | - | - | - | 0.0119 ± 0.0021* | 0.0100 ± 0.0039 | 0.0126 ± 0.0024* | 0.20 | 0.17 | 0.19 | |
| 1 | L | V | G | - | - | - | - | - | - | |||||||||
| 48 | 21 | 21 | L | V | - | - | - | - | - | - | - | 0.0104 ± 0.0019 | 0.0095 ± 0.0038 | 0.0107 ± 0.0020 | 0.27 | 0.46 | 0.27 | |
| Overall | 0.0123 ± 0.0013 | 0.0103 ± 0.0029 | 0.0129 ± 0.0023 | 0.32 | 0.44 | 0.28 | ||||||||||||
| 3158 | 0 | 22 | 22 | - | - | N | - | R | V | - | - | Q | 0.0061 ± 0.0010 | 0.0082 ± 0.0023 | 0.0053 ± 0.0011 | 0.14 | 0.28 | 0.10 |
| 24 | 24 | 15 | - | - | N | - | R | - | - | - | Q | 0.0089 ± 0.0010 | 0.0127 ± 0.0031 | 0.0076 ± 0.0015 | 0.16 | 0.37 | 0.11 | |
| 8 | - | - | N | - | - | - | - | - | Q | |||||||||
| 1 | - | - | N | - | S | - | - | - | Q | |||||||||
| Overall | 46 | 0.0087 ± 0.0013 | 0.0113 ± 0.0029 | 0.0073 ± 0.0013 | 0.24 | 0.36 | 0.15 | |||||||||||
For each plasma sample, the products of five separate reverse transcription-PCRs were pooled and purified. Five separate nested PCRs were then performed with each pooled reverse transcription-PCR product using primers OOPF2 and OOR3 (18) and cloned within the TOPO-TA 2.1 vector (Invitrogen Corporation, Carlsbad, CA).
Subject 3005 interrupted treatment with zidovudine/3TC and continued indinavir/ritonavir; subject 3040 interrupted treatment with d4T/3TC and continued lopinavir/ritonavir; subject 3057 interrupted treatment with d4T/3TC and continued indinavir; subject 3158 interrupted treatment with d4T/3TC and did not continue any other treatment. Amino acid residues are indicated by single-letter abbreviations.
The HIV-1 clade B consensus sequence is shown for comparison. Dashes indicate identity with the consensus B sequence.
Clonal sequence diversity and the nonsynonymous/synonymous nucleotide substitution (Ka/Ks) ratio are shown per each subject in whom M184V viruses reverted back to 184M. Data are presented by week of clonal sequence sampling, as well as considering all patient clones together (overall). Distances (d) were estimated with MEGA, version 3.1, using a Tamura-Nei model for nucleotide substitution, assuming a gamma distribution with an alpha of 0.5. Standard errors (SE) were estimated using 1,000 bootstrap replicates. *, P < 0.05, by a two-tailed P value (one-sample t test), compared with baseline.
The nonsynonymous/synonymous Ka/Ks ratio was also estimated with MEGA, version 3.1, using the Nei-Gojobori method and a Jukes-Cantor model for nucleotide substitution. NA, calculation not available because the rate of synonymous substitutions (dS) is 0.
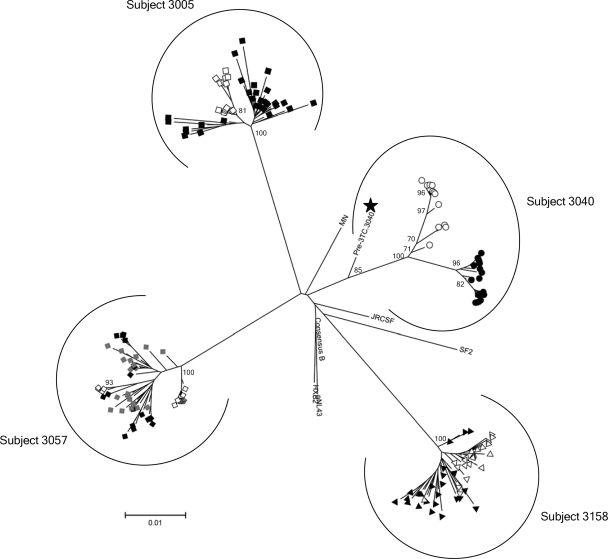
Phylogenetic trees reconstructed in PAUP, version 4.0b (Hasegawa-Kishino-Yano model), and MEGA, version 3.1 (Tamura-Nei model) (13), using both maximum-likelihood and neighbor-joining (NJ) approaches plus 1,000 NJ bootstrapping replicates showed that in subjects 3057, 3005, and 3158, sequences from T0 and T1 were intermingled and shared a most-recent common ancestor (MRCA) (Fig. 2), as expected from back-mutated sequences. Interestingly, in these three subjects there was an increase in sequence diversity in both the PR and RT genes, suggesting ongoing exploration of sequence space by the virus population (Table 2). Conversely, viral sequences from subject 3040 at T0 and T1 clustered separately with different MRCAs. In addition, in this subject sequence diversity increased in the PR gene but decreased in the RT gene (Table 2). The T1 sequences were more closely related to the M184V variants at T0 (genetic distance, 0.019) than to a pre-3TC consensus sequence (genetic distance, 0.034) obtained in 1995, providing further support for the inference that the 184M variant arose from back mutation of the M184V variant as opposed to reemergence of archived viruses. Analyses of the rate of nonsynonymous and synonymous substitutions for samples from all patients and time points, using MEGA software, version 3.1 (13), indicated that purifying selection (i.e., reduction of alleles with a deleterious effect on the phenotype) was the main mechanism for M184V reversion in all subjects (Table 2).
FIG. 2.
Continued evolution of pol (and back mutation) as a major mechanism of MDR-184M variant emergence after RTI therapy interruption. This is an unrooted NJ phylogenetic tree generated with MEGA, version 3.1. Data were derived from a multiple sequence alignment including nonidentical clonal pol sequences from all subjects plus the laboratory and patient-derived HIV-1 reference sequences. We assumed a Tamura-Nei (TN93) model of nucleotide evolution including transitions and transversions and a gamma-distributed variability rate among sites with an alpha value of 0.8. The node reliability was assessed using 1,000 bootstrap replications. Bootstrap values of >70% are presented. Additional analyses using different models of nucleotide evolution, maximum-likelihood tree reconstruction approaches, separate alignments per each subject, or separate analyses of the PR and the RT coding regions of pol yielded identical results. In subjects 3005, 3057, and 3158, MDR-184M variants emerging after treatment interruption (black symbols, T1; gray symbols, T2) and baseline MDR-M184V viruses (T0, white symbols) did not have different MRCAs. In subject 3040, 184M viruses emerging after treatment interruption (black circles) derived from a significantly different MRCA than baseline M184V variants. However, a WT consensus sequence obtained before the initiation of 3TC (black star) treatment was more closely related to the baseline M184V MRCA (genetic distance ± standard error, 0.0162 ± 0.0038) than to the 184M MRCA (0.0336 ± 0.0057).
This study showed that the M184V mutation disappeared quickly after a variable lag phase that lasted as long as 32 weeks. Differences between individual subjects in the time to the disappearance of the M184V mutation seemed related to the length of the initial lag phase rather than to differences in the rate of exponential decay. The estimates of the relative fitness of MDR HIV-1 carrying the M184V (MDR-M184V) mutation were in close agreement with previous data, indicating a fitness cost of the M184V mutation of approximately 10% relative to the WT (14).
In contrast to observations made when all drugs are discontinued (9), we did not observe evidence for the escape of a preexisting MDR virus with the wild type at codon 184 (MDR-184M), nor did we find evidence of the emergence of viruses with a WT RT and a mutant PR through recombination of actively replicating and archived variants (16). Several findings suggested that 184M viruses emerged through continuous evolution of pol and back mutation (12). First, NRTI mutations decayed in serial clonal and population-based sequences in an ordered, stepwise fashion at a rate roughly proportional to their associated fitness costs, as estimated in vitro in the absence of drug (7). Second, there was clonal evidence for back mutation through partial revertants at codon 215 (GAC [Asp] and AAC [Asn] derived from the mutant TAC [Tyr] in subject 3005). Third, the MDR-M184V and MDR-184M variants shared an MRCA in three out of four subjects, and even in the fourth subject (3040), emerging MDR-184M variants were more closely related to on-treatment MDR-M184V mutants than to a pre-3TC WT sequence.
The opposing selective pressures exerted on different coding regions of pol by interrupting RTI therapy and continuing PI treatment likely favored the loss of RT mutations that conferred a fitness cost in the absence of RTI therapy while favoring the persistence of PI resistance mutations. Given the high mutation rate of HIV-1 (6, 11), however, 184M revertants should have been generated frequently in the setting of ongoing virus replication. Thus, it appears that the probability that the 184M revertants would become fixed in the quasispecies was low. This finding could be explained by the following: (i) the existence of lower than expected levels of viral replication (i.e., a limited effective population size despite relatively high viral loads); (ii) mutations or recombination occurring outside the PR and RT genes, modifying the overall fitness of the 184M revertants (5); and (iii) the continued competition of MDR-M184V viruses (not actively inhibited by treatment) with the MDR-184M variants after treatment interruption (17). In addition, the MDR-184M population present at the time of RTI interruption might have been quite small in the subjects who had initiated 3TC prior to or together with PI therapy since all PI-resistant mutants would be linked to M184V, and few, if any, MDR variants with a WT 184 codon would exist in the quasispecies.
Other factors such as defective cytotoxic T-lymphocyte responses in subjects with advanced HIV disease (10, 19, 20) and the presence of other RTI resistance mutations could also have modulated the fitness cost of the M184V mutation and influenced the rate of reversion (7).
In conclusion, withdrawal of RTI therapy and continuation of PI treatment was associated with slow decay of the M184V mutation in MDR HIV-1-infected subjects. The time to back mutation appeared to be the rate-limiting step in replacement of 184V by 184M. The challenge for the virus of generating variants with a WT RT while maintaining PI resistance likely contributed to the observed delay. WT RT variants eventually emerged due to continued evolution of pol and back mutation in the context of negative selection.
Acknowledgments
This work was supported in part by the following: U.S. Public Health Service grants from the National Institutes of Health (R01 AI42567, R01 AI052745, R37 AI055357, K24 RR16482), a Virology Support Laboratory contract from the Adult ACTG (U01 AI068636), a grant from the Harvard Medical School Center for AIDS Research Virology Core (P30 AI060354) to D.R.K, and grants from the UCSF/Gladstone CFAR (P30 MH59037) and the UCSF Clinical and Translational Science Institute (UL1 RR024131-01) to S.G.D. Roger Paredes received the La Caixa Fellowship Grant for Post-Graduate Studies, Caixa d'Estalvis i Pensions de Barcelona, La Caixa, Barcelona, Catalonia, Spain.
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Bonhoeffer, S., A. D. Barbour, and R. J. De Boer. 2002. Procedures for reliable estimation of viral fitness from time-series data. Proc. Biol. Sci. 2691887-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonhoeffer, S., G. A. Funk, H. F. Gunthard, M. Fischer, and V. Muller. 2003. Glancing behind virus load variation in HIV-1 infection. Trends Microbiol. 11499-504. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, T. B., N. S. Shulman, S. C. Johnson, A. R. Zolopa, R. K. Young, L. Bushman, C. V. Fletcher, E. R. Lanier, T. C. Merigan, and D. R. Kuritzkes. 2005. Antiviral activity of lamivudine in salvage therapy for multidrug-resistant HIV-1 infection. Clin. Infect. Dis. 41236-242. [DOI] [PubMed] [Google Scholar]
- 4.Castagna, A., A. Danise, S. Menzo, L. Galli, N. Gianotti, E. Carini, E. Boeri, A. Galli, M. Cernuschi, H. Hasson, M. Clementi, and A. Lazzarin. 2006. Lamivudine monotherapy in HIV-1-infected patients harbouring a lamivudine-resistant virus: a randomized pilot study (E-184V study). AIDS 20795-803. [DOI] [PubMed] [Google Scholar]
- 5.Charpentier, C., T. Nora, O. Tenaillon, F. Clavel, and A. J. Hance. 2006. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J. Virol. 802472-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267483-489. [DOI] [PubMed] [Google Scholar]
- 7.Cong, M. E., W. Heneine, and J. G. Garcia-Lerma. 2007. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J. Virol. 813037-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deeks, S. G., R. Hoh, T. B. Neilands, T. Liegler, F. Aweeka, C. J. Petropoulos, R. M. Grant, and J. N. Martin. 2005. Interruption of treatment with individual therapeutic drug classes in adults with multidrug-resistant HIV-1 infection. J. Infect. Dis. 1921537-1544. [DOI] [PubMed] [Google Scholar]
- 9.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344472-480. [DOI] [PubMed] [Google Scholar]
- 10.Harrer, E., T. Harrer, P. Barbosa, M. Feinberg, R. P. Johnson, S. Buchbinder, and B. D. Walker. 1996. Recognition of the highly conserved YMDD region in the human immunodeficiency virus type 1 reverse transcriptase by HLA-A2-restricted cytotoxic T lymphocytes from an asymptomatic long-term nonprogressor. J. Infect. Dis. 173476-479. [DOI] [PubMed] [Google Scholar]
- 11.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373123-126. [DOI] [PubMed] [Google Scholar]
- 12.Kitchen, C. M., J. Lu, M. A. Suchard, R. Hoh, J. N. Martin, D. R. Kuritzkes, and S. G. Deeks. 2006. Continued evolution in gp41 after interruption of enfuvirtide in subjects with advanced HIV type 1 disease. AIDS Res. Hum. Retrovir. 221260-1266. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 14.Lu, J., and D. R. Kuritzkes. 2001. A novel recombinant marker virus assay for comparing the relative fitness of hiv-1 reverse transcriptase variants. J. Acquir. Immune Defic. Syndr. 277-13. [DOI] [PubMed] [Google Scholar]
- 15.Maree, A. F., W. Keulen, C. A. Boucher, and R. J. De Boer. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 7411067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nora, T., C. Charpentier, O. Tenaillon, C. Hoede, F. Clavel, and A. J. Hance. 2007. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J. Virol. 817620-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowak, M. A., S. Bonhoeffer, G. M. Shaw, and R. M. May. 1997. Anti-viral drug treatment: dynamics of resistance in free virus and infected cell populations. J. Theor. Biol. 184203-217. [DOI] [PubMed] [Google Scholar]
- 18.Paredes, R., V. C. Marconi, T. B. Campbell, and D. R. Kuritzkes. 2007. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J. Virol. Methods 146136-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samri, A., G. Haas, J. Duntze, J. M. Bouley, V. Calvez, C. Katlama, and B. Autran. 2000. Immunogenicity of mutations induced by nucleoside reverse transcriptase inhibitors for human immunodeficiency virus type 1-specific cytotoxic T cells. J. Virol. 749306-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt, M., E. Harrer, A. Goldwich, M. Bauerle, I. Graedner, J. R. Kalden, and T. Harrer. 2000. Specific recognition of lamivudine-resistant HIV-1 by cytotoxic T lymphocytes. AIDS 14653-658. [DOI] [PubMed] [Google Scholar]
- 21.Wainberg, M. A. 2004. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev. Anti Infect. Ther. 2147-151. [DOI] [PubMed] [Google Scholar]