Abstract
The nuclear magnetic resonance (NMR) structure of a central segment of the previously annotated severe acute respiratory syndrome (SARS)-unique domain (SUD-M, for “middle of the SARS-unique domain”) in SARS coronavirus (SARS-CoV) nonstructural protein 3 (nsp3) has been determined. SUD-M(513-651) exhibits a macrodomain fold containing the nsp3 residues 528 to 648, and there is a flexibly extended N-terminal tail with the residues 513 to 527 and a C-terminal flexible tail of residues 649 to 651. As a follow-up to this initial result, we also solved the structure of a construct representing only the globular domain of residues 527 to 651 [SUD-M(527-651)]. NMR chemical shift perturbation experiments showed that SUD-M(527-651) binds single-stranded poly(A) and identified the contact area with this RNA on the protein surface, and electrophoretic mobility shift assays then confirmed that SUD-M has higher affinity for purine bases than for pyrimidine bases. In a further search for clues to the function, we found that SUD-M(527-651) has the closest three-dimensional structure homology with another domain of nsp3, the ADP-ribose-1"-phosphatase nsp3b, although the two proteins share only 5% sequence identity in the homologous sequence regions. SUD-M(527-651) also shows three-dimensional structure homology with several helicases and nucleoside triphosphate-binding proteins, but it does not contain the motifs of catalytic residues found in these structural homologues. The combined results from NMR screening of potential substrates and the structure-based homology studies now form a basis for more focused investigations on the role of the SARS-unique domain in viral infection.
Severe acute respiratory syndrome (SARS) is a highly contagious disease caused by the SARS-associated coronavirus (SARS-CoV) (26, 48), for which the complete genome sequence was first reported in 2003 (23, 32). The genome of SARS-CoV is a single strand of positive-sense RNA 29.7 kb in length. The viral proteins have been classified as “structural proteins” that act at the level of the virion, “nonstructural proteins” (nsp) associated with RNA replication and transcription, and “accessory proteins” that perform functions that are dispensable in cell culture (38). The nsp are of particular interest, since they mediate the replication and processing of the SARS-CoV genome by forming a membrane-associated replicase complex (55). The nsp are initially expressed as two large polyproteins, pp1a and pp1ab, with sizes of about 500 kDa and 800 kDa, respectively (55). The longer form of the polyprotein is expressed via a ribosomal (−1) frameshift event (2). Open reading frames 1a and 1b make up about two-thirds of the SARS-CoV genome, starting from the 5′ end of the viral RNA (43). The polypeptides pp1a and pp1ab are processed by two proteases, the picornavirus 3C-like protease (3CLpro, also known as nsp5) and the papain-like protease (PL2pro, a domain of nsp3), yielding 16 mature nonstructural proteins, nsp1 to nsp16 (29, 38).
Although the SARS outbreak has been contained by public health measures, a vaccine against the virus is still elusive, and the continued search for effective drug treatments is tightly linked to ongoing research on the virus and the proteins associated with it. We determine atomic resolution three-dimensional (3D) structures of proteins encoded by the SARS viral genome to provide a basis for the design of biochemical assays that might unravel some or all of the protein functions and establish structure-function relationships for SARS-CoV proteins. A special focus is on the 213-kDa protein nsp3 (27), which is the largest nonstructural SARS-CoV protein, with 1,922 amino acid residues that correspond to the segment 819 to 2740 of pp1a (GenBank accession number NP_828862; gi:34555776) (44). Based on considerations of phylogenetic conservation and amino-acid-sequence-based secondary structure prediction, SARS-CoV nsp3 has been annotated as a multidomain protein (27) consisting of a minimum of seven domains, nsp3a to nsp3g (27, 38). So far, three SARS-CoV nsp3 domains have been structurally and biochemically characterized: nsp3a (residues 1 to 183) has a ubiquitin-like fold and is an RNA-binding protein with affinity for single-stranded RNA (ssRNA) (37), nsp3b (residues 184 to 351) is a poly(ADP-ribose)-binding protein and has ADP-ribose-1"-phosphatase activity (7, 33), and nsp3d (residues 723 to 1037) contains a ubiquitin-related fold and is a papain-like protease involved in the proteolytic processing of the polyproteins pp1a and pp1ab (30). This paper describes a nuclear magnetic resonance (NMR) structure determination and a preliminary functional annotation for part of the region described as the “SARS-unique domain” (SUD), nsp3c.
Nsp3c, which is the polypeptide segment of the nsp3 residues 366 to 722, has been termed the SUD to reflect its apparent uniqueness to the SARS-CoV (38). From previous work, there have been indications that the SUD may comprise more than one structural domain (4, 42), and nucleic acid-binding activity has been attributed to the carboxy-terminal region of SUD, which is conserved among several bat coronaviruses (27, 42, 51). In this paper, we describe the structure of a globular domain, SUD-M, in the center of the SUD, which has been shown to fold independently and has long-term stability in aqueous solution (4). This sequence segment has less than 30% amino acid identity with known proteins, except for the corresponding polypeptide segments in SARS-like and HKU3-like bat coronaviruses (27), which have more than 90% sequence identity but for which no 3D structures have as yet been determined. It is, however, of interest that SARS-CoV SUD-M shows 16 to 28% sequence identity with homologous regions in group IIc and group IId bat coronaviruses, as such an identity, although small, might indicate an evolutionary development of the SUD. The NMR structure determination of SUD-M residues 513 to 651 [SUD-M(513-651)] now reveals that this polypeptide forms a globular domain of residues 528 to 648, which is flanked by a flexibly extended N-terminal tail of residues 513 to 527 and a C-terminal flexible tail of residues 649 to 651. To investigate possible effects of the unstructured N-terminal tail on the globular domain, we then also determined the NMR structure of the construct SUD-M(527-651). A search of the Protein Data Bank (PDB) for folds homologous to SUD-M and NMR screening of likely reaction partners of SUD-M were then performed for an initial functional annotation.
MATERIALS AND METHODS
Protein preparation.
The preparation of the SUD-M(513-651) protein was described previously (4). The construct encoding SUD-M(527-651) was expressed in Escherichia coli strain BL21(DE3) (Stratagene). Vector pET-28b was used, which encodes an N-terminal His6 tag followed by a thrombin cleavage site that leaves the tag-related N-terminal tetrapeptide segment GSHM. Cells were grown at 37°C, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm of 0.8, and then grown for another 3 h at 37°C. The protein purification was done by a procedure similar to that described previously for SUD-M (4) except that the thrombin cleavage used to remove the His6 tag was pursued for 1 h. Isotope labeling was accomplished by growing cultures in minimal medium containing either 1 g/liter of 15NH4Cl as the sole nitrogen source, yielding the uniformly 15N-labeled protein, or 1 g/liter of 15NH4Cl and 4 g/liter of [13C6]-d-glucose (Cambridge Isotope Laboratories), yielding the uniformly 13C,15N-labeled protein. Growth in M9 minimal medium yielded about 20 mg of pure SUD-M(527-651) from 1 liter of culture. In the 550-μl NMR samples, the protein concentration was adjusted to 1.4 mM, since higher concentrations led to precipitation.
NMR data acquisition and chemical shift assignment.
NMR measurements were performed at a temperature of 298 K with Bruker Avance 600, DRX 700, and Avance 800 spectrometers equipped with TXI-HCN-z- or TXI-HCN-xyz-gradient probe heads. The NMR experiments acquired for obtaining the sequence-specific resonance assignments of SUD-M(513-651) were described previously (4). For the SUD-M(527-651) protein, the new automated projection spectroscopy (APSY) technology was used. Four-dimensional APSY-HNCOCA, four-dimensional APSY-HACANH, five-dimensional APSY-CBCACONH, and five-dimensional APSY-HACACONH data sets were recorded and analyzed with the software GAPRO (9, 13, 14). The resulting peak lists were used as input for the software MATCH (45) for automated polypeptide backbone assignments. The side-chain assignments for the nonaromatic residues were based on 3D 15N-resolved [1H,1H]-total correlation spectroscopy (TOCSY) (τm = 60 ms), 3D HC(C)H-TOCSY (35), 3D 15N-resolved [1H,1H]-nuclear Overhauser effect spectroscopy (NOESY) (τm = 60 ms) (41), and 3D 13C-resolved [1H,1H]-NOESY (τm = 60 ms) (24) experiments. The assignment of the aromatic side-chain resonances was based on 3D 13C-resolved [1H,1H]-NOESY (τm = 60 ms) and two-dimensional (2D) [13C,1H]-heteronuclear single quantum coherence spectroscopy (HSQC) experiments (24, 52). Proton chemical shifts were referenced to internal 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt (DSS). The 13C and 15N chemical shifts were referenced indirectly to DSS using the absolute frequency ratios (49).
Steady-state 15N{1H}-nuclear Overhauser enhancements (NOEs) for studies of high-frequency dynamics were measured using transverse relaxation optimized spectroscopy (TROSY)-based experiments (31, 54) with a Bruker Avance 600 spectrometer with a saturation period of 3.0 s and a total interscan delay of 5.0 s.
The interaction of SUD-M(527-651) with ADP-ribose and ssRNA was evaluated by comparison of the 2D [15N,1H]-HSQC spectra of SUD-M(527-651) recorded in the presence and absence of ssRNA or ADP-ribose using the uniformly 15N-labeled protein at a 0.4 mM concentration. The ssRNAs used were the homodecamers of uridine [poly(U10)], guanosine [poly(G10)], and adenosine [poly(A10)].
Structure calculation from the NMR data.
The structure calculations were performed with the software ATNOS/CANDID/DYANA (10-12). The standard protocol of seven cycles of automated NOESY peak picking and NOE cross-peak identification with ATNOS (12), automated NOE assignment with CANDID (11), and structure calculation with the torsion angle dynamics algorithm contained in CYANA (10) were performed. In the second and subsequent cycles, the intermediate protein structure was used as an additional guide for the interpretation of the NOESY spectra (11, 12). Backbone ϕ and ψ dihedral angle constraints derived from the 13Cα chemical shifts were used as supplementary data for the NOE upper distance constraints in the input for the structure calculation (22, 39). The 20 conformers with the lowest residual CYANA target function values obtained from the seventh ATNOS/CANDID/CYANA cycle were energy minimized in a water shell with the program OPALp (17, 21) using the AMBER force field (6). The program MOLMOL (18) was used to analyze the ensemble of 20 energy-minimized conformers. The stereochemical quality of the models was analyzed using the Joint Center for Structural Genomics validation central suite (http://www.jcsg.org) and the PDB validation server (http://deposit.pdb.org/validate).
Enzyme assays.
The NTPase activity was evaluated by monitoring the phosphate released when ATP or GTP was added to SUD-M(527-651) by using an Enzchek assay (Molecular Probes Inc., Eugene, OR) according to the manufacturer's instructions. This assay uses a method described previously by Webb (47), in which the release of inorganic phosphate is monitored by the coupling of the phosphatase reaction with the purine nucleoside phosphorylase conversion of the substrate 2-amino-6-mercapto-7-methyl purine riboside (MESG) to 2-amino-6-mercapto-7-methyl purine and ribose-1-phosphate. MESG has an absorbance maximum of 330 nm, whereas that of the product is 360 nm. The reaction mixture contained 50 mM Tris (pH 7.5), 1 mM MgCl2, 0.1 mM sodium azide, 200 mM MESG, 1 U purine nucleoside phosphorylase, and 5 μM SUD-M(527-651). It was checked for activity by adding variable amounts of ATP or GTP. No phosphate release was detected by monitoring the absorbance at 360 nm.
Bioinformatics.
The following amino acid sequences were used for alignments: transmissible gastroenteritis virus (GenBank accession number NP_840002), bat coronavirus BtCoV-HKU8 (accession number YP_001718611), BtCoV-1B (accession number YP_001718596), BtCoV-HKU2 (accession number YP_001552234), porcine epidemic diarrhea virus (accession number NP_598309), BtCoV-512/2005 (accession number YP_001351683), human coronavirus HCoV-229E (accession number NP_073549), HCoV-NL63 (accession number YP_003766), SARS-CoV (accession number AAP41036), BtCoV-Rm1 (accession number YP_001382397), BtCoV-HKU5 (accession number YP_001039961), BtCoV-HKU9-1 (accession number YP_001039970), and BtCoV-HKU9-3 (accession number ABN10926). Homology searches were carried out using BLASTP 2.2.18+ (40). Alignments were performed using ClustalW 2.0 (19) and displayed using JalView (5). Coronavirus naming and abbreviation follow ICTV conventions where possible or follow the abbreviation proposed in the first publication of each virus otherwise.
Electrophoretic mobility shift assays.
Purified SUD-M(527-651) was mixed with ssRNA substrate in an assay buffer containing either 150 mM NaCl (physiological salt concentration) or 56 mM NaCl (low salt) in addition to 50 mM sodium phosphate at pH 6.5, 7% glycerol, and 4 mM MgCl2. The following custom-synthesized RNA oligomers (Integrated DNA Technologies, Inc., San Diego, CA) were tested: (ACUG)5; the homopolymers A10, A15, C10, and U10; 5′-CCCGAUACCC-3′, which contains the core GAUA sequence that was shown to bind to nsp3a (37); 5′-CUAAACGAAC-3′, which is the leader transcription regulatory sequence from the SARS-CoV genome [TRS(+)]; and 5′-GUUCGUUUAG-3′, which is the leader transcription regulatory sequence from the SARS-CoV antigenome [TRS(−)]. Protein-nucleic acid mixtures were incubated for 45 min either at room temperature or at 37°C and then analyzed by native electrophoresis on precast 6% acrylamide DNA retardation gels (Invitrogen). Nucleic acid was detected by SYBR gold poststain (Invitrogen) and photographed using a UV light source equipped with a digital camera. SYBR gold was rinsed out, and protein was subsequently detected by SYPRO ruby poststain (Invitrogen).
Protein structure accession numbers.
The chemical shifts of SUD-M(527-651) have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 15618. The atomic coordinates of the two ensembles of 20 conformers used to represent the SUD-M(513-651) and SUD-M(527-651) structures have been deposited in the PDB (http://www.rcsb.org/pdb) under accession numbers 2RNK and 2JZD, respectively, and a single representative conformer for each protein (the conformer with the lowest root mean square deviation [RMSD] from the mean coordinates of the ensemble) have been deposited under accession numbers 2JZF and 2JZE, respectively.
RESULTS
NMR structure determination of SUD-M(513-651) and SUD-M(527-651).
The construct SUD-M(513-651) was identified by Edman degradation analysis of a stable 15.5-kDa fragment obtained by spontaneous proteolysis of a polypeptide comprising the residues 451 to 651 of nsp3. The details of this construct optimization were described previously (4).
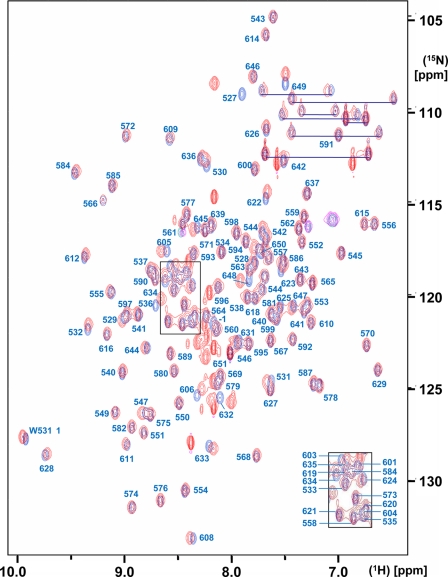
For each of the two proteins, the input for the NMR structure determination consisted of a 3D 15N-resolved [1H,1H]-NOESY spectrum and two 3D 13C-resolved [1H,1H]-NOESY spectra optimized for the aliphatic and the aromatic spectral regions and of the chemical shift lists taken from the previously reported sequence-specific resonance assignments for SUD-M(513-651) (4) and from the presently obtained assignments for SUD-M(527-651) (BioMagResBank accession number 15618). The near identity of the overlapping parts in the two sets of chemical shifts is visualized in Fig. 1 by the [15N,1H]-HSQC spectra of SUD-M(513-651) and SUD-M(527-651).
FIG. 1.
Superposition of the 2D [15N,1H]-HSQC spectra of SUD-M(513-651) (red) and SUD-M(527-651) (blue). The protein concentrations were 1.2 mM and 1.4 mM for SUD-M(513-651) and SUD-M(527-651), respectively. The solvent contained 25 mM sodium phosphate buffer at pH 6.5, 150 mM NaCl, and 2 mM NaN3. The spectra were recorded at a 1H frequency of 600 MHz and a temperature of 25°C, with 256 increments in the 15N dimension and 4 scans/increment. The resonance assignments for SUD-M(527-651) are marked in blue, where the assignments for the crowded central region are shown as an insert in the lower right corner. Residue −1 indicates the methionine residue of the tetrapeptide segment −4GSHM−1 that is left after thrombin cleavage (see the text). The side-chain amide resonances of asparagine and glutamine are connected by blue horizontal lines.
Automated peak picking and NOE assignment by the standalone ATNOS/CANDID program package gave 2,606 and 2,738 meaningful distance restraints for SUD-M(513-651) and SUD-M(527-651), respectively, which represented the core of the input for the CYANA structure calculation (Table 1). Although SUD-M(513-651) is larger than SUD-M(527-651), we observed a slightly larger number of middle-range and long-range NOE restraints for SUD-M(527-651), which is due to the higher-quality NMR spectra obtained for the shorter construct. The total numbers of distance restraints per residue were 23 and 24 for SUD-M(513-651) and SUD-M(527-651), respectively. The residual CYANA target function values, the RMSDs relative to the mean coordinates, and other statistics shown in Table 1 indicate that we achieved high-quality NMR structure determinations for both proteins.
TABLE 1.
Input for NMR structure calculations of the SUD-M(513−651) and SUD-M(527−651) proteins, statistics of the convergence of the CYANA structure calculations, and characterization of the bundle of 20 conformers used to represent the NMR structures
| Parameter | Valuea
|
|
|---|---|---|
| SUD-M(513−651) | SUD-M(527−651) | |
| NOE upper distance limits | 2,606 | 2,738 |
| Intraresidual | 679 | 668 |
| Short range | 648 | 660 |
| Medium range | 528 | 567 |
| Long range | 751 | 843 |
| Dihedral angle constraints | 138 | 142 |
| Avg residual target function value (Å2) ± SD | 2.05 ± 0.48 | 1.52 ± 0.47 |
| Residual NOE violation (Avg ± SD) | ||
| No. > 0.1 Å | 10 ± 4 | 7 ± 3 |
| Maximum (Å) | 0.57 ± 0.20 | 0.56 ± 0.14 |
| Residual dihedral angle violation (Avg ± SD) | ||
| No. > 2.5° | 3 ± 1 | 2 ± 0 |
| Maximum (°) | 62.85 ± 0.91 | 61.62 ± 1.25 |
| Amber energy (kcal/mol) (Avg ± SD) | ||
| Total | −4,982.26 ± 132.41 | −4,653.20 ± 78.81 |
| van der Waals | −414.24 ± 22.99 | −433.64 ± 20.43 |
| Electrostatic | −5,737.30 ± 115.17 | −5,241.40 ± 77.31 |
| RMSD from ideal geometry (Avg ± SD) | ||
| Bond length (Å) | 0.0078 ± 0.0001 | 0.0076 ± 0.0002 |
| Bond angle (°) | 1.988 ± 0.049 | 1.930 ± 0.056 |
| Avg RMSD to the mean coordinates (Å) ± SD (range)b | ||
| bb (residues 528-648) | 0.49 ± 0.08 (0.37-0.67) | 0.49 ± 0.10 (0.35-0.67) |
| ha (residues 528-648) | 0.92 ± 0.07 (0.76-1.04) | 0.93 ± 0.07 (0.83-1.14) |
| Ramachandran plot statistic (%)c | ||
| Most favored regions | 75.8 | 84.4 |
| Additional allowed regions | 20.8 | 14.2 |
| Generously allowed regions | 2.2 | 1.4 |
| Disallowed regions | 1.2 | 0.0 |
The top five entries refer to the 20 CYANA conformers with the lowest residual target function values; the remaining entries refer to the same conformers after energy minimization with OPALp (17, 21). The ranges indicate the minimum and maximum values. Where applicable, the average value for the bundle of 20 conformers and the standard deviations are given; numbers in parentheses indicate the range of values for the given quantity.
bb indicates the backbone atoms N, Cα, and C′; ha stands for “all heavy atoms.” The numbers in parentheses indicate the residues for which the RMSD was calculated.
As determined by PROCHECK (20).
SUD-M(513-651) and SUD-M(527-651) NMR structures.
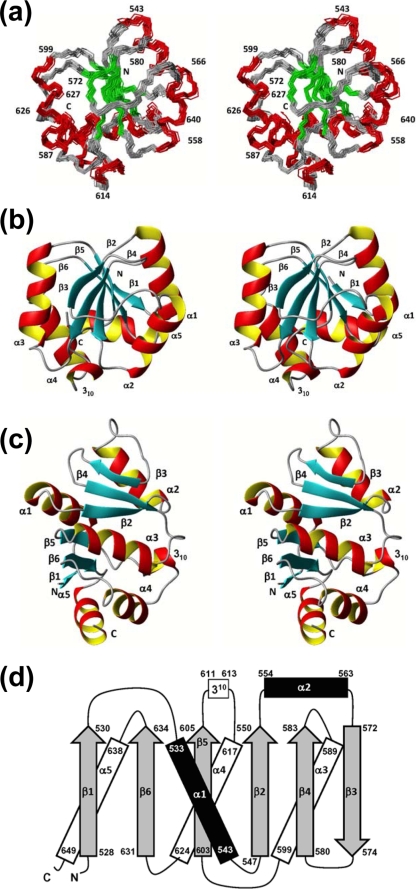
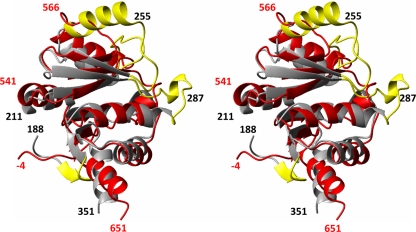
Both proteins exhibit a globular domain involving residues 528 to 648, which form six β-strands, five α-helices, and one 310-helix in the sequential order β1-α1-β2-α2-β3-β4-α3-β5-310-α4-β6-α5 (Fig. 2). The first regular secondary structure element is the short-strand β1 formed by residues 528 to 530, which is in SUD-M(513-651), preceded by a flexible N-terminal tail of residues 513 to 527 (Fig. 3). The lengths of all regular secondary structures are marked in Fig. 2d. It is interesting that the structure contains two well-defined long loops with nonregular secondary structure, containing residues 564 to 571 between helix α2 and strand β3 and residues 606 to 616 between strand β5 and helix α4.
FIG. 2.
NMR structure of SUD-M(527-651). (a) Stereo view of the polypeptide backbone of a bundle of 20 energy-minimized conformers superimposed for the minimal RMSD value of the backbone atoms of residues 528 to 648. The helical regular secondary structures are red, the β-strands are green, and the polypeptide segments with no regular secondary structure are gray. Selected sequence positions are identified by numerals. (b) Stereo view in the same orientation as described above (a), of a ribbon presentation of the closest conformer of SUD-M(527-651) to the mean coordinates of the bundle above (a). The regular secondary structures are identified. (c) Same as above (b) after a 90° rotation about a horizontal axis. (d) Topology of the regular secondary structures in SUD-M(527-651). β-Strands are shown as gray arrows, helices in the front of the β-sheet are in black, and helices behind the β-sheet are represented by white rectangles. The numbers represent the starts and the ends of the individual regular secondary structure elements.
FIG. 3.
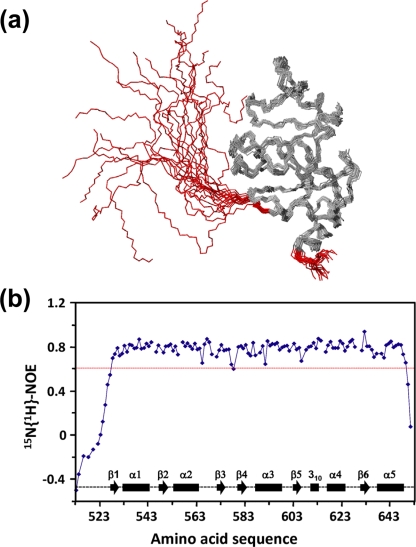
(a) NMR structure of SUD-M(513-651). The polypeptide backbone of a bundle of 20 energy-minimized conformers has been superimposed for the minimal RMSD value calculated for the backbone atoms of residues 528 to 648. The flexibly extended N-terminal tail of residues 513 to 527 and the C-terminal flexible tail of residues 649 to 651 are red. (b) Relative 15N{1H}-NOE intensities plotted versus the sequence of SUD-M(513-651). Diamonds represent the experimental measurements, which are linked in sequential order by straight lines. Gaps represent either proline residues or residues for which the 15N-1H correlation peak could not be integrated because of spectral overlap. The experiment was recorded at a 1H frequency of 600 MHz using a saturation period of 3.0 s and a total interscan delay of 5.0 s. The red line represents a cutoff at 0.6; residues with values below this cutoff value are identified as having high-frequency intramolecular mobility. Positions of the regular secondary structures are indicated at the bottom of the figure.
Superposition of the mean coordinates of the 3D structures of SUD-M(513-651) and SUD-M(527-651) yielded a backbone RMSD of 0.78 Å for residues 528 to 648, which form the globular domain of the protein. Since the flexible region of residues 513 to 527 has no apparent contact with the globular domain, the RMSD calculated for residues 528 to 648 between the independently determined structures of SUD-M(513-651) and SUD-M(527-651) provides a meaningful estimate for the accuracy of the solution NMR structure determination.
Internal mobility in SUD-M(513-651).
The intramolecular flexibility on the subnanosecond time scale was characterized for the polypeptide backbone of SUD-M(513-651) by 15N{1H}-NOE measurements (Fig. 3b). For residues 528 to 648, positive NOE values of 0.6 or larger show that the mobility of the backbone 15N-1H moieties is essentially limited to the overall tumbling of the molecule. For residues 513 to 527 and 649 to 651, NOE values in the range of −0.6 to 0.5 indicate increased high-frequency mobility. These results were interpreted to indicate that the central SUD-M region with residues 528 to 648 forms a compact globular domain with flexibly extended polypeptide segments attached at both chain ends.
SUD-M(527-651) is an RNA-binding protein with affinity for purine bases.
As part of a search for a functional annotation of SUD-M(527-651), we performed NMR chemical shift perturbation experiments by comparing the [15N,1H]-HSQC spectrum of SUD-M(527-651) in the absence of potential ligands to that in the presence of potential ligands, such as ssRNA, ATP, GTP, and ADP-ribose. The motivation for choosing ssRNAs came from recent studies by Neuman et al. (27) and Tan et al. (42), which showed that the SUD binds RNA. Nucleoside triphosphates (NTPs) and ADP-ribose were selected based on the observation of structural homology between SUD-M and various NTPases, with the closest structural similarity to SARS-CoV nsp3b, which displays ADP-ribose-binding activity (7, 33).
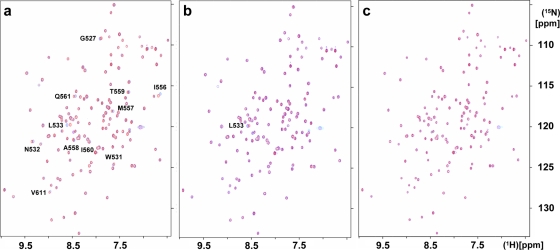
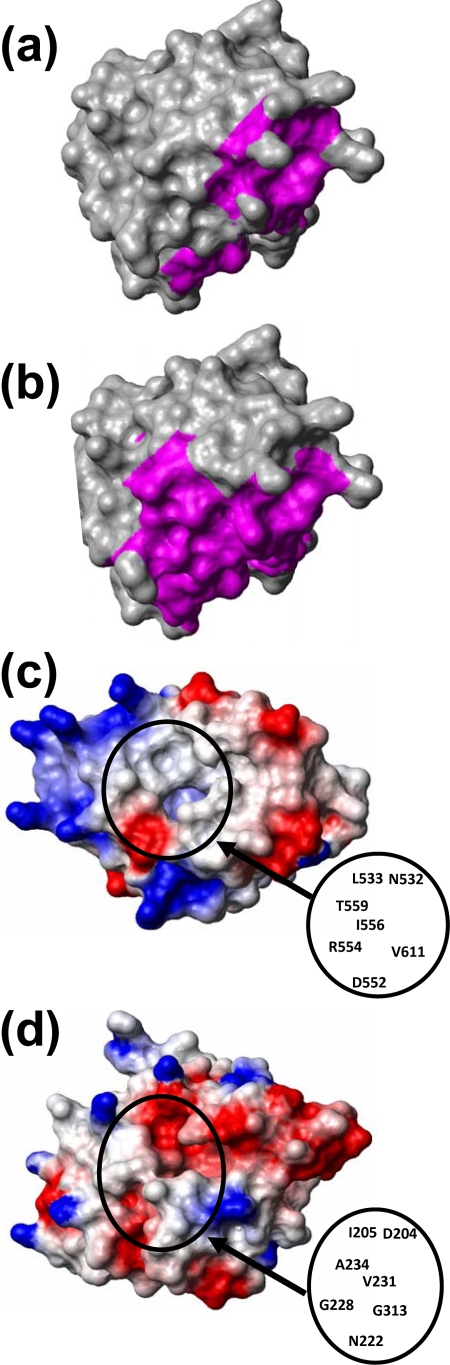
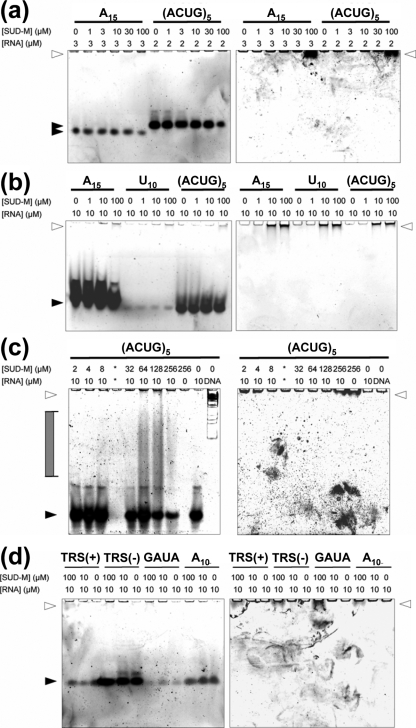
The addition of the ssRNA poly(G10) led to extensive precipitation of the protein, which may be rationalized by the fact that poly(G) ssRNA is much less water soluble than single-stranded poly(A) or poly(U). The addition of poly(U10) had a measurable effect only on the chemical shifts of residue L533 (Fig. 4b). The addition of poly(A10) resulted in significant shifts of 11 peaks corresponding to the residues G527, W531 to L533, I556 to Q561, and V611 (Fig. 4a). These residues are marked by green lines above the sequence in Fig. 8 and highlighted in magenta in the space-filling model of the structure shown in Fig. 5a. It is seen that all the perturbed residues are located at or near a putative ligand-binding cleft (see also below), with residues N532, L533, I556, T559, and V611 within the cleft and residues M557, A558, I560, and Q561 in helix α2, adjacent to the cleft. None of the 15N-1H correlation peaks in the HSQC spectrum of SUD-M(527-651) showed significant chemical shift changes upon the addition of ATP, GTP, or ADP-ribose (Fig. 4c). From these data, we conclude that SUD-M(527-651) is a poly(A) ssRNA-binding protein and does not bind either NTPs or ADP-ribose.
FIG. 4.
Superposition of pairs of 2D [15N,1H]-HSQC spectra of 0.4 mM SUD-M(527-651) (solvent composed of 25 mM sodium phosphate buffer at pH 6.5, 150 mM NaCl, and 2 mM NaN3) recorded in the absence (red peaks) and presence (blue peaks) of 0.4 mM of three different ligands: poly(A10) ssRNA (a), poly(U10) ssRNA (b), and ADP-ribose (c). (a and b) The peaks that show chemical shift changes after the addition of the ligand are identified. The spectra were recorded on a Bruker DRX 700 spectrometer with a 1.7-mm TXI HCN z-gradient probehead at a temperature of 25°C; 256 increments in the 15N dimension were accumulated, with 32 scans per increment.
FIG. 8.
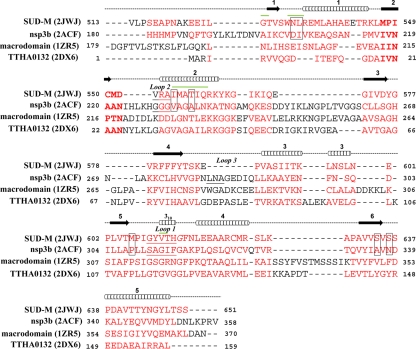
3D structure-based sequence alignment of the SUD-M protein with its closest structural homologues, as identified through a DALI (16) search of the PDB with SUD-M(527-651). PDB accession numbers are given in parentheses. Above the sequence, the locations of the regular secondary structures in SUD-M are indicated by cylinders for helices and by arrows for β-strands. The aligned residues are highlighted in red, and conserved sequence motifs described in the text are indicated in boldface type. The residues that form the adenosine-binding cleft in nsp3b and the corresponding residues in SUD-M are boxed. The loops that form the ribose-binding cleft in nsp3b (see the text) and the corresponding regions in SUD-M are underlined (SUD-M has a three-residue deletion at the location corresponding to loop 3). The residues that show chemical shift changes upon the addition of poly(A10) ssRNA (see the text and Fig. 4a) are marked by green lines above the sequence.
FIG. 5.
Space-filling models of the NMR structure of SUD-M(527-651). (a) Regions affected by poly(A10) ssRNA binding (data from Fig. 4a) are highlighted in magenta. (b) The residues in positions structurally corresponding to those that contact the ADP-ribose ligand in nsp3b are highlighted. (c) Display of the electrostatic surface potential, with positive and negative electrostatic charges represented in blue and red, respectively. (d) Nsp3b (PDB accession number 2ACF). Shown is the same presentation of the electrostatic surface potential as that in panel c. (c and d) The black circle surrounds the ligand-binding clefts discussed in the text. Selected residues within the cleft of SUD-M(527-651) and in the active site of nsp3b are identified.
In order to extend these initial observations on RNA binding by SUD-M(527-651) to a larger array of potential RNA substrates (see Materials and Methods), we performed electrophoretic mobility shift assay experiments. Figure 6 shows the results for the binding of SUD-M(527-651) to single-stranded poly(A15), poly(U10), poly(A10), (ACUG)5, TRS(+), TRS(−), and the nsp3a-binding sequence 5′-CCCGAUACCC-3′ (GAUA). Weak binding was evidenced for poly(A15) and (ACUG)5 by the reduced intensity of the RNA band at higher protein concentrations (Fig. 6a and b). This result corroborates our NMR observations that SUD-M(527-651) binds poly(A) ssRNA (Fig. 4a). Moreover, the absence of an observable effect of increasing concentrations of SUD-M(527-651) on the poly(U10) band indicates that there is at most minimal binding of this pyrimidine ssRNA (Fig. 6b), which again corroborates the NMR observation (Fig. 4b). Similarly, we could not detect binding to poly(C10) (data not shown). In addition, we observed that SUD-M binds weakly to TRS(−) ssRNA, but there was no detectable binding to the TRS(+) ssRNA or to GAUA (Fig. 6d). Increased binding was observed for (ACUG)5 and poly(A15) when the incubation was conducted at 37°C (Fig. 6b), compared to the incubation at 25°C (Fig. 6a). In Fig. 6c, we further present evidence for the binding of SUD-M(527-651) to (ACUG)5 at physiological salt concentrations (150 mM NaCl). The last two experiments suggest that the binding of SUD-M to ssRNA also prevails under physiological conditions of temperature and ionic strength.
FIG. 6.
Investigation of RNA binding by electrophoretic mobility shift assay (see the text). Data are given for poly(A15), poly(U10), poly(A10), (ACUG)5, TRS(+), TRS(-), and 5′-CCCGAUACCC-3′ (GAUA). These single-stranded oligonucleotides were incubated with various concentrations of SUD-M either at room temperature (a and d) or at 37°C (b and c) before analysis by native polyacrylamide gel electrophoresis. Lane designations indicate the final concentration of protein and RNA or the presence of a double-stranded DNA marker (DNA). The binding assays in a and b were carried out in low-salt buffer (50 mM phosphate at pH 6.5 containing 56 mM NaCl, 7% glycerol, and 4 mM MgCl2), and those in c and d were carried out using buffer containing physiological salt concentrations (50 mM phosphate at pH 6.5 with 150 mM NaCl, 7% glycerol, and 4 mM MgCl2). Nucleic acid was detected by SYBR gold staining (left), and protein was detected by SYPRO ruby staining (right). White arrowheads indicate the electrophoretic mobility of SUD-M, and black arrowheads indicate free nucleic acid. Complexes of intermediate mobility are indicated by a gray filled bracket.
DISCUSSION
The NMR structure determinations of the two constructs SUD-M(513-651) and SUD-M(527-651) showed that the central part of the SUD forms a self-folding globular domain, which is flanked by two flexibly extended polypeptide segments. It has further been shown that the C-terminally adjoining polypeptide segment of the SUD forms another independently folding globular domain (M. Johnson et al., unpublished data). In view of these structural data, we assume as a working hypothesis that the isolated SUD-M(527-651) globular domain is also an independent functional domain, leaving open that it might function in concert with other proteins, either from SARS-CoV or from the host organism. In the context of possible intra-SARS-CoV concerted multidomain functionality, it is of interest that at least three among the seven initially annotated SARS-CoV nsp3 domains exhibit RNA-binding activity, i.e., nsp3a (37), the SUD (nsp3c) (42), and nsp3e (27). This would be compatible with a role of either or all of these proteins in viral replication. In this section, we describe a search for possible further leads to the function of SUD-M based on homology considerations with structurally related proteins.
SUD-M(527-651) forms a macrodomain fold.
In a search of the PDB for proteins with 3D structural similarity to SUD-M(527-651), the program DALI (15, 16) identified more than 300 structures with a z score larger than 2.0, which is a value that indicates “overall fold similarity” (16). The closest match was found for macrodomains, and DALI z scores of ≥5 were also obtained for various helicases and NTP-binding proteins (Table 2). As a first result from our homology studies, we thus found that the polypeptide fold of SUD-M(527-651) corresponds to a macrodomain fold (1, 3, 25): the six β-strands in the arrangement 165243 form the protein core, whereby the third β-strand is oriented antiparallel to the other strands (25) and the α-helices form an outer layer of the protein architecture (Fig. 2b and c).
TABLE 2.
Protein structures with z values of ≥5 identified by a DALI search of the PDB with SUD-M(527−651)
| PDB accession no.a | z scoreb | RMSDc | % Iseqd | Descriptione |
|---|---|---|---|---|
| 2ACF | 10.2 | 2.9 | 5 | ADP-ribose-1"-phosphatase from SARS-CoV (nsp3b)* |
| 2DX6 | 9.8 | 2.5 | 11 | Conserved hypothetical protein; TTHA0132 from Thermus thermophilus HB8* |
| 1YD9 | 9.4 | 2.9 | 10 | Structural protein; nonhistone domain of the histone variant macroH2A1.1 of human macrodomain* |
| 1SPV | 8.8 | 2.9 | 11 | Putative phosphatase of E. coli* |
| 1GYT | 8.7 | 2.6 | 11 | Aminopeptidase A from E. coli |
| 1VHU | 8.6 | 3.1 | 7 | Putative phosphoesterase; hypothetical protein AF1521 from Archaeoglobus fulgidus* |
| 1ZR5 | 8.4 | 3.1 | 8 | Gene regulation; macrodomain of human core histone variant macroH2A1.2, residues 161-372, forms A and B* |
| 2JYC | 7.8 | 2.8 | 11 | Human protein C6orf130; putative macrodomain* |
| 2FG1 | 7.7 | 2.7 | 11 | Hypothetical protein BT1257 from Bacteroides thetaiotaomicron |
| 1NLF | 5.9 | 3.6 | 11 | Human interleukin-19; helical cytokine |
| 1OYW | 5.2 | 3.4 | 8 | ATP-dependent DNA helicase; E. coli RecQ catalytic core |
| 1W4R | 5.1 | 3.0 | 3 | Type II thymidine kinase from Homo sapiens |
| 2Q6T | 5.0 | 3.3 | 8 | Hydrolase; Thermus aquaticus DnaB helicase monomer |
PDB accession numbers from http://www.rcsb.org/pdb.
The z score gives a quantitative measure for structural similarity and is defined in terms of equivalent intramolecular distances (16).
RMSD calculated for the Cα atoms of residues in structurally equivalent positions of SUD-M(527−651).
% Iseq, Percentage of sequence identity for the aligned residues with SUD-M(527−651).
Proteins labeled with an asterisk form a macrodomain fold.
3D structure homology of SUD-M(527-651) with SARS-CoV nsp3b.
SUD-M(527-651) has a DALI z score of 10.2 with the crystal structure of SARS-CoV nsp3b (PDB accession number 2ACF), which comprises residues 184 to 365 of nsp3 and is thus located immediately N terminal to the SUD domain. Figure 7 shows a superposition of the NMR solution structure of SUD-M(527-651) with the X-ray crystal structure of nsp3b. Compared to SUD-M(527-651), nsp3b has an extra β-strand at the N terminus, an α-helix inserted between helix α2 and strand β3, and a 310-helix inserted between strands β4 and α3. These differences are highlighted in Fig. 7 with yellow coloring of the sequence insertions in nsp3b (Fig. 8) that have no matching residues in SUD-M(527-651). The close 3D structure homology visualized in Fig. 7 is remarkable considering that the sequence homology in the matching segments amounts to only 5% (Fig. 8).
FIG. 7.
Stereo view of a ribbon presentation showing a superposition of the NMR structure of SUD-M(527-651) (red) and the X-ray structure of nsp3b (33) (gray). The following residues for the superposition were identified with the software DALI (15), yielding an RMSD value of 2.9 Å for the Cα atoms of these residues: residues 527 to 541, 544 to 552, 553 to 556, 557 to 566, 567 to 570, 572 to 575, 576 to 579, 580 to 586, 588 to 597, 599 to 625, and 626 to 649 in SUD-M(527-651) and residues 199 to 213, 214 to 222, 227 to 230, 232 to 241, 244 to 247, 263 to 266, 268 to 271, 274 to 280, 290 to 299, 300 to 326, and 328 to 351 in nsp3b. The insertions in the sequence of nsp3b are highlighted in yellow (see the text). Selected sequence positions are identified by black numerals for nsp3b and red numerals for SUD-M(527-651).
Evolution of tandem macrodomains in coronavirus nsp3.
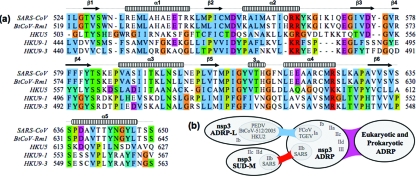
After the unexpected finding of two macrodomains located in the SARS-CoV nsp3b-nsp3c region, we scanned the corresponding regions of other coronavirus nsp3's for further evidence of macrodomain homology. In addition to the conserved ADP-ribose-1"-phosphatase homologs, two genetic clusters related to validated macrodomains were identified: a group of SUD-M-like domains was identified in nsp3 of the BtCoV-HKU5 and BtCoV-HKU9 lineages (Fig. 9a), and a second group of ADP-ribose-1"-phosphatase-like domains was identified in nsp3 of viruses related to HCoV-229E and BtCoV-HKU2 (Fig. 9b) (amino acids 1444 to 1609 in 229E pp1a under GenBank accession number NP_073549). The group I ADP-ribose-1"-phosphatase-like domain family differs from both ADP-ribose-1"-phosphatase and SUD-M proteins at the sites predicted to form the substrate-binding pocket, suggesting that a different substrate may be bound. If these findings are taken in the context of the duplicate ubiquitin-related and papain-related domains of coronavirus nsp3 (27) and also in the context of the evidence for variable numbers of short direct amino acid repeats near the amino terminus of HCoV-HKU1 nsp3 (50), it becomes apparent that the sequence of the amino-terminal half of nsp3 has been shaped by ancestral sequence duplication events. Therefore, while we are unable to rule out hypotheses of convergent or divergent evolution in nsp3 per se, we favor the explanation that SUD-M diverged from a duplicated SARS-CoV ADP-ribose-1"-phosphatase domain.
FIG. 9.
Conservation of SUD-M in bat coronavirus lineages. (a) Multiple-sequence alignment of domains homologous to SARS-CoV SUD-M. Homologies are highlighted with clustalx conservation coloring, and sequences are numbered from the first residue of nsp3. (b) Schematic representation of the homology between macrodomains found in coronavirus nsp3 and eukaryotic and prokaryotic organisms. Genetic homology (BLAST) (blue), structural homology (DALI) (red), and combined homology (violet) are indicated. Coronavirus subgroup nomenclature was taken from http://www.ncbi.nlm.nih.gov/Taxonomy/Browser. PEDV, porcine epidemic diarrhea virus; TGEV, transmissible gastroenteritis virus; FCoV, feline coronavirus.
Putative substrate-binding site in SUD-M(527-651).
Crystal structures are available for nsp3b in the free form (33) and in a complex with the substrate ADP-ribose (7). Starting from the thus unambiguously identified nsp3b substrate-binding site, examination of the corresponding surface region in SUD-M(527-657) revealed that the SUD contains a cleft in a homologous location (Fig. 5c), which seemed worthy of further investigation as a candidate for a ligand-binding site. In nsp3b, an important polypeptide sequence in the active site is 217IVNAAN222 (Fig. 8), which corresponds to the characteristic “hhNAAN” motif (where “h” can be any hydrophobic residue) of macrodomains (7, 33). The C-terminal asparagine residue at position 222 in this motif (Fig. 8) plays a pivotal role in the ADP-ribose-1"-phosphatase activity of nsp3b, where it is involved in hydrogen bonding to a water molecule and thus assists the nucleophilic attack on the phosphate group of the ADP-ribose-1"-monophosphate (7). Egloff et al. (7) previously found that the replacement of this asparagine residue with alanine abrogated the phosphatase activity. The residues homologous to this motif in the 3D structure of SUD-M are 547MPICMD552 (Fig. 8). Thus, there is a coincidence with nsp3b only for the first two hydrophobic residues, and the position of the key catalytic residue N222 in nsp3b is occupied by D552 in SUD-M.
In the complex with nsp3b, the adenosine moiety of ADPR is located in a cleft surrounded by residues D204, I205, V231, A234, P307, A336, and N338 (7). The corresponding residues in SUD-M (marked by boxes in Fig. 8) lead to a quite different surface topology than that of the corresponding area in nsp3b. It was further observed that the ribose-binding site adjacent to the adenosine-binding area in the catalytic center of nsp3b is surrounded by three loops, with residues 310SAGIF314, 228GGG230, and 282LNA284 (underlined in Fig. 8). These three loops form a groove that accommodates the ribose moiety. In SUD-M, the residues corresponding to the first two loops are 609GYVTH613 and 553VRA555, and the third loop is deleted (Fig. 8), which again contributes to differences in the protein surface topology compared to that of nsp3b. Furthermore, in nsp3b, the hexapeptide segment consists of the second loop and the following residues: 228GGGVAG233, which is reminiscent of a Walker A motif (46), and GX4GK[T/S] (X can be any residue), which forms the NTP-binding site in P-loop NTPases (34). In SUD-M, it is the residues 609GYVTHG614 and 548PICMD552 that are reminiscent of Walker A and Walker B motifs (46), but although these sequence motifs are part of the presently discussed putative SUD-M active site, they are not in the same relative positions as in P-loop NTPases (34).
Overall, it is interesting to note that the comparisons with the well-characterized nsp3b and its substrate complex revealed the presence of potentially functional sequence motifs in SUD-M(527-651), although these potentially functional elements are not properly arranged in the 3D structure to confer nsp3b-like enzymatic activity to the SUD. The outcome of this part of the homology investigations is fully compatible with data for the experimental functional assays, which showed that SUD-M does not have either NTPase activity or affinity for the binding of ADP-ribose.
3D structure homology of SUD-M(527-651) with non-SARS-CoV proteins.
Although SARS-CoV nsp3b is its closest structural homologue, SUD-M also shows significant similarity to other classes of NTP-binding proteins. Thus, for example, comparison with the hepatitis C virus helicase (PDB accession number 1HEI) (53) yielded a DALI z score of 3.5 and revealed similarity to the catalytic domain of the helicase. However, a comparison of the sequences and the 3D structures of the two proteins shows that SUD-M lacks the characteristic “DEXH” (where X can be any residue) helicase sequence (36). Similar conclusions resulted from comparisons with other proteins so that a putative functional assignment for SUD-M remains elusive also on the basis of comparisons with non-SARS-CoV proteins.
Progress with the structural coverage of nsp3.
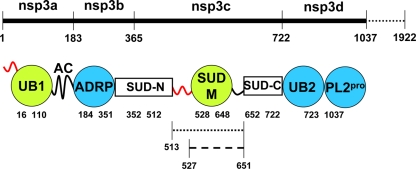
The data in this paper are yet another step toward a complete structural characterization of SARS-CoV nsp3, which was initially annotated as consisting of seven domains, nsp3a to nsp3g (27, 38). Figure 10 summarizes the current structural coverage of the N-terminal half of nsp3, which includes globular domain structures solved by NMR and by X-ray crystallography, as well as flexibly disordered linker segments characterized by NMR 15N{1H}-NOE measurements. Two white boxes in Fig. 10 indicate polypeptide segments with unknown folds that are currently under investigation.
FIG. 10.
Structural coverage of the N-terminal half of the 1,922-residue nsp3. Initially annotated domains are marked above the thick line, and the numbers below this line represent the residues that bound the individual domains. Circles indicate globular folds, with the NMR structures in green and the crystal structures in blue. The curved thick lines represent flexibly disordered segments that were characterized by NMR 15N{1H}-NOE measurements either as an N-terminal attachment of the nearest globular domain (red) or as a C-terminal tail (black). The white boxes indicate domains that are currently being investigated by NMR. The residues binding the individual structural entities are indicated below the circles and the boxes. The dotted line represents SUD-M(513-651), and the broken line represents SUD-M(527-651). UB1 and UB2 are ubiquitin-like folds. AC is a region rich in acidic residues. ADRP is an ADP-ribose-1"-phosphatase. SUD-N (MBD) (27), SUD-M, and SUD-C represent three structural regions of the SARS-unique domain. PL2pro is a papain-like protease.
Although the structural data surveyed in Fig. 10 have so far resulted in functional annotations of only two of the nsp3 domains (see above), they further provide a foundation for hypotheses on additional functional annotations. Thus, it was previously observed that the globular domain of nsp3a (UB1) and the ubiquitin-like domain of nsp3d (UB2) exhibit structural similarity (37). Combined with the present finding that SUD-M is structurally similar to nsp3b, this may be interpreted as an indication that the nsp3 is composed of small “cassettes” with structure similarity but low sequence homology. Recent work by Oostra et al. (28) showed that nsp3 has two transmembrane domains, and hence a cytosolic location is expected for the SARS-CoV nsp3a-to-nsp3e and nsp3g regions. This topology would place the bulk of nsp3 on the same side of the membrane as all of the viral replicase enzymes and proteinase cleavage sites (28). SUD-M is separated from the first predicted transmembrane region, which begins at residue 1319 (27), by 668 amino acids, which comprise the globular domains SUD-C (Johnson et al., unpublished), PLpro (two domains) (30), and nsp3e (P. Serrano et al., unpublished data) and one uncharacterized domain (residues 1226 to 1318, or G2M [27]). With the transmembrane regions of nsp3 functioning as an anchor to the double-membrane vesicles, the nsp3 domains are nonetheless likely to participate in the assembly of the membrane-associated replicase complex. This complex is thought to involve many of the other nonstructural proteins, such as the RNA-dependent RNA polymerase, the helicase, exo- and endonucleases, and other membrane proteins of currently unknown function, such as nsp4 and nsp6. The observation of multiple different RNA-binding activities in the N-terminal region of nsp3 (27, 37, 42; Serrano et al., unpublished; Johnson et al., unpublished) actually suggests such an involvement in the replicase complex. The modular construction and the two pairwise structure homologies seen within the N-terminal region of nsp3 (nsp3b and SUD-M, and UB1 and UB2) might facilitate the proper assembly of this complex, where each domain would contribute its specific activity toward the full functional repertoire of nsp3. With respect to functions within the replicase complex, the observation of polyadenosine ssRNA binding to SUD-M(527-651) is of particular interest, considering that coronavirus replication is initiated by a base-pair-scanning step involving the poly(A) tail (8, 56). There is thus an indication that SUD-M might be involved in an initiation step involving binding to the poly(A) tail.
Acknowledgments
This study was supported by NIAID/NIH contract no. HHSN266200400058C (Functional and Structural Proteomics of the SARS-CoV) and by the Joint Center for Structural Genomics through NIH/NIGMS grant no. U54-GM074898. Additional support was obtained for M.A.J., P.S., and B.P. through fellowships from the Canadian Institutes of Health Research, the Spanish Ministry of Science and Education, and the Swiss National Science Foundation (PA00A-109047/1), respectively, and by the Skaggs Institute for Chemical Biology.
K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at the Scripps Research Institute.
This is the Scripps Research Institute manuscript 19411.
Footnotes
Published ahead of print on 3 December 2008.
REFERENCES
- 1.Allen, M. D., A. M. Buckle, S. C. Cordell, J. Löwe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330503-511. [DOI] [PubMed] [Google Scholar]
- 2.Brierly, I., and F. J. Dos Ramos. 2006. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 11929-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarthy, S., S. K. Gundimella, C. Caron, P. Y. Perche, J. R. Pehrson, S. Khochbin, and K. Luger. 2005. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 257616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee, A., M. A. Johnson, P. Serrano, B. Pedrini, and K. Wüthrich. 2007. NMR assignment of the domain 513-651 from the SARS-CoV nonstructural protein nsp3. Biomol. NMR Assign. 1191-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java Alignment Editor. Bioinformatics 20426-427. [DOI] [PubMed] [Google Scholar]
- 6.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, J. Merz, K. M. Ferguson, D. M. Ferguson, D. C. Spellmyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1175179-5197. [Google Scholar]
- 7.Egloff, M.-P., H. Malet, A. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 808493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enjuanes, L., F. Almazán, I. Sola, and S. Zuñiga. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 60211-230. [DOI] [PubMed] [Google Scholar]
- 9.Fiorito, F., S. Hiller, G. Wider, and K. Wüthrich. 2006. Automated resonance assignment of proteins: 6D APSY-NMR. J. Biomol. NMR 3527-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Güntert, P., C. Mumenthaler, and K. Wüthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273283-298. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann, T., P. Güntert, and K. Wüthrich. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319209-227. [DOI] [PubMed] [Google Scholar]
- 12.Herrmann, T., P. Güntert, and K. Wüthrich. 2002. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24171-189. [DOI] [PubMed] [Google Scholar]
- 13.Hiller, S. 2006. NMR methods for studies of folded and unfolded forms of globular proteins. ETH Diss. no. 16729. Ph.D. dissertation. Swiss Federal Institute of Technology, Zurich, Switzerland.
- 14.Hiller, S., F. Fiorito, K. Wüthrich, and G. Wider. 2005. Automated projection spectroscopy (APSY). Proc. Natl. Acad. Sci. USA 10210876-10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233123-138. [DOI] [PubMed] [Google Scholar]
- 16.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20478-480. [DOI] [PubMed] [Google Scholar]
- 17.Koradi, R., M. Billeter, and P. Güntert. 2000. Point-centered domain decomposition for parallel molecular dynamics simulation. Comp. Phys. Commun. 124139-147. [Google Scholar]
- 18.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1451-55. [DOI] [PubMed] [Google Scholar]
- 19.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. ClustalW and ClustalX version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 21.Luginbühl, P., P. Güntert, M. Billeter, and K. Wüthrich. 1996. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR 8136-146. [DOI] [PubMed] [Google Scholar]
- 22.Luginbühl, P., T. Tszyperski, and K. Wüthrich. 1995. Statistical basis for the use of 13Cα chemical shifts in protein structure determination. J. Magn. Reson. 109229-233. [Google Scholar]
- 23.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 3001399-1404. [DOI] [PubMed] [Google Scholar]
- 24.Muhandiram, D. R., N. A. Farrow, G.-Y. Xu, S. H. Smallcombe, and L. E. Kay. 1993. A gradient 13C NOESY-HSQC experiment for recording NOESY spectra of 13C-labeled proteins dissolved in H2O. J. Magn. Reson. B 102317-321. [Google Scholar]
- 25.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247536-540. [DOI] [PubMed] [Google Scholar]
- 26.Navas-Martin, S. R., and S. Weiss. 2004. Coronavirus replication and pathogenesis: implications for the recent outbreak of severe acute respiratory syndrome (SARS), and the challenge for vaccine development. J. Neurovirol. 1075-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuman, B. W., J. S. Joseph, K. S. Saikatendu, P. Serrano, A. Chatterjee, M. A. Johnson, L. Liao, J. P. Klaus, J. R Yates III, K. Wüthrich, R. C. Stevens, M. J. Buchmeier, and P. Kuhn. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 825279-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oostra, M., M. C. Hagemeijer, M. van Gent, C. P. Bekker, E. G. te Lintelo, P. J. M. Rottier, and C. A. de Haan. 8 October 2008. Topology and membrane anchoring of the coronavirus replication complex: not all of the hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed]
- 29.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 789977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 1035717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renner, C., M. Schleicher, L. Moroder, and T. A. Holak. 2002. Practical aspects of the 2D 15N-[1H]-NOE experiment. J. Biomol. NMR 2323-33. [DOI] [PubMed] [Google Scholar]
- 32.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 3001394-1399. [DOI] [PubMed] [Google Scholar]
- 33.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure 131665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15430-434. [DOI] [PubMed] [Google Scholar]
- 35.Sattler, M., J. Schleucher, and C. Griesinger. 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magnet. Reson. Spectrosc. 3493-158. [Google Scholar]
- 36.Schmid, S. R., and P. Linder. 1992. D-E-A-D protein family of putative RNA helicases. Mol. Microbiol. 6283-291. [DOI] [PubMed] [Google Scholar]
- 37.Serrano, P., M. A. Johnson, M. S. Almeida, R. Horst, T. Herrmann, J. S. Joseph, B. W. Neuman, V. Subramanian, K. S. Saikatendu, M. J. Buchmeier, R. C. Stevens, P. Kuhn, and K. Wüthrich. 2007. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 8112049-12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spera, S., and A. Bax. 1991. Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 1135490-5492. [Google Scholar]
- 40.Stephen, F. A., L. M. Thomas, A. S. Alejandro, Z. Jinghui, Z. Zheng, M. Webb, and J. L. David. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talluri, S., and G. Wagner. 1996. An optimized 3D NOESY-HSQC. J. Magn. Reson. B 112200-205. [DOI] [PubMed] [Google Scholar]
- 42.Tan, J., Y. Kusov, D. Mutschall, S. Tech, K. Nagarajan, R. Hilgenfeld, and C. L. Schmidt. 2007. The “SARS-unique domain” (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem. Biophys. Res. Commun. 364877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 756676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 842305-2315. [DOI] [PubMed] [Google Scholar]
- 45.Volk, J., T. Herrmann, and K. Wüthrich. 2008. Automated sequence-specific protein NMR assignment using the memetic algorithm MATCH. J. Biomol. NMR 41127-138. [DOI] [PubMed] [Google Scholar]
- 46.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webb, M. R. 1992. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. USA 894884-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wishart, D. S., C. G. Bigam, J. Yao, F. Abildgaard, H. J. Dyson, E. Oldfield, J. L. Markley, and B. D. Sykes. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6135-140. [DOI] [PubMed] [Google Scholar]
- 50.Woo, P. C., S. K. Lau, C. C. Yip, Y. Huang, H. W. Tsoi, K. H. Chan, and K. Y. Yuen. 2006. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 807136-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo, P. C., M. Wang, S. K. Lau, H. Xu, R. W. Poon, R. Guo, Wong, B. H., K. Gao, H. W. Tsoi, Y. Huang, K. S. Li, C. S. Lam, K. H. Chan, B. J. Zheng, and K. Y. Yuen. 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 811574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York, NY.
- 53.Yao, N., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4463-467. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, G., Y. Xia, L. K. Nicholson, and K. H. Sze. 2000. Protein dynamics measurements by TROSY-based NMR experiments. J. Magn. Reson. 143423-426. [DOI] [PubMed] [Google Scholar]
- 55.Ziebuhr, J. 2004. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 7412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuñiga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]