Abstract
Nuclear receptors have a unique role in governing hepatitis B virus (HBV) transcription and replication. Hepatocyte nuclear factor 4α (HNF4α) and retinoid X receptor α (RXRα) plus peroxisome proliferator-activated receptor α (PPARα) have been shown to support viral biosynthesis in nonhepatoma cells in the absence of additional liver-enriched transcription factors. However, the in vivo importance of these nuclear receptors in HBV biosynthesis has been investigated only to a limited extent. Fasting has been shown to activate gluconeogenesis, in part, by activating PPARγ coactivator 1 α, which in turn leads to activation of HNF4α- and RXRα/PPARα-mediated transcription. As HBV pregenomic RNA synthesis is primarily believed to be regulated by HNF4α under normal physiological conditions, it was of interest to determine the effect of fasting on the levels of HBV RNA and DNA synthesis. Fasting was shown to rather modestly increase the levels of viral proteins, transcripts, and replication intermediates in the HBV transgenic mouse model of chronic viral infection, suggesting that caloric restriction may modulate viremia to some extent during natural infection.
Hepatitis B virus (HBV) infects hepatocytes and replicates by the reverse transcription of the pregenomic 3.5-kb RNA synthesized from the nucleocapsid or core promoter (25). Consequently, controlling the level of expression of the pregenomic RNA is a major regulatory step in the viral replication cycle (20). A variety of liver-enriched transcription factors have been shown to modulate nucleocapsid promoter activity, as determined by using reporter gene constructs (8, 15, 19, 26). However, in the context of viral replication in nonhepatoma cells, only the nuclear receptors hepatocyte nuclear factor 4α (HNF4α) and retinoid X receptor α plus peroxisome proliferator-activated receptor α (PPARα) have been shown to support pregenomic RNA synthesis and viral replication (20). These observations suggest that nuclear receptors have a unique role in governing viral biosynthesis. However, the role of nuclear receptors in regulating HBV biosynthesis in vivo has not been extensively studied (4).
The in vivo role of PPARα has been examined using the HBV transgenic mouse model of chronic viral infection (4). This study demonstrated that under normal physiological conditions, PPARα did not influence HBV transcription and replication. However, activation of PPARα by synthetic ligands did lead to enhanced viral biosynthesis, demonstrating that PPARα can modulate HBV RNA and DNA synthesis under conditions where PPARα is activated by an appropriate small molecule (4).
The potential importance of HNF4α in the regulation of HBV biosynthesis in vivo was investigated indirectly by examining the effect of fasting on HBV transcription in mice in which viral DNA was introduced into the liver by hydrodynamic injection (17). This study first demonstrated that PPARγ coactivator 1 α (PGC1α) could enhance HNF4α-mediated HBV transcription from the nucleocapsid promoter in cell culture. Consequently, the effect of PGC1α activation in response to fasting on HBV transcription in the liver was examined in mice subjected to hydrodynamic injection of HBV DNA. These studies suggested that viral transcription from the nucleocapsid promoter was modestly increased in response to fasting (17). These findings appear consistent with the anticipated results, given the detailed analysis of the induction of gluconeogenesis by PGC1α and HNF4α in response to fasting (10-12, 27). However, the extensive variability associated with the hydrodynamic injection technique and the limited analysis of viral antigen synthesis and replication performed in this study suggested that this issue should be investigated further using alternative in vivo models of HBV infection. Additionally, the potential role of PPARα in the modulation of HBV transcription was not considered, despite the known induction of PPARα expression and modulation of some PPARα-responsive genes during fasting (7, 23).
In this study, the effect of fasting on HBV biosynthesis has been investigated using the HBV transgenic mouse model of chronic viral infection (5). This analysis demonstrated that secreted HBV e antigen (HBeAg) encoded by the precore 3.5-kb transcript is increased approximately 40% as a consequence of fasting for 48 h. Consistent with this observation, fasting is associated with approximately the same increase in viral 3.5-kb RNA synthesis. An increase in viral replication intermediates in the liver is less apparent, particularly in the male HBV transgenic mice. Overall, these results suggest that fasting enhances HBV RNA and DNA synthesis to a modest extent. Although the effects observed in the HBV transgenic mice in response to fasting are relatively minor, they may be very important over time in a natural infection. A minor increase in replication efficiency per replication cycle could produce major effects over time as this effect would be amplified by the number of replication cycles occurring during the period of caloric restriction. In many parts of the world where chronic HBV infections are endemic and food supplies are unreliable, the consequences of caloric restriction for disease progression should be considered.
MATERIALS AND METHODS
Transgenic mice.
The production and characterization of the HBV transgenic mouse lineage 1.3.32 has been described previously (5). These HBV transgenic mice contain a single copy of the terminally redundant, 1.3-genome length copy of the HBV ayw genome integrated into the mouse chromosomal DNA. High levels of HBV replication occur in the livers of these mice. The mice used in the breeding experiments were homozygous for the HBV transgene and were maintained on the SV129 genetic background (9).
The production and characterization of the FoxA3-null mice has been described previously (6, 16). These mice do not express FoxA3 (also called HNF3γ), which is required for maintenance of glucose homeostasis during a prolonged fast (16). The mice used in the breeding experiments were homozygous null for FoxA3 (FoxA3−/−) and maintained on the SV129 genetic background.
FoxA3-null HBV transgenic mice were generated by mating the HBV transgenic mice with the FoxA3−/−mice. The resulting FoxA3 heterozygous (Fox+/−) HBV transgenic F1 mice were subsequently mated with the FoxA3+/− mice, and the F2 mice were screened for the HBV transgene and FoxA3-null allele by PCR analysis of tail DNA. Tail DNA was prepared by incubating 1 cm of tail in 500 μl of 100 mM Tris hydrochloride (pH 8.0), 200 mM NaCl, 5 mM EDTA, and 0.2% (wt/vol) sodium dodecyl sulfate (SDS) containing 100 μg/ml proteinase K for 16 to 20 h at 55°C. Samples were centrifuged at 14,000 rpm in an Eppendorf 5417C microcentrifuge for 10 min, and the supernatant was precipitated with 500 μl of isopropanol. DNA was pelleted by centrifugation at 14,000 rpm in an Eppendorf 5417C microcentrifuge for 10 min and subsequently dissolved in 100 μl of 5 mM Tris hydrochloride (pH 8.0) and 1 mM EDTA. The HBV transgene was identified by PCR analysis using the oligonucleotides 5′-TCGATACCTGAACCTTTACCCCGTTGCCCG-3′ (oligonucleotide XpHNF4-1; HBV coordinates 1133 to 1159) and 5′-TCGAATTGCTGAGAGTCCAAGAGTCCTCTT-3′ (CpHNF4-2; HBV coordinates 1683 to 1658) and 1 μl of tail DNA. A PCR product of 551 bp indicated the presence of the HBV transgene. The FoxA3 wild-type and null alleles were identified by PCR analysis using the oligonucleotides 5′-GGCAGTGCTTCCGGGTATGTA-3′ (oligonucleotide Foxa3-6), 5′-GGGAAGAGGTCCATGATCCAT-3′ (Foxl1-7), and 5′-CAAAGCGCCATTCGCCATTCA-3′ (LacZ-3) and 1 μl of tail DNA. A PCR product of 196 bp indicated the wild-type FoxA3 allele, whereas a PCR product of 290 bp indicated the FoxA3-null allele. The samples were subjected to 42 amplification cycles consisting of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension from the primers at 72°C for 2 min. The 20-μl reaction mixtures used were as described by the manufacturer (Gene Choice) and contained 2 units of Taq DNA polymerase.
Fasting was performed for 48 h, essentially as described previously (16). Water was available ad libitum. Mice were sacrificed, and liver tissue was frozen in liquid nitrogen and stored at −70°C prior to DNA and RNA extraction.
HBV DNA and RNA analysis.
Total DNA and RNA were isolated from liver of HBV transgenic mice as described previously (2, 14). DNA (Southern) filter hybridization analyses were performed using 20 μg of HindIII-digested DNA (14). Filters were probed with 32P-labeled HBV ayw genomic DNA (3) to detect HBV sequences. RNA (Northern) filter hybridization analyses were performed using 10 μg of total cellular RNA as described previously (14). Filters were probed with 32P-labeled HBV ayw genomic DNA to detect HBV sequences, rat cytochrome P450 4A1 (CYP4A1) cDNA to detect the CYP4A transcripts (9), and mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA to detect the GAPDH transcript used as an internal control (13).
RNase protection assays were performed using an Ambion RPA III kit, and riboprobes were synthesized using an Ambion Maxiscript kit as described by the manufacturer. Transcription initiation sites for the HBV 3.5-kb transcripts were examined using 10 μg of total cellular RNA and a 333-nucleotide (HBV coordinates 1990 to 1658) 32P-labeled HBV riboprobe. The riboprobe contained additional flanking vector sequences that are not protected by HBV transgenic mouse RNA. Filter hybridization and RNase protection analyses were quantitated by phosphorimaging using a Packard Cyclone Storage Phosphor System.
Reverse transcription-quantitative PCR (RT-qPCR) was used to measure the level of PGC1α transcripts in mouse liver RNA. After DNase I treatment, 1 μg of RNA was used for cDNA synthesis using the TaqMan RT reagents (Applied Biosystems, Foster City, CA), followed by real-time PCR quantification using SYBR Green and an Applied Biosystems 7300 real-time thermocycler (Applied Biosystems). Thermal cycling consisted of an initial denaturation step for 10 min at 95°C followed by 40 cycles of denaturation (15 s at 95°C) and annealing/extension (1 min at 60°C). The relative PGC1α RNA expression levels were estimated using the ΔΔCT method (where C[r]T is threshold cycle) with normalization to mouse GAPDH RNA. The PCR primers used were 5′-AACAATGAGCCTGCGAACAT-3′ (PGC1α exon 1 sense primer), 5′-AAATGAGGGCAATCCGTCTT-3′ (PGC1α exon 2 antisense primer), 5′-TCTGGAAAGCTGTGGCGTG-3′ (mouse GAPDH sense primer), and 5′-CCAGTGAGCTTCCCGTTCAG-3′ (mouse GAPDH antisense primer).
HBV Ag analysis.
HBeAg analysis was performed using 2 μl of mouse serum and an HBe enzyme-linked immunosorbent assay as described by the manufacturer (Epitope Diagnostics). The level of antigen was determined in the linear range of the assay. Immunohistochemical detection of HBcAg in paraffin-embedded mouse liver sections was performed as previously described (5).
RESULTS
Effect of fasting on serum glucose levels and HBeAg synthesis in HBV transgenic mice.
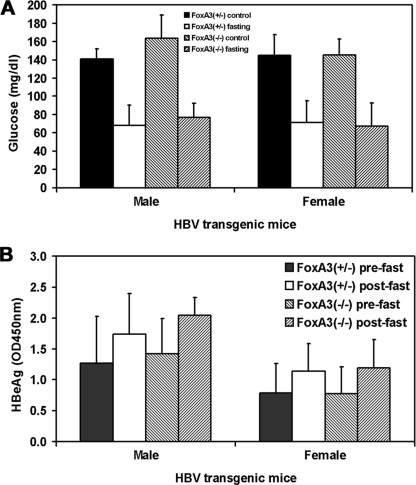
HBV transgenic mice were bred with FoxA3-null mice, and HBV transgenic mice hemizygous for the HBV transgene and heterozygous for the wild-type FoxA3 allele (FoxA3+/−) or homozygous for the FoxA3-null allele (FoxA3−/−) were identified in the F2 generation. In these studies, both HBV FoxA3+/− and FoxA3−/− transgenic mice were fasted for 48 h. Male and female mice of each genotype were assayed for the levels of glucose and HBeAg in their sera before and after fasting (Fig. 1). The serum glucose levels in the fasted mice were approximately half those observed in the fed mice (Fig. 1A). This demonstrated that the mice were fasted and that glucose levels in the serum were being maintained by gluconeogenesis. Somewhat surprisingly, the serum glucose levels in the fasted animals were similar in the FoxA3+/− and FoxA3−/− HBV transgenic mice. Previously, it had been reported that FoxA3-null mice were moderately less able to maintain serum glucose levels under fasting conditions, probably due to reduced glucose efflux from the liver (16). The differences observed between this analysis and the previous study may reflect differences in the mouse strains used or additional unidentified factors.
FIG. 1.
Effect of fasting on serum glucose levels and HBeAg synthesis in HBV transgenic mice. (A) Mice were fed (control) or fasted for 48 h (fasting). The mean serum glucose levels plus standard deviations are shown for the following groups of HBV transgenic mice: five control male FoxA3+/−, six fasted male FoxA3+/−, eight control male FoxA3−/−, eight fasted male FoxA3−/−, seven control female FoxA3+/−, five fasted female FoxA3+/−, nine control female FoxA3−/−, and seven fasted female FoxA3−/− mice. The levels of serum glucose in each group of fasted HBV transgenic mice decreased in a statistically significantly manner, as determined by a Student's t test (P < 0.05). (B) Serum HBeAg levels were measured before (prefast) and after (postfast) fasting (optical density [OD] at 450 nm). The mean HBeAg levels plus standard deviations are shown for the following groups of HBV transgenic mice: six male FoxA3+/−, eight male FoxA3−/− HBV, five female FoxA3+/−, and seven female FoxA3−/− mice. The levels of HBeAg in each group of HBV transgenic mice increased after fasting in a statistically significantly manner, as determined by a paired Student's t test (P < 0.05).
HBV transgenic mice displayed a statistically significant increase of 40 to 50% in the level of serum HBeAg after fasting compared to the level of serum HBeAg immediately prior to the caloric restriction (Fig. 1B). Twenty-five of the 26 individual mice examined demonstrated an increase in serum HBeAg as a consequence of fasting, whereas one mouse failed to demonstrate an alteration in serum HBeAg level. The increase in serum HBeAg was independent of the FoxA3 genotype of the HBV transgenic mice. As HBeAg is translated from the HBV 3.5-kb precore RNA (24), these observations suggest that fasting in these HBV transgenic mice may have a modest effect on the synthesis of the HBV 3.5-kb precore RNA.
Effect of fasting on PGC1α and CYP4A1 transcription in HBV transgenic mice.
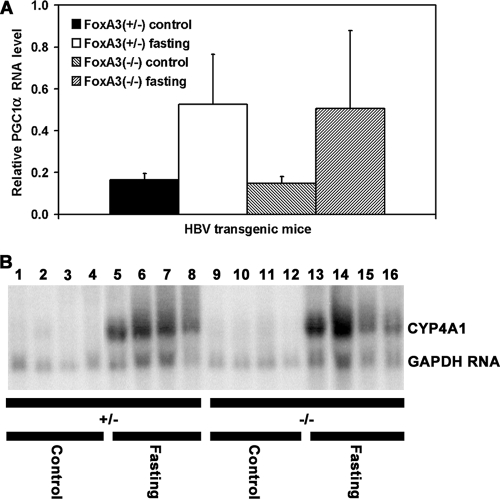
Given the modest effect of fasting on serum HBeAg levels, it was important to establish that gene expression patterns modulated by fasting were occurring normally in the HBV transgenic mice. Consequently, the levels of the PGC1α and CYP4A transcripts in the control and fasted mice were examined. PGC1α transcripts were measured relative to the GAPDH transcripts by RT-qPCR analysis. In male HBV transgenic mice, the level of PGC1α transcripts was induced approximately threefold by fasting (Fig. 2A). This level of induction is similar to previous reports (12, 27) and indicated that the expected alterations in liver transcription patterns associated with fasting induced gluconeogenesis were occurring in the HBV transgenic mice in the presence or absence of FoxA3 expression. Similarly, the level of the mouse CYP4A transcripts was significantly induced by fasting (Fig. 2B), as previously reported (23). As CYP4A induction had previously been shown to be dependent on PPARα (23), these observations indicate that PPARα is activated by fasting in the HBV transgenic mice in the presence or absence of FoxA3 expression.
FIG. 2.
Analysis of PGC1α and CYP4A1 transcripts in the livers of HBV transgenic mice. Mice were fed (control) or fasted for 48 h (fasting). (A) Quantitative analysis of the PGC1α transcript by RT-qPCR in the HBV transgenic mice. The GAPDH transcript was used as an internal control for the quantitation of the PGC1α RNA. The mean relative PGC1α transcript levels plus standard deviations are shown for the following groups of HBV transgenic mice: five control male FoxA3+/−, six fasted male FoxA3+/−, eight control male FoxA3−/−, and seven fasted male FoxA3−/− mice. The levels of the PGC1α transcript in the fasted HBV transgenic mice are statistically significantly different from their levels in the control HBV transgenic mice by a Student's t test (P < 0.05). (B) RNA (Northern) filter hybridization analysis of CYP4A1 transcripts in the livers of HBV transgenic mice. Groups of four representative male mice of each genotype are shown. The GAPDH transcript was used as an internal control for the quantitation of the CYP4A1 RNA. The probes used were CYP4A1 plus GAPDH cDNA.
Effect of fasting on viral transcription in HBV transgenic mice.
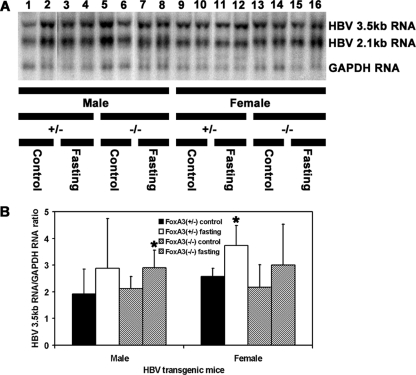
HBV transgenic mice that were heterozygous or homozygous for the FoxA3-null allele were examined for their steady-state levels of HBV transcripts by analysis of the total liver RNA (Fig. 3). The steady-state levels of the HBV 3.5- and 2.1-kb transcripts in the livers of the HBV transgenic mice with and without FoxA3 were not greatly influenced by fasting (Fig. 3A). Measurement of the levels of the HBV 3.5-kb transcripts indicated that they were increased about 40% based on this RNA filter hybridization analysis (Fig. 3B). This observation is consistent with the similar increase in serum HBeAg (Fig. 1). The effect of fasting on the steady-state levels of HBV transcripts in these mice is less than previously reported but supports the suggestion that fasting increases viral RNA synthesis to a limited extent (17). Interestingly, the absence of FoxA3 did not appear to modulate HBV transcription. This contrasts with the effect of FoxA1 (HNF3α) and FoxA2 (HNF3β), which can inhibit HBV transcription in cell culture and HBV transgenic mice (1, 20, 21). This suggests that either FoxA3 does not inhibit HBV transcription in vivo or FoxA1 and FoxA2 can compensate for the loss of FoxA3 in the FoxA3-null HBV transgenic mouse.
FIG. 3.
RNA (Northern) filter hybridization analysis of HBV transcripts in the livers of HBV transgenic mice. Groups of two representative mice of each sex and genotype are shown. Mice were fed (control) or fasted for 48 h (fasting). (A) The GAPDH transcript was used as an internal control for the quantitation of the HBV 3.5- and 2.1-kb RNAs. The probes used were HBV ayw genomic DNA plus GAPDH cDNA. +/−, FoxA3+/− mice; −/−, FoxA3−/− mice. (B) Quantitative analysis of the HBV 3.5-kb transcript in the HBV transgenic mice. The mean HBV 3.5-kb transcript levels plus standard deviations are shown for the following groups of HBV transgenic mice: five control male FoxA3+/−, six fasted male FoxA3+/−, eight control male FoxA3−/−, eight fasted male FoxA3−/−, seven control female FoxA3+/−, five fasted female FoxA3+/−, nine control female FoxA3−/−, and 7 fasted female FoxA3−/− mice. The levels of the HBV 3.5-kb transcript in the fasted HBV transgenic mice are statistically significantly different from their levels in the control HBV transgenic mice as determined by a Student's t test (P < 0.05) where indicated by an asterisk.
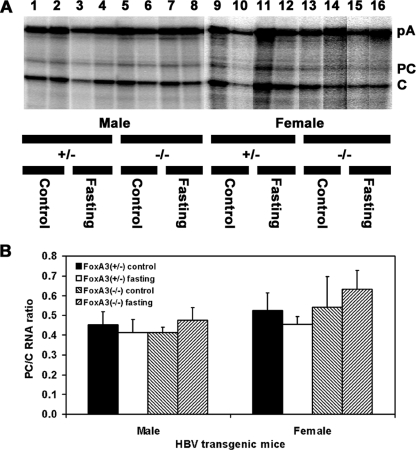
The effect of fasting on the relative levels of the precore and pregenomic HBV 3.5-kb RNAs was examined by RNase protection analysis (Fig. 4). The ratio of these two transcripts was not significantly altered by fasting in the presence or absence of FoxA3 (Fig. 4B). These observations indicate that the precore and pregenomic RNAs are coordinately expressed under these conditions and support the suggestion that the increase in serum HBeAg associated with fasting (Fig. 1) is due to the modest increase in the steady-state level of the HBV 3.5-kb precore RNA present in the livers of the fasted mice (Fig. 3 and 4).
FIG. 4.
RNase protection analysis mapping the transcription initiation sites of the precore (PC) and pregenomic (C) transcripts from the livers of HBV transgenic mice. Mice were fed (control) or fasted for 48 h (fasting). (A) The 3′ ends of the all the HBV transcripts corresponding to the polyadenylation site (pA) of these RNAs also generated a protected fragment in this analysis. The riboprobe used included the HBV ayw sequence spanning nucleotide coordinates 1990 to 1658. The 3.5-kb HBV RNAs protect fragments of 283 (pA), 206 (PC), and 175 (C) nucleotides. +/−, FoxA3+/− mice; −/−, FoxA3−/− mice. (B) Quantitative analysis of the HBV precore to pregenomic RNA ratio in HBV transgenic mice. The mean HBV precore to pregenomic (PC/C) RNA ratios plus standard deviations are shown for the following groups of HBV transgenic mice: four control male FoxA3+/−, four fasted male FoxA3+/−, four control male FoxA3−/−, four fasted male FoxA3−/−, four control female FoxA3+/−, four fasted female FoxA3+/−, four control female FoxA3−/−, and four fasted female FoxA3−/− mice. The HBV precore to pregenomic (PC/C) RNA ratios in the fasted HBV transgenic mice are not statistically significantly different from the ratios in the control HBV transgenic mice, as determined by a Student's t test (P > 0.05).
Effect of fasting on viral replication intermediates in HBV transgenic mice.
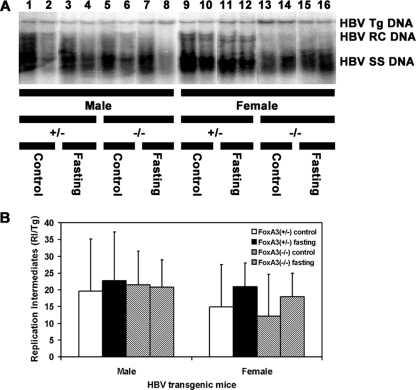
The level of replication intermediates in the livers of FoxA3-null HBV transgenic mice was similar to that observed in FoxA3-expressing HBV transgenic mice (Fig. 5). This indicated that FoxA3 expression did not affect the level of viral replication in HBV transgenic mice. This observation is consistent with the finding that FoxA3 did not affect HBV transcription (Fig. 3 and 4). Additionally, the effect of fasting on viral replication was quite limited (Fig. 5). In the male HBV transgenic mice, there was no apparent difference in the level of viral replication intermediates between the fed and fasted mice (Fig. 5B). In contrast, fasting increased viral replication by approximately 40% in the female mice. However, these changes in viral replication upon fasting were not statistically significant, reflecting the relatively large degree of variation in viral replication intermediates found within each group compared with the rather modest effect of fasting.
FIG. 5.
DNA (Southern) filter hybridization analysis of HBV DNA replication intermediates in the livers of HBV transgenic mice. Groups of two representative mice of each sex and genotype are shown. Mice were fed (control) or fasted for 48 h (fasting). (A) The HBV transgene (Tg) was used as an internal control for the quantitation of the HBV replication intermediates. The probe used was HBV ayw genomic DNA. RC, HBV relaxed circular replication intermediates; SS, HBV single-stranded replication intermediates. The probe used was HBV ayw genomic DNA. +/−, FoxA3+/− mice; −/−, FoxA3−/− mice. (B) Quantitative analysis of the HBV DNA replication intermediate (RI) levels in HBV transgenic mice. The mean HBV DNA replication intermediate levels plus standard deviations are shown for the following groups of HBV transgenic mice: five control male FoxA3+/−, six fasted male FoxA3+/−, eight control male FoxA3−/−, eight fasted male FoxA3−/−, seven control female FoxA3+/−, five fasted female FoxA3+/−, nine control female FoxA3−/−, and seven fasted female FoxA3−/− mice. The levels of the HBV replication intermediates in the fasted HBV transgenic mice are not statistically significantly different from their levels in the control HBV transgenic mice, as determined by a Student's t test (P > 0.05).
Effect of fasting on viral HBcAg distribution within the livers of HBV transgenic mice.
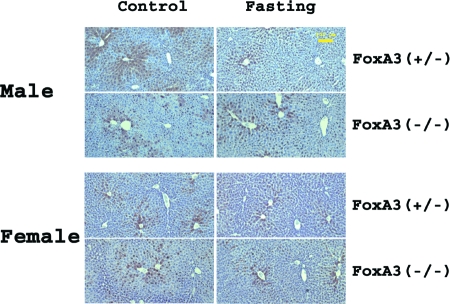
Immunohistochemical analysis of the livers of HBV transgenic mice demonstrated that fasting and FoxA3 expression do not appear to influence greatly the distribution of HBcAg within the liver lobule (Fig. 6). In both male and female FoxA3-expressing HBV transgenic mice, the HBcAg staining is both nuclear and cytoplasmic in the hepatocytes located around the central vein. HBcAg staining is limited to the nuclei of hepatocytes located further from the central vein and is considerably reduced within hepatocytes surrounding the portal vein. The same pattern of staining was apparent in FoxA3-null HBV transgenic mice, whether they were fed or fasted. These findings are consistent with the modest effect fasting has on HBV RNA and DNA synthesis (Fig. 3 to 5).
FIG. 6.
Immunohistochemical staining of HBcAg in the livers of HBV transgenic mice. Mice were fed (control) or fasted for 48 h. Nuclear staining of HBcAg is observed throughout the liver, whereas cytoplasmic staining is located primarily in the centrolobular hepatocytes in the livers of all mice. The yellow size bar is 100 μm.
DISCUSSION
Fasting induces PGC1α expression in the liver, leading to the activation of HNF4α activity (12, 27). In turn, HNF4α increases the level of transcription of the key gluconeogenic genes, phosphoenolpyruvate carboxykinase, and G6Pase (12, 27). In this manner, glucose homeostasis is maintained during caloric restriction. FoxA3 appears to contribute to this process, in part, through its effects on GLUT2 expression (16). Importantly, HNF4α and FoxA transcription factors are also major regulators of HBV biosynthesis (20). Nuclear receptors such as HNF4α can support viral replication in nonhepatoma cells, whereas FoxA1 and FoxA2 are efficient inhibitors of HBV RNA and DNA synthesis (1, 20, 21). Furthermore, fasting activates PPARα, which can also modulate HBV biosynthesis (4, 20, 23). Therefore, it was of some interest to determine the potential role these factors might have in regulating HBV transcription and replication under conditions where the activities of these transcription factors are modulated. Consequently, the effect of fasting on HBV biosynthesis in the HBV transgenic mouse model of chronic infection was examined.
Initially, it was observed that serum HBeAg levels were increased approximately 40% during fasting (Fig. 1). This increase in serum HBeAg was associated with a concomitant increase in viral transcripts, suggesting that the increase in viral transcription was directly responsible for the elevated level of circulating viral antigen (Fig. 3 and 4). Presumably, this modest increase in transcription was mediated by the activation of HNF4α, and possibly PPARα, by PGC1α, which is induced by fasting (12, 17, 22, 23, 27). Similarly, the increase in viral transcripts was associated with about a 40% increase in viral replication in the female HBV transgenic mice (Fig. 5). The failure to observe a similar increase in the male mice may simply reflect the limited effect fasting has on viral replication or could indicate that although transcription is somewhat affected under these conditions, it does not always translate into a similar effect on replication. As a consequence of the complex metabolic alterations resulting from fasting, additional factors may also contribute to the modulation of HBV transcription and replication in both a positive and negative manner, ultimately producing the observed modest effects on viral biosynthesis (7, 16).
Therefore, it is apparent that the effect of fasting is rather limited in the HBV transgenic mouse model of chronic viral infection. However, this does not mean it is not important. Assuming fasting increases viral biosynthesis approximately 40% in each replication cycle, this would translate into about a 10-fold increase in viral load every seven cycles of synthesis, secretion, and infection of new hepatocytes. Indeed, such findings linking the levels of viral transcription and replication to the metabolic state of the hepatocyte have led to the suggestion that HBV might be considered a “metabolovirus” (18). Consequently, it may be worth considering metabolic interventions that might limit HBV biosynthesis as an additional therapeutic approach to chronic HBV treatment (18).
Acknowledgments
We thank Luca G. Guidotti and Francis V. Chisari (The Scripps Research Institute, La Jolla, CA) for providing the HBV transgenic mice and Bruno Sainz, Jr., and Susan L. Uprichard (University of Illinois at Chicago, Chicago, IL) for assistance with the RT-qPCR analysis.
This work was supported by Public Health Service grant AI30070 from the National Institutes of Health.
Footnotes
Published ahead of print on 10 December 2008.
REFERENCES
- 1.Banks, K. E., A. L. Anderson, H. Tang, D. E. Hughes, R. H. Costa, and A. McLachlan. 2002. Hepatocyte nuclear factor 3β inhibits hepatitis B virus replication in vivo. J. Virol. 7612974-12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 3.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281646-650. [DOI] [PubMed] [Google Scholar]
- 4.Guidotti, L. G., C. M. Eggers, A. K. Raney, S. Y. Chi, J. M. Peters, F. J. Gonzalez, and A. McLachlan. 1999. In vivo regulation of hepatitis B virus replication by peroxisome proliferators. J. Virol. 7310377-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 696158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaestner, K. H., H. Hiemisch, and G. Schütz. 1998. Targeted disruption of the gene encoding hepatocyte nuclear factor 3γ results in reduced transcription of hepatocyte-specific genes. Mol. Cell. Biol. 184245-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersten, S., J. Seydoux, J. M. Peters, F. J. Gonzalez, B. Desvergne, and W. Wahli. 1999. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J. Clin. Investig. 1031489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosovsky, M. J., I. Qadri, and A. Siddiqui. 1998. The regulation of hepatitis B virus gene expression: an overview of the cis- and trans-acting components, p. 21-50. In R. Koshy and W. H. Caselmann (ed.), Hepatitis B virus: molecular mechanisms in disease and novel strategies for therapy. Imperial College Press, London, United Kingdom.
- 9.Lee, S. S. T., T. Pineau, J. Drago, E. J. Lee, J. W. Owens, D. L. Kroetz, P. M. Fernandez-Salguero, H. Westphal, and F. J. Gonzalez. 1995. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 153012-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, J., C. Handschin, and B. M. Spiegelman. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1361-370. [DOI] [PubMed] [Google Scholar]
- 11.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2478-90. [DOI] [PubMed] [Google Scholar]
- 12.Rhee, J., Y. Inoue, J. C. Yoon, P. Puigserver, M. L. Fan, F. J. Gonzalez, and B. M. Spiegelman. 2003. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): Requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc. Natl. Acad. Sci. USA 1004012-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabath, D. E., H. E. Broome, and M. B. Prystowsky. 1990. Glyceraldehyde-3-phosphate dehydrogenase mRNA is a major interleukin 2-induced transcript in a cloned T-helper lymphocyte. Gene 91185-191. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Schaller, H., and M. Fischer. 1991. Transcriptional control of hepadnavirus gene expression. Curr. Top. Microbiol. Immunol. 16821-39. [DOI] [PubMed] [Google Scholar]
- 16.Shen, W., L. M. Scearce, J. E. Brestelli, N. J. Sund, and K. H. Kaestner. 2001. Foxa3 (hepatocyte nuclear factor 3γ) is required for the regulation of hepatic GLUT2 expression and the maintenance of glucose homeostasis during a prolonged fast. J. Biol. Chem. 27642812-42817. [DOI] [PubMed] [Google Scholar]
- 17.Shlomai, A., N. Paran, and Y. Shaul. 2006. PGC-1α controls hepatitis B virus through nutritional signals. Proc. Natl. Acad. Sci. USA 10316003-16008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shlomai, A., and Y. Shaul. 2008. The “metabolovirus” model of hepatitis B virus suggests nutritional therapy as an effective anti-viral weapon. Med. Hypotheses 7153-57. [DOI] [PubMed] [Google Scholar]
- 19.Tang, H., K. E. Banks, A. L. Anderson, and A. McLachlan. 2001. Hepatitis B virus transcription and replication. Drug News Perspect. 14325-334. [PubMed] [Google Scholar]
- 20.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 981841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang, H., and A. McLachlan. 2002. Mechanisms of inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by hepatocyte nuclear factor 3β. J. Virol. 768572-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vega, R. B., J. M. Huss, and D. P. Kelly. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 201868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan, Y. J. Y., Y. Cai, W. Lungo, P. Fu, J. Locker, S. French, and H. M. Sucov. 2000. Peroxisome proliferator-activated receptor α-mediated pathways are altered in hepatocyte-specific retinoid X receptor α-deficient mice. J. Biol. Chem. 27528285-28290. [DOI] [PubMed] [Google Scholar]
- 24.Weimer, T., J. Salfeld, and H. Will. 1987. Expression of the hepatitis B virus core gene in vitro and in vivo. J. Virol. 613109-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Will, H., W. Reiser, T. Weimer, E. Pfaff, M. Buscher, R. Sprengle, R. Cattaneo, and H. Schaller. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yen, T. S. B. 1993. Regulation of hepatitis B virus gene expression. Semin. Virol. 433-42. [Google Scholar]
- 27.Yoon, J. C., P. Puigserver, G. X. Chen, J. Donovan, Z. D. Wu, J. Rhee, G. Adelmant, J. Stafford, C. R. Kahn, D. K. Granner, C. B. Newgard, and B. M. Spiegelman. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413131-138. [DOI] [PubMed] [Google Scholar]