Abstract
A tight junction (TJ) protein, claudin-1 (CLDN1), was identified recently as a key factor for hepatitis C virus (HCV) entry. Here, we show that another TJ protein, occludin, is also required for HCV entry. Mutational study of CLDN1 revealed that its tight junctional distribution plays an important role in mediating viral entry. Together, these data support the model in which HCV enters liver cells from the TJ. Interestingly, HCV infection of Huh-7 hepatoma cells downregulated the expression of CLDN1 and occludin, preventing superinfection. The altered TJ protein expression may contribute to the morphological and functional changes observed in HCV-infected hepatocytes.
Recently, considerable progress has been made in elucidating the molecular mechanisms by which hepatitis C virus (HCV) infects human liver cells. The current accepted model of HCV infection is that virus particles associated with lipoproteins, found circulating in the bloodstream, use glycosaminoglycans and/or the LDL receptor on host cells as initial attachment factors. After binding, the HCV particle interacts with SR-BI and CD81 and is subsequently relocalized to the tight junction (TJ) protein claudin-1 (CLDN1) (6). Next, the HCV particle becomes internalized via clathrin-mediated endocytosis, followed by viral fusion, which likely occurs in early endosomes. Some critical information, however, is missing in such a model with regard to the role of CLDN1. (i) The interaction between CLDN1 and incoming HCV virions has yet to be verified experimentally; (ii) the precise site of viral entry needs to be determined given that CLDN1 predominantly localizes to TJs in polarized cells; and (iii) the potential involvement of other TJ proteins in HCV entry remains untested. We have shown previously that the TJ-like CLDN1 distribution correlates with cellular permissiveness to HCV infection (19). In the current study, we intend to define the importance of junctional CLDN1 and other TJ proteins in HCV entry.
TJ protein OCLN is required for HCV entry.
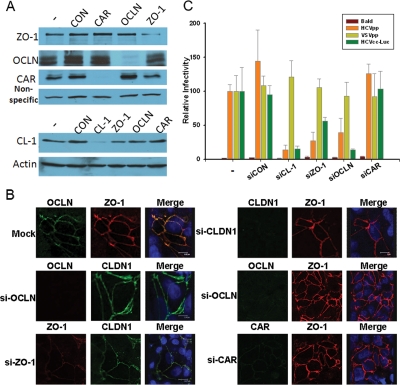
As hepatocytes are highly polarized in vivo, we first sought to investigate whether HCV entry mimics the major group B coxsackievirus (CVB) entry, in which CVB enters polarized epithelial cells through TJs by a complex mechanism requiring attachment to occludin (OCLN) and the induction of caveolar endocytosis (3). To this end, we utilized synthetic interference RNA (siRNA) or packaged retroviruses to deliver short-hairpin-based RNA (shRNA) to knock down the expression of TJ proteins CLDN1, OCLN, ZO-1, JAM-1, and CAR (CVB receptor) to examine the roles of each of the proteins during HCV infection. Targeted sequences of the siRNAs and shRNAs are presented in the supplemental material. As shown in Fig. 1A and B, depletion of OCLN affected neither the expression level nor the localization of CLDN1; however, depletion of ZO-1 by siRNA modestly reduced the CLDN1 level (Fig. 1A). We then performed the infection assay according to a previously established procedure (19). Reduction of CLDN1, OCLN, and ZO-1 expression inhibited entry of human immunodeficiency virus (HIV)-HCV pseudotypes (HCVpp), but not vesicular stomatitis virus G pseudotypes (VSVpp), into Huh7.5.1 (Fig. 1C; see Fig. S1 in the supplemental material). Similar results were observed using cell culture-grown HCV (HCVcc) encoding firefly luciferase (Fig. 1C). Notably, depletion of ZO-1 by shRNA targeting a different region of ZO-1 had minimal effect on HCVpp entry (see Fig. S1 in the supplemental material), suggesting that ZO-1 is unlikely to be directly involved in HCV entry. The observed effect of ZO-1 knockdown on viral entry in Fig. 1C could be due to the modest reduction in CLDN1 level, as ZO-1 is known to determine where claudins are polymerized in TJ strand formation (18).
FIG. 1.
Depletion of OCLN blocks HCV entry. Huh7.5.1 cells (2 × 105) were transfected with the indicated siRNA oligonucleotides (made by Dharmacon, Ambion, and IDTDNA) at the final concentration of 40 nM by the RNAiMax reagent (Invitrogen) for 24 h. Cells were incubated for an additional 24 h, and the specific knockdown of each protein was verified by immunoblotting (A) and confocal microscopy (B). (C) Cells derived from the steps in panel A were infected with the indicated viruses for 2 h and further incubated for 24 to 48 h prior to the luciferase assay. Data plotted are the means ± standard deviations of the results. Bald virus was packaged using the same HIV core construct without the Env-encoding gene, and HCVcc-Luc represents an HCVcc virus with a luciferase gene inserted into its genome. The results shown are representative of three independent experiments.
Next, we examined whether any portion of OCLN, a four-transmembrane protein with a relatively long C-terminal tail, may replace CLDN1 in mediating HCV entry. Specifically, we generated chimeric proteins containing the two extracellular loops from one of these proteins and the C-terminal domain from the other. Chimeric proteins containing the two CLDN1 extracellular loops and the OCLN C-terminal domain were still able to render 293T cells susceptible to HCVpp, but this was not the case for the alternative chimeric protein (see Fig. S2 in the supplemental material). Together, these results imply that CLDN1 and OCLN function distinctly in mediating HCV entry.
OCLN coprecipitates with HCV E2 in HCVcc-infected hepatoma cells.
HCV entry is dependent upon clathrin-mediated endocytosis (1). By screening a pool of pharmacological inhibitors, we found that dynasore, an inhibitor of dynamin, which interacts with OCLN-based cellular structure (10), nearly abolished HCVpp entry (see Table S1 in the supplemental material). Dynamin plays an essential role in receptor-mediated endocytosis via clathrin-coated pits and caveolae (4). Since Huh7 and its derivatives are known to be naturally deficient in caveolin-1 and caveolin-2 (5), perhaps HCV gains entry by first interacting with a primary receptor and then inducing OCLN-dynamin-clathrin-dependent internalization. Interestingly, a recent study using recombinant HCV E1E2 revealed a relocalization of the HCV E2/CD81 complex to cell-cell contact areas where CD81 comes into contact with the TJ proteins OCLN, ZO-1, and CLDN1 (2). To explore the possible interaction between OCLN and HCV envelope proteins, we carried out a coimmunoprecipitation study with Huh7.5.1 cells infected by a recently described JFH1 variant, JFH1-AM2, which produced more infectious virions than did the wild type (15). An equal number of cells were also infected by the JFH1-FlagE2-AM2 virus, in which a Flag tag was inserted into the hypervariable region 1 of E2 of JFH1-AM2. Such a tagging strategy affects neither virus replication nor infection (12) (data not shown) but allows specific pull-down of FlagE2-associated proteins. Indeed, OCLN, but not CAR, specifically precipitated with FlagE2 (Fig. 2). Thus, perhaps at some point during infection, OCLN interacts with HCV E2 to facilitate viral entry. It must be noted, however, that the captured interactions in our system can be derived from either an intracellular pool of the OCLN-E2 complex or a complex formed only during viral entry. An assay that allows direct study of the interaction between surface-bound infectious virions and HCV receptors should be developed in the future in order to distinguish between the two possibilities.
FIG. 2.
OCLN coprecipitates with HCV E2 from HCVcc-infected Huh7.5.1 cells. Huh-7.5.1 cells (2 × 109) were infected by JFH1-AM2 (sample 1) and JFH1-FlagE2-AM2 (sample 2) viruses. On day 3 postinfection, cells were trypsinized and washed with ice-cold phosphate-buffered saline and then Dounce homogenized in 10 ml of immunoprecipitation (IP) buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 0.5% NP-40, 5 mM β-glycerophosphate) supplemented with a protease inhibitor cocktail (Sigma). Centrifugation-cleared lysates were then subjected to IP with 50 μl anti-Flag M2 affinity resin (Sigma). After four washes with the IP buffer, bound proteins were eluted with Flag peptide (100 μg/ml; Sigma) in 100 μl Tris-buffered saline. A total of 0.1% of input samples and 7.5 μl of pull-down products were loaded for Western blot analysis. Flag-E2, OCLN, and CAR were detected by their corresponding antibodies.
Extrajunctional CLDN1 is less efficient in mediating HCV entry.
Previously, we reported a close correlation between TJ-like localization of CLDN1 and CLDN1's ability to mediate viral entry (19); however, conflicting results suggest that the nonjunctional distributed CLDN1 may be required for its function (11). In particular, a recent report indicates that both tight-junctional (apical-canalicular) and extrajunctional (basolateral-sinusoidal) forms of CLDN1 exist in hepatocytes (14) in our cultured Huh7 cell system, although we observed that CLDN1 and OCLN concentrated exclusively at the sites of cell-cell contacts of a confluent monolayer (19) (data not shown). To address this matter, we performed site-directed mutagenesis and created a panel of CLDN1 mutants containing single amino acid substitutions in its first or second extracellular loops (EL) as a recent report suggests that the conserved residues on the second EL of CLDNs contribute to the trans interactions of TJ formation (13). Strikingly, the I32M mutant (located in the N-terminal third of the first EL) exclusively localized to the cytosol (Fig. 3A and B). Consistent with the previous report (6), this substitution completely abolished CLDN1's ability to mediate HCVpp entry (Fig. 3C). Western blot analysis further revealed that this mutant was expressed at much lower level than was the wild type, implying that the mislocalized protein is less stable (data not shown). Moreover, F148A and R158A mutants, which appeared to localize on the cell surface but not be concentrated at cell junctions (extrajunction type), were much less efficient in rendering 293T cells susceptible to HCVpp infection than was the wild-type protein (Fig. 3A to C). Furthermore, when confluent 293T cells transfected with wild-type CLDN1 were split and reseeded to avoid the formation of cell-cell contact, they became far less permissive to HCVpp infection despite cell surface expression of CLDN1 (unpublished results). Together, these data suggest that the tight-junctional localization of CLDN1 is critical for viral entry and that HCV entry may require a delicate molecular architecture of multiple proteins, occurring only at TJs.
FIG. 3.
Junctional CLDN1 is important for HCV entry. (A) 293T cells, free of endogenous CLDN1 (19), were transfected with the indicated CLDN1 constructs (all fused with green fluorescent protein [GFP]). The subcellular distribution of each fusion protein (green) was examined by confocal microscopy. Nuclei were stained with Draq5 (blue). Images shown are XY, XZ, and YZ planes derived from the three-dimensional scanning. (B) A total of 1 × 106 293T cells were transfected with 1 μg of the constructs that were used in panel A. Twenty-four hours after transfection, cells were stained with an anti-CLDN1 antibody (MAB4618; R&D Systems) which specifically recognizes surface CLDN1. Total CLDN1 expression was first determined by measuring the GFP expression on a flow cytometer. Gated GFP-positive cells (blue) were then quantified for surface CLDN1 expression (magenta). The percentage of positive cells and the mean fluorescence intensity are indicated in the figure. (C) 293T cells were transfected with the indicated constructs and further spin-infected by HCVpp for the luciferase assay. Normalized luciferase activity was plotted. Data shown are the means ± standard deviations of the results. The results shown are representative of four independent experiments.
HCV infection downregulates CLDN1 and OCLN expressions, preventing superinfection.
Next, we examined the expression levels of CLDN1 and OCLN during HCV infection and found that they were downregulated following infection (Fig. 4A). Flow cytometric analysis confirmed the downregulation of cell surface CLDN1, but not CD81, following infection (Fig. 4B and data not shown). Consequently, HCVcc-infected cells became refractory to HCVpp infection (Fig. 4C). In order to elucidate which viral protein may be causing this downregulation, we coexpressed individual HCV proteins with CLDN1 in 293T cells. Here, expression of HCV structural proteins, Core and E1E2, significantly inhibited the expression of CLDN1 from a cotransfected plasmid. Deletion of the first 50 amino acids of the Core protein nullified this inhibition (Fig. 4D).
FIG. 4.
HCV infection downregulates CLDN1/OCLN expression. (A) Naïve Huh7 cells were infected with JFH1 HCVcc (MOI, 0.1) for 5 days or left uninfected. Total cellular CLDN1 and OCLN were determined by Western blotting. (B) The live cells from panel A were analyzed for surface CLDN1 expression by flow cytometry. The rat anti-CLDN1 antibody (MAB4618; R&D Systems) that specifically recognizes surface CLDN1 was used. (C) The cells from panel A were infected with HCVpp to determine the susceptibility to superinfection. (D) 293T cells seeded in a 12-well plate were transfected with combinations of 0.5 μg of CLDN1- or OCLN-expressing plasmid with 0.5 μg of plasmids expressing the indicated viral proteins. Forty-eight hours after transfection, the levels of all proteins were quantified by Western blotting. β-Actin was determined for loading control. FL Core represents full-length HCV Core protein. The truncated Core protein does not have the first 50 amino acids. The position of each individual viral protein is indicated with a star sign. Except for E1 and E2, all proteins were N-terminally Flag-tagged for simple detection by anti-Flag antibody. E1 and E2 were detected by A4 and H52 antibodies, respectively. E1 and E2 are derived from genotype 1a (H77), and other viral proteins were derived from HCV genotype 2a (JFH1).
The downregulation of CLDN1/OCLN following HCV infection provides an appealing explanation for the exclusion of HCV superinfection, a state in which infected cells become resistant to future infection, allowing the host cells to contain the infection. It has been documented that HCV infection does result in the exclusion of superinfection, and this was demonstrated to be not due to reduction in CD81 or SR-BI level on cell surface but, rather, mediated primarily by interference at the level of HCV RNA translation and subsequent viral replication (16). While our findings certainly add more pieces to the understanding of the mechanisms by which exclusion of superinfection occurs, they are directly conflicting with a recent report in which Reynolds et al. reported that CLDN1 expression was slightly upregulated in HCV JFH1-infected Huh7.5 cells using confocal microscopy (14). We are unable to explain the discrepancy except that different detection methods, and more importantly, different incubation periods, were noted between the two studies. In fact, it appears that HCV infection may lead to a rather global reduction in TJ proteins, as even the CAR protein level decreased in HCVcc-infected Huh7.5.1 cells (see Fig. S3 in the supplemental material). It has been reported that loss of CLDN1 expression correlates with malignancy and the development of hepatocellular carcinoma (8). Expression of HCV structural proteins Core, E1, and E2 significantly suppressed the expression of CLDN1 from a murine stem cell virus/cytomegalovirus promoter-driven plasmid, arguing that the downregulation of CLDN1 presumably occurs at the posttranscriptional level. The downregulation of OCLN, on the other hand, was only slightly affected by coexpression of NS4A. Because TJ proteins are critical in maintaining the polarity of hepatocytes and polarity is required for the directional trafficking of many proteins and the bile secretion (9), altered expressions of TJ proteins during the course of HCV infection may lead to morphological changes on hepatocytes, resulting in many reported symptoms, such as cholestatic disorders (17). Indeed, the lack of CLDN1 has been linked to neonatal sclerosing cholangitis syndrome (7). Future work should evaluate hepatocellular CLDN1/OCLN expression levels in well-controlled groups during HCV disease progression.
Supplementary Material
Acknowledgments
This work was supported by University of Pittsburgh Central Research Development Funds (to T.W.) and NIHDK 061931 (to J.R.T.).
We gratefully acknowledge T. Wakita, C. Rice, F. Chisari, F. Cosset, G. Luo, R. Bartenschlager, J. Dubuisson, H. Greenberg, S. Duncan, R. Purcell, and S. Emerson for providing cell lines and reagents. We thank Q. Han, N. Biswas, and S. Chadwick for the help with cloning and confocal microscopy.
Footnotes
Published ahead of print on 3 December 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 806964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazzoli, M., A. Bianchi, S. Filippini, A. Weiner, Q. Zhu, M. Pizza, and S. Crotta. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 828316-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coyne, C. B., L. Shen, J. R. Turner, and J. M. Bergelson. 2007. Coxsackievirus entry across epithelial tight junctions requires occludin and the small GTPases Rab34 and Rab5. Cell Host Microbe 2181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damke, H., T. Baba, D. E. Warnock, and S. L. Schmid. 1994. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 127915-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damm, E. M., L. Pelkmans, J. Kartenbeck, A. Mezzacasa, T. Kurzchalia, and A. Helenius. 2005. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 168477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446801-805. [DOI] [PubMed] [Google Scholar]
- 7.Hadj-Rabia, S., L. Baala, P. Vabres, D. Hamel-Teillac, E. Jacquemin, M. Fabre, S. Lyonnet, Y. De Prost, A. Munnich, M. Hadchouel, and A. Smahi. 2004. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology 1271386-1390. [DOI] [PubMed] [Google Scholar]
- 8.Higashi, Y., S. Suzuki, T. Sakaguchi, T. Nakamura, S. Baba, H. C. Reinecker, S. Nakamura, and H. Konno. 2007. Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J. Surg. Res. 13968-76. [DOI] [PubMed] [Google Scholar]
- 9.Lazaridis, K. N., and N. F. LaRusso. 2003. Bile formation: do not ignore the role of plasma membrane-cytoskeleton linking proteins. Hepatology 37218-220. [DOI] [PubMed] [Google Scholar]
- 10.Lie, P. P., W. Xia, C. Q. Wang, D. D. Mruk, H. H. Yan, C. H. Wong, W. M. Lee, and C. Y. Cheng. 2006. Dynamin II interacts with the cadherin- and occludin-based protein complexes at the blood-testis barrier in adult rat testes. J. Endocrinol. 191571-586. [DOI] [PubMed] [Google Scholar]
- 11.Mee, C. J., J. Grove, H. J. Harris, K. Hu, P. Balfe, and J. A. McKeating. 2008. Effect of cell polarization on hepatitis C virus entry. J. Virol. 82461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 13.Piontek, J., L. Winkler, H. Wolburg, S. L. Muller, N. Zuleger, C. Piehl, B. Wiesner, G. Krause, and I. E. Blasig. 2008. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 22146-158. [DOI] [PubMed] [Google Scholar]
- 14.Reynolds, G. M., H. J. Harris, A. Jennings, K. Hu, J. Grove, P. F. Lalor, D. H. Adams, P. Balfe, S. G. Hubscher, and J. A. McKeating. 2008. Hepatitis C virus receptor expression in normal and diseased liver tissue. Hepatology 47418-427. [DOI] [PubMed] [Google Scholar]
- 15.Russell, R. S., J. C. Meunier, S. Takikawa, K. Faulk, R. E. Engle, J. Bukh, R. H. Purcell, and S. U. Emerson. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. USA 1054370-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 814591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trauner, M., P. J. Meier, and J. L. Boyer. 1998. Molecular pathogenesis of cholestasis. N. Engl. J. Med. 3391217-1227. [DOI] [PubMed] [Google Scholar]
- 18.Umeda, K., J. Ikenouchi, S. Katahira-Tayama, K. Furuse, H. Sasaki, M. Nakayama, T. Matsui, S. Tsukita, and M. Furuse. 2006. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126741-754. [DOI] [PubMed] [Google Scholar]
- 19.Yang, W., C. Qiu, N. Biswas, J. Jin, S. C. Watkins, R. C. Montelaro, C. B. Coyne, and T. Wang. 2008. Correlation of the tight junction-like distribution of claudin-1 to the cellular tropism of HCV. J. Biol. Chem. 2838643-8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.