Abstract
Pneumonia virus of mice (PVM) strain 15 causes fatal pneumonia in mice and provides a convenient model for human respiratory syncytial virus pathogenesis and immunobiology. We prepared PVM mutants lacking the genes for nonstructural proteins NS1 and/or NS2. In Vero cells, which lack type I interferon (IFN), deletion of these proteins had no effect on the efficiency of virus growth. In IFN-competent mouse embryo fibroblasts, wild-type (wt) PVM and the ΔNS1 virus grew efficiently and strongly inhibited the IFN response, whereas virus lacking NS2 was highly attenuated and induced high levels of IFN and IFN-inducible genes. In BALB/c mice, intranasal infection with wt PVM caused overt disease that began on day 6 and was lethal by day 9 postinoculation. In comparison, ΔNS1 induced transient, reduced disease, and ΔNS2 and ΔNS12 caused no disease. Thus, NS1 and NS2 are virulence factors, with NS2 being a major antagonist of the type I IFN system. The pulmonary titers of wt PVM and ΔNS1 were high on day 3 and increased further by day 6; in addition, expression of IFN and representative proinflammatory cytokines/chemokines and T lymphocyte-related cytokines was undetectable on day 3 but increased dramatically by day 6 coincident with the onset of disease. The titers of ΔNS2 and ΔNS12 were somewhat lower on day 3 and decreased further by day 6; in addition, these viruses induced a more circumscribed set of cytokines/chemokines (IFN, interleukin-6 [IL-6], and CXCL10) that were detected on day 3 and had largely subsided by day 6. Lung immunohistology revealed abundant PVM-positive pneumocytes and bronchial and bronchiolar epithelial cells in wt PVM- and ΔNS1-infected mice on day 6 compared to few PVM-positive foci with ΔNS2 and ΔNS12. These results indicate that severe PVM disease is associated with high, poorly controlled virus replication driving the expression of high levels of pulmonary IFN and a broad array of cytokines/chemokines. In contrast, in the absence of NS2, there was an early, transient innate response involving moderate levels of IFN, IL-6, and CXCL10 that restricted virus replication and prevented disease.
Pneumonia virus of mice (PVM) is an enveloped nonsegmented negative-strand RNA virus of genus Pneumovirus, family Paramyxoviridae. The Pneumovirus genus also includes the respiratory pathogens bovine respiratory syncytial virus (BRSV) and human respiratory syncytial virus (HRSV), the latter being the genus type species. HRSV is the most important viral respiratory pathogen of infants worldwide. Pneumoviruses have genomes of approximately 15 kDa that contain 10 genes encoding 11 to 12 proteins. The gene order and constellation of proteins are conserved between the viruses. Although its natural history is poorly understood, PVM was first recovered and identified from laboratory mice and causes respiratory tract disease in mice and other rodents. Infection of mice with PVM provides a convenient surrogate in vivo model for HRSV immunobiology and pathogenesis.
The first two genes in the Pneumovirus gene order encode the nonstructural proteins NS1 and NS2, which for HRSV were shown to inhibit the induction of type 1 interferon (IFN) in human and mouse cells by inhibiting activation of the transcription factor IFN regulatory factor 3 (IRF3) (20, 33, 34). In addition, NS1 and NS2 interfere with type I IFN-induced signal transduction through the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway. Specifically, NS1 and NS2 cooperate to decrease intracellular levels of STAT2 (26), NS2 inhibits the phosphorylation of STAT1 (29), and NS1 functions as an E3 ligase and targets STAT2 for ubiquitination and degradation (11).
Type I IFNs are part of the first line of defense against virus infection (30, 31, 35, 36). Detection of viral macromolecules by host cell pattern recognition receptors results in the induction of IFN-β and IFN-α4 (in mice) as one of the first responses to infection (27, 42). These IFNs are secreted and bind to the IFN-α/β receptor in an autocrine and paracrine fashion to induce JAK/STAT signaling, leading to changes in expression of hundreds of IFN-regulated genes (17, 30, 36). Prominent among these is IRF7, whose activation in conjunction with IRF3 leads to the expression of the full panel of type I IFNs (15). Other IFN-stimulated genes encode proteins involved in establishing the IFN-induced antiviral state and in enhancing the innate and adaptive immune response. Thus, in addition to their protective effects against virus infections (30, 31, 35, 36), IFN-α and IFN-β and the genes they control, enhance, and link innate and adaptive immunity by enhancing natural killer (NK) cells and T-lymphocyte-mediated responses, promoting leukocyte chemotaxis, and enhancing antibody production and class switching (4, 24).
Recombinant HRSV and BRSV with deletions of NS1 and/or NS2 have been described previously (2, 19, 32, 38). In addition to their use in analyzing NS1 and NS2 and their effects on the host IFN system, these mutants have been evaluated as attenuated live vaccine candidates for prevention of respiratory virus infections in infants (6, 46) and calves (41). HRSV mutants lacking NS1 or NS2 were shown to be attenuated and immunogenic in preclinical tests in nonhuman primates (7, 18, 39, 45). HRSV vaccine candidates based on deletion of NS2 combined with attenuating point mutations in other proteins were subsequently tested in clinical phase 1 studies in seropositive and seronegative children of 6 to 24 months of age but appeared to be overattenuated, most likely because the NS2 deletion had been combined with too many attenuating point mutations (46). In addition, a recombinant HRSV live vaccine candidate whose attenuation is based solely on deletion of NS1 is currently being prepared for clinical trial (unpublished data). Unfortunately, we lack a comprehensive understanding of the effects of deletion of NS1 and NS2 on HRSV immunobiology and pathogenesis in vivo. Studies with the chimpanzee model are limited due to the scarcity of animals, expense, and ethical considerations against invasive sampling. Other nonhuman primates are less permissive and thus less suitable. Rodent and other small animal models are not highly permissive to HRSV; in addition, their evolutionary distance from the natural human host makes them inauthentic surrogates. In contrast, mice appear to be a natural host for PVM and provide a convenient system that can employ genetically identical animals with a multitude of genetic and immunologic reagents. Using a recently developed PVM reverse genetics system (22), we prepared PVM recombinants with deletions of NS1 and NS2, individually or together, in order to evaluate the effects of these deletions in a convenient permissive host.
MATERIALS AND METHODS
Virus and cells.
PVM strain 15 was obtained from the ATCC (ATCC VR-25) and was propagated on BHK-21 cells. BHK-21 cells (ATCC CCL-10) and BHK21 BSR T7/5 cells that constitutively express T7 RNA polymerase (2) were maintained in Glasgow minimal essential medium (Glasgow MEM; Invitrogen, Carlsbad, CA) with 4 mM l-glutamine, MEM amino acids (Invitrogen), and 10% fetal bovine serum. Vero cells (ATCC CCL-81) were grown in OptiProSFM (Invitrogen) supplemented with 4 mM l-glutamine. Mouse embryo fibroblast (MEF) cells generated from CF-1 mice (Millipore, Billerica, MA) were maintained in Dulbecco MEM with 10% fetal bovine serum, 2 mM l-glutamine, and 1% 100× nonessential amino acids (Invitrogen).
Generation of PVM NS gene deletion mutants.
We previously constructed a complete cDNA copy of the PVM strain 15 genome (21) in a plasmid under the control of a T7 promoter (pPVM) (22). The sequence of this cDNA differed from the consensus sequence of the biological virus (GenBank accession no. AY729016) by the presence of two restriction sites that had been introduced as markers: an AgeI restriction site at the SH-G intergenic region (AY729016 nucleotide [nt] 4509, involving the substitution of one nucleotide and the addition of 3 nt), and a BstBI site at the M2/liter intergenic region (AY729016 nt 8458, involving two nucleotide substitutions and the addition of one nucleotide) (22).
Versions of pPVM lacking NS1, NS2, or NS1 and NS2 together (pPVMΔNS1, ΔNS2, and ΔNS12) were constructed, taking advantage of (i) a unique XmaI site present in vector sequence in pPVM immediately upstream of the T7 promoter and viral leader region, (ii) a naturally occurring AvrII site (AY729016 nt 1052) present immediately downstream of the N gene start signal, and (iii) a naturally occurring Acc65I site (AY729016 nt 1654) present further downstream in the N gene (Fig. 1). For each mutant, an XmaI-AvrII (ΔNS1 and ΔNS2) or XmaI-Acc65I (ΔNS12) fragment containing the desired deletion was produced by PCR using pPVM as a template and was used to construct the corresponding mutant pPVM derivative. The PCR fragment for pΔNS1 was made using a mutagenic forward primer that fused the leader region directly to the gene start signal of the NS2 gene (5′-aaacccgggtaatacgactcactatagggACGCGAAAAAATGCATAACAAAACTATCAACCTGAAAAAAGTTAGGACAAGTCCTAATGTCCACAGC TATG-3′; the XmaI restriction site is underlined and in lowercase and is followed by the T7 promoter in lowercase, the PVM sequence is in uppercase and begins with the leader region, the NS2 gene start signal is in boldface, and the NS2 coding sequence is italicized); the reverse primer was located downstream of the above-mentioned AvrII site. The PCR fragment for pΔNS2 was made using a forward primer containing the XmaI restriction site and T7 promoter followed by PVM nt 1 to 71 and a mutagenic reverse primer containing the AvrII restriction site and N gene start signal fused to the NS1-NS2 intergenic region and downstream end of the NS1 gene (5′-aaacccgggCCTAGGATGTGTATTTATCCTACCCTTTGTTTTAATTAACTACATAATGTGCTGTCATCTTAACC-3′; the AvrII site is underlined, the PVM sequence is in uppercase, the complement of the N gene start signal is in boldface, the complement of the NS1 gene end signal is in boldface and underlined, and the NS1 translation stop codon and upstream codons is italicized). Deletion of the NS1 and NS2 genes together employed a mutagenic forward primer that placed the N gene immediately downstream of the leader region (5′-aaacccgggtaatacgactcactatagggACGCGAAAAAATGCATAACAAAACTATCAACCTGAAAAAAGTTAGGATAAATACACATCCTAGGCCGGGCC-3′; the XmaI site is in lowercase and underlined and is followed by the T7 promoter sequence, the PVM sequence is in uppercase, and the N gene start signal is in boldface) and a reverse primer located downstream of the Acc65I restriction site. To make the full-length pΔNS12 construct, the XmaI/Acc65I-digested PCR fragment was cloned directly into the XmaI/Acc65I window of pPVM. In the case of the pΔNS1 and pΔNS2 derivatives, the AvrII site was not unique in pPVM and thus a three-fragment ligation was performed in which the XmaI/AvrII-digested PCR fragment was inserted into the XmaI/Acc65I window of pPVM, together with an AvrII/Acc65I restriction fragment that had been generated separately from pPVM.
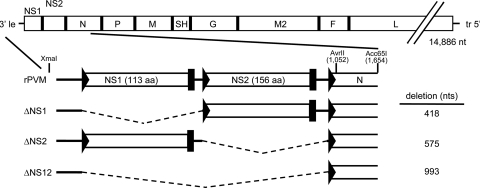
FIG. 1.
Schematic overview of the PVM NS gene-deletion mutants. The wt rPVM genome (based on the strain 15 consensus sequence, GenBank accession no. AY729016) is shown at the top, drawn approximately to scale. The XmaI, AvrII, and Acc651 sites used in the constructions (see Materials and Methods) are shown. The XmaI site is outside of the PVM sequence, directly upstream the T7 promoter (not shown) and 3′ leader sequence (le). In the enlargements, NS1 and NS2 are shown with gene start signals represented by triangles and gene end signals represented by rectangles. The number of deleted nucleotides is shown on the right. tr, 5′ trailer sequence.
rPVM recovery.
Confluent BSR T7/5 cells in six-well dishes were transfected with (per well) 5 μg of the respective pPVM derivative, together with support plasmids expressing the indicated PVM proteins: 2 μg each of pT7-N and pT7-P and 1 μg each of pT7-M2-1 and pT7-L, expressing the respective PVM proteins. Transfections were done with Lipofectamine 2000 (Invitrogen) in OptiMEM without serum and were maintained overnight at 32°C. The transfection medium was removed the next day and replaced with Glasgow MEM containing 3% serum. On day 6, cell-medium mixtures were harvested and passaged onto confluent BHK21 cells. The cells and supernatant were harvested when the cytopathic effect was extensive. The cells were collected by low-speed centrifugation and freeze-thawed three times on dry ice. The suspension was clarified by a second low-speed centrifugation step, and the supernatants were combined with the supernatants from the first centrifugation step, divided into aliquots, and stored at −80°C. Where indicated, viruses were grown in Vero cells, harvested as described above, and subjected to centrifugation through 30/60% (wt/vol) sucrose gradients in order to remove cytokines and other cell-derived material. Titers were determined by plaque assay on Vero or BHK-21 cells as described previously (22). To confirm the presence of the correct mutations, RNA was extracted (RNeasy; Qiagen, Valencia, CA) on day 10 from the second passage in BHK-21 cells and was analyzed by reverse transcription-PCR (RT-PCR) with primers complementary to the PVM leader region and N gene, followed by direct sequencing of the PCR products. This confirmed that the sequences were correct. Biologically derived PVM was analyzed in parallel. Control negative reactions were made that lacked RT enzyme, and the absence of PCR products in these controls showed that the products depended on RNA.
qRT-PCR and ELISA.
The expression of IFN-α, IFN-β, CXCL10, IRF7 mRNA, and PVM genomic RNA in MEF cells was detected by quantitative RT-PCR (qRT-PCR). Triplicate cultures in six-well dishes were mock infected or infected with Vero cell-grown sucrose-purified rPVM, ΔNS1, ΔNS2, or ΔNS12 at an input multiplicity of infection (MOI) of 3 PFU/cell. UV-inactivated ΔNS2 as a control was prepared by irradiation with 240 kJ using a UV Stratalinker 1800 (Stratagene, La Jolla, CA), and the absence of infectious virus was confirmed in Vero cells (not shown). After 2 h of adsorption, the monolayers were washed with medium. At 24 and 60 h postinfection, supernatants were harvested and stored for protein detection assays, and total cellular RNA was extracted from infected cells by using an RNeasy total RNA isolation kit (Qiagen). RNA was treated with DNase I (Qiagen) to remove any contaminating DNA, the absence of which was confirmed in control experiments in which reverse transcriptase was omitted (data not shown). In the case of cellular mRNAs, RT (using Superscript II reverse transcriptase [Invitrogen]) was performed with random primers; in the case of PVM genomic RNA, RT was performed with positive-sense primers hybridizing to the N and F genes. qRT-PCRs were done by using the Brilliant QPCR plus core kit (Stratagene) with PCR primers and TaqMan probes for mouse IFN-β, IFN-α4, CXCL10, IRF7, PVM N and F, and the housekeeping mRNA β-actin, which were designed by using Primer3 (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) software (Table 1). The probe for β-actin was labeled with the reporter dye 5-carboxyfluorescein (FAM) at the 5′ end and black-hole quencher (BHQ1) at the 3′ end. The probes for the genes of interest were labeled with the reporter dye 5′HEX at the 5′ end and BHQ1 at the 3′ end. PCRs for MEF genes of interest were done as duplex PCRs with β-actin as the housekeeping gene (FAM-labeled probe) and gene of interest (HEX-labeled probe). Each result was normalized to the housekeeping mRNA control. In addition, each set of PCRs was run in parallel with a single-standard cDNA preparation made from RNA from MEF cells infected with ΔNS2 and known to contain significant mRNA levels of the genes of interest; this cDNA was run as a 10-fold dilution series that was used to make a standard curve for quantitation. The PVM specific PCRs were done as single reactions (i.e., not duplex). 10-fold dilution standard curves were used for quantitation, and the results were calculated as the fold differences to replicas harvested 24 h postinoculation with UV-inactivated ΔNS2. Commercial ELISA kits were used to determine the concentrations of IFN-α and IFN-β (Invitrogen) and IP-10/CXCL10 (R&D Systems, Minneapolis, MN).
TABLE 1.
qPCR primers and probes
| Primer or probea | Sequence (5′-3′) | Amplicon size (bp) | Accession no. | Probe labelb |
|---|---|---|---|---|
| β-actin f | ACTGCTCTGGCTCCTAGCAC | 115 | NM_007393 | |
| β-actin r | ACATCTGCTGGAAGGTGGAC | |||
| β-actin probe | TCATTGCTCCTCCTGAGCGCAAGTA | FAM | ||
| CXCL10 f | AAGTGCTGCCGTCATTTTCT | 129 | NM_021274 | |
| CXCL10 r | CCTATGGCCCTCATTCTCAC | |||
| CXCL10 probe | ATCCTGCTGGGTCTGAGTGGGACTC | HEX | ||
| IRF7 f | AGCTCCCAGATCAGAAGCAG | 138 | NM_016850 | |
| IRF7 r | TGCCTACCTCCCAGTACACC | |||
| IRF7 probe | CACAGAGACGCTTCTCCAGCATGTG | HEX | ||
| IFN-β f | GCACTGGGTGGAATGAGACT | 112 | NM_010510 | |
| IFN-β r | TCCCACGTCAATCTTTCCTC | |||
| IFN-β probe | CTCCTGGATGAACTCCACCAGCAGA | HEX | ||
| IFN-α4 f | TTCTGCAATGACCTCCATCA | 101 | NM_010504 | |
| IFN-α4 r | TATGTCCTCACAGCCAGCAG | |||
| IFN-α4 probe | TGTGATGCAGGAACCTCCTCTGACC | HEX | ||
| PVM N f† | AGGACACTCGGCATGTTCTT | 132 | AY729016 | |
| PVM N r | GTCCTTGAGCTGTGTGTCCA | |||
| PVM N probe | CAACCGTTGCGAAGAAGTGGCA | HEX | ||
| PVM F f† | AGGACTCTGCCAGATGGTTG | 156 | AY729016 | |
| PVM F r | CAGGGAAACTCAAAGGGTCA | |||
| PVM F probe | CCAACAAAGGTGTGGACAGGGTTCA | HEX |
f, forward; r, reverse. †, Also used as specific primer in the first-strand reaction.
That is, a 5′ fluorophore probe label.
Replication and pathogenicity in BALB/c mice.
Groups of 6-week-old BALB/c mice (Charles River Laboratories, Wilmington, MA) were infected intranasally, under light methofane anesthesia, with 80 μl of L15 medium containing 200 PFU of rPVM, ΔNS1, ΔNS2, or ΔNS12. Clinical observations were conducted daily over a period of 14 days and on day 17 postinoculation (n = 10 mice per group). Disease signs were quantified by using a modification of a clinical scoring system described by Cook et al. (8) (Table 2), and the body weight was measured. Animals were euthanized if weight loss was more than 25% or if severe clinical signs were present. To evaluate virus replication, lungs were harvested on days 3 and 6 postinfection (n = 6 mice per group), and the virus titers of the individual specimens were quantified by titration of tissue homogenates on BHK21 cells.
TABLE 2.
Scores for clinical symptoms of PVM infection in micea
| Scorea | Clinical sign(s) |
|---|---|
| 1 | Healthy with no signs of illness |
| 2 | Consistently ruffled fur, especially on neck |
| 3 | Ruffled fur, breathing may be deeper |
| 4 | Ruffled fur, hunched back, abnormal gait, labored breathing, less alert |
| 5 | Reduced mobility, labored breathing, may show cyanosis of tail and ears, lethargy |
| 6 | Death |
Modified from Cook et al. (8).
Histopathological and immunohistochemical analysis of lung tissues.
Lungs were inflated with 10% buffered formalin, fixed by immersion in formalin, and embedded in paraffin, and 4- to 6-μm sections were processed for staining with hematoxylin and eosin. Lung sections were stained for PVM antigen with a PVM G specific rabbit antiserum (22) at a dilution of 1:8,000 or for F4/80 (a macrophage marker, 1:200) or CD3 (a pan-T-cell marker, 1:400) using rat anti-mouse F4/80 (Serotec, Inc., Raleigh, NC) or rabbit anti-human CD3 (Dako, Carpinteria, CA; note that this polyclonal human-specific antibody preparation cross-reacts efficiently with murine CD3 and was used because it has good reactivity in paraffin sections) as the primary antibodies (44). For F4/80, a secondary biotinylated goat anti-rat (Biocare Medical, Concord, CA) was used as secondary antibody, followed by a horseradish peroxidase-streptavidin complex (Biocare Medical). For CD3 and the PVM G-specific antiserum, a biotinylated goat anti-rabbit secondary antibody (Vector Laboratories, Burlingame, CA) was applied, followed by R.T.U. Vectastain Elite ABC reagent (Vector). Staining was completed with 3,3′-diaminobenzidine chromogen. Antigen retrieval was used prior to PVM, CD3, and F4/80 immunostaining with Diva Solution (Biocare Medical, Inc., Concord, CA) in a food steamer.
RESULTS
Deletion of NS2, but not NS1, reduces replication in IFN-competent MEFs.
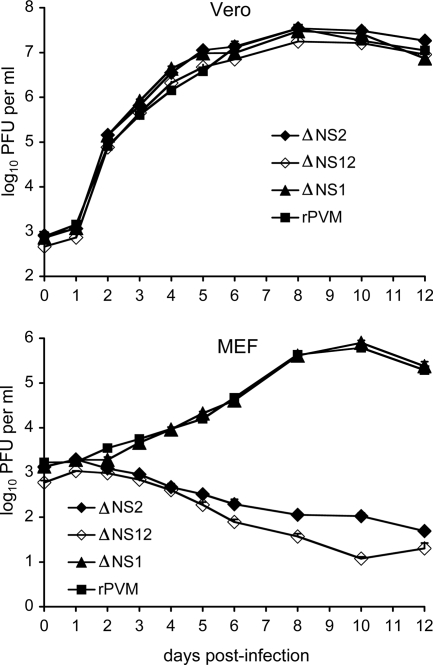
Recombinant PVM mutants were designed in which the NS1 and NS2 genes were deleted individually or together, involving deletions of 418 (ΔNS1), 575 (ΔNS2), or 993 nt (ΔNS12) (Fig. 1). The viruses were readily recovered and propagated. Multicycle growth was evaluated in Vero cells, an African green monkey cell line that lacks functional genes for type I IFN, and in MEF cells, which are competent for producing type I IFN (Fig. 2). The wild-type (wt) and gene deletion viruses replicated in both cell lines with minimal cytopathic effect. In Vero cells, the kinetics and magnitude of replication of the gene deletion mutants were indistinguishable from that of wt rPVM, reaching peak titers on day 8 postinoculation. In MEF cells, viruses lacking the NS2 gene (ΔNS2 and ΔNS12) were highly restricted for replication, such that the titers decreased rather than increased during the course of the infection. In contrast, deletion of NS1 alone had no effect on the replication of PVM in MEF cells. However, ΔNS12 consistently replicated less efficiently than ΔNS2, suggesting that there was a modest additive effect of deleting NS1 in the context of the NS2 deletion.
FIG. 2.
Comparison of the multistep growth kinetics of the PVM gene deletion mutations in Vero cells (top) and CF-1 MEFs (bottom). Monolayer cultures of Vero cells or MEFs were infected at an input MOI of 0.1 PFU per cell with the indicated virus. Supernatant aliquots (0.5 ml out of a total medium volume of 2 ml per well) were taken on the indicated days postinfection and replaced by an equivalent volume of fresh medium. The samples were flash frozen and analyzed later in parallel by plaque assay. Each time point was represented by two wells, and each virus titration was done in duplicate. Means are shown. The standard errors were calculated, but at most time points the bars are not visible because the errors were very small and the bars are obscured by the symbols.
NS2 strongly inhibits type I IFN induction in MEFs.
To investigate possible effects on the host cell type I IFN response, MEF cells were inoculated with the deletion mutants or wt rPVM at an input MOI of 3 PFU/cell or with UV-inactivated ΔNS2 as a control. Medium supernatants and total cellular RNA were harvested 24 and 60 h postinfection. These time points were determined in preliminary experiments in which type I IFN mRNA and protein were not detected significantly at 6 and 12 h postinfection (data not shown).
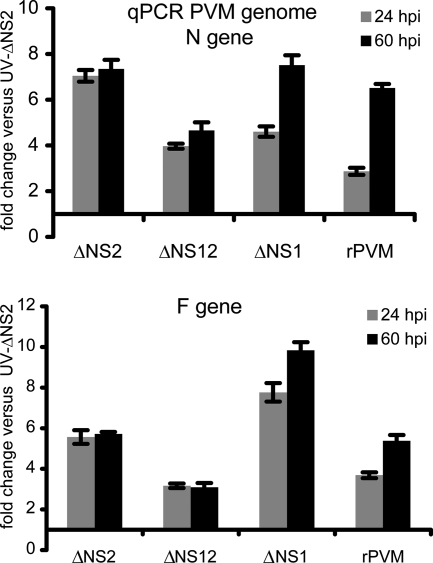
Viral RNA synthesis was monitored by a real-time qRT-PCR assay that used positive-sense RT primers based on the N and F genes and thus was designed to measure genomic RNA (Fig. 3). The level of cell-associated genomic RNA was expressed relative to that of UV-inactivated ΔNS2, which serves as a control to indicate the background level of input genomic RNA contributed by particles adsorbed to cells or internalized by fusion or endocytosis. The amount of intracellular genomic RNA in cells infected with each of the gene deletion viruses was consistently higher than that of cells infected with UV-NS2, indicating that each of the mutants was competent for RNA replication despite the apparent lack of replication of the ΔNS2 and ΔNS12 viruses in MEFs. In cells infected with rPVM or ΔNS1, the amount of genomic RNA increased between 24 and 60 h, indicative of ongoing RNA replication, and the amount of genomic RNA produced by ΔNS1 equaled or exceeded that of rPVM. In contrast, the amount of genomic RNA in cells infected with PVM lacking the NS2 gene (ΔNS2 and ΔNS12) did not increase between 24 and 60 h.
FIG. 3.
qRT-PCR analysis of intracellular PVM genomic RNA from MEFs infected with rPVM or the indicated gene deletion mutants. Replicate wells of MEFs were infected with the indicated virus at an input MOI of 3 PFU/cell and total cell-associated RNA was extracted 24 and 60 h postinfection. Experiments were done in triplicate. UV-inactivated ΔNS2 was used as a negative control. RT-PCR was primed with a positive-sense PVM-specific primer derived from the N or F gene (Table 1). PCR results were normalized to those from wells inoculated UV-inactivated ΔNS2.
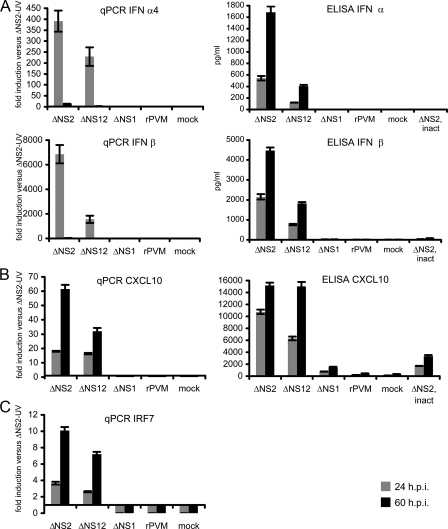
Next, we used qRT-PCR to evaluate the expression of IFN-α4 and IFN-β, which constitute the initial type I IFN response in mice, as well as the expression of CXCL10 and IRF7, which are representative IFN-stimulated genes dependent on JAK/STAT signaling from the type I IFN receptor (3, 14, 15, 17, 28). This was done by duplex qRT-PCR assays with primers and probe specific for the gene of interest together with β-actin specific primers and probe. The results for the genes of interest were normalized to mouse β-actin and are expressed as the fold increase compared to replicate samples from UV-ΔNS2-inoculated cells harvested 24 h postinoculation (Fig. 4A). The levels of expression in UV-ΔN2-infected cells used for comparison were essentially the same as in mock-treated cells (data not shown). Increased expression of IFN-α4 or IFN-β mRNA was not detected at any time point in MEF cells infected with rPVM or ΔNS1. In contrast, high levels of both mRNAs were detected by 24 h postinfection in cells infected with ΔNS2 and ΔNS12. Compared to replicas inoculated with UV-ΔNS2, ca. 400- and 7,000-fold higher levels of IFN-α4 and IFN-β mRNA, respectively, were detected in cells infected with ΔNS2. In cells inoculated with the double deletion mutant ΔNS12, ca. 250- and 1,600-fold higher levels of IFN-α4 and IFN-β mRNA, respectively, were detected. By 60 h postinfection, the levels of both IFN mRNA species had returned to baseline.
FIG. 4.
Expression of IFN-α4, IFN-β, CXCL10, and IRF-7 in MEF cells. Expression of mouse IFN-α and IFN-β (A), the early IFN response gene CXCL10 (B), and the transcription factor IRF7 that functions as amplifier of the type I IFN response (C) was quantified on the mRNA (left panel) and protein (right panel) level. MEFs were infected with the indicated virus at an input MOI of 3 PFU/cell, and cell culture medium supernatants and total cell-associated RNA were harvested 24 and 60 h postinfection (from the experiment shown in Fig. 3). UV-inactivated ΔNS2 served as a negative control. Total RNA was subjected to qRT-PCR using the specific primers and probes listed in Table 1, and the results were normalized to mouse β-actin. The fold increase in mRNA expression was expressed relative to the level of expression in MEFs infected with UV-inactivated ΔNS2 (left panel). The concentrations of IFN-α, IFN-β, and CXCL10 protein in cell culture supernatants were determined by ELISA (right panel).
The secretion of IFN-α and IFN-β from infected MEF cells was monitored by ELISA. Essentially, no type I IFN was detected in medium supernatants from cells that had been mock treated or infected with rPVM, ΔNS1, or the UV-ΔNS2 control. In contrast, medium supernatants from cells infected with ΔNS2 or ΔNS12 contained 550 and 120 pg/ml, respectively, of IFN-α at 24 h. These levels increased strongly by 60 h postinfection, a time when the accumulation of intracellular type I IFN mRNA had returned to background levels. Similarly, the secretion of IFN-β was detected only for infectious virus lacking the NS2 gene, and detection of the protein lagged behind the mRNA. The finding that UV-inactivated ΔNS2 induced only baseline levels of type I IFN indicates a dependence on viral RNA replication.
As representative IFN-stimulated genes, the expression of the CXCL10 and IRF7 mRNAs was measured by qRT-PCR, and the secretion of CXCL10 protein was confirmed by ELISA (Fig. 4B and C). The accumulation of CXCL10 and IRF7 mRNA remained at baseline levels in cells infected with rPVM and ΔNS1 but was strongly induced in response to ΔNS2 and ΔNS12 (Fig. 4B and C). As was the case with IFN-α4 and IFN-β mRNAs, the magnitude of expression was higher in ΔNS2-infected cells than in ΔNS12-infected cells. Compared to IFN-α4 and IFN-β mRNAs, the accumulation of CXCL10 and IRF7 mRNA was increased and prolonged: whereas levels of the two IFN mRNAs had returned to baseline levels by 60 h postinfection, the levels of CXCL10 and IRF7 mRNAs increased ∼3-fold from 24 to 60 h postinfection (Fig. 4B and C). This pattern probably reflects the sequential timing of the induction of the initial type I IFN species versus that of the IFN-stimulated genes. The slightly weaker induction of IFN and IFN-stimulated genes by ΔNS12 compared to ΔNS2 may be a reflection of its somewhat lower level of RNA replication as detected by qPCR (Fig. 3).
NS2 and NS1 are pathogenicity factors in mice.
Groups of BALB/c mice were infected intranasally with 200 PFU of each virus and animals were sacrificed on days 3 and 6 to measure pulmonary virus titers (Table 3). wt rPVM reached a titer of 5.6 log10 PFU per g of lung on day 3 postinoculation. Interestingly, the titers of ΔNS1 was nearly as high, at 5.4 log10 PFU per g, whereas the titers of ΔNS2 and ΔNS12 were somewhat lower (up to 10-fold) at 4.8 and 4.6 log10 PFU per g, respectively. All of the mice appeared to be healthy until the final sacrifice day (day 6 postinoculation), when clinical signs (ruffled fur, hunched backs) were detected in four of six mice inoculated with rPVM but not in any of the animals inoculated with the gene deletion viruses. Between days 3 and 6, the pulmonary titer of rPVM and ΔNS1 increased approximately 40-fold (to 7.2 log10) and 13-fold (to 6.5 log10), respectively, whereas the titers of the recombinants lacking NS2 were three- to fourfold lower on day 6 than on day 3. The day 6 titers of ΔNS2 and ΔNS12 were approximately 1,000-fold lower than those of rPVM.
TABLE 3.
Level of replication of gene deletion rPVMs in mouse lungs
| Virusa | Day p.i.b | Mean titer (log10 PFU/g of tissue ± SD) and statistical significancec | Reduction of mean titerd (log10 PFU/g of tissue) |
|---|---|---|---|
| ΔNS2 | 3 | 4.8 ± 0.3AB | 0.8 |
| ΔNS12 | 3 | 4.6 ± 0.1B | 1.0 |
| ΔNS1 | 3 | 5.4 ± 0.2AC | 0.2 |
| rPVM | 3 | 5.6 ± 0.4C | |
| ΔNS2 | 6 | 4.3 ± 0.2D | 2.9 |
| ΔNS12 | 6 | 4.0 ± 0.3D | 3.2 |
| ΔNS1 | 6 | 6.5 ± 0.1E | 0.7 |
| rPVM | 6 | 7.2 ± 0.2F |
Mice in groups of six were administered recombinant PVMs intranasally under light anesthesia on day 0.
Animals were sacrificed on the indicated days postinfection (p.i.).
Nasal turbinates and lungs were harvested, and virus titers were determined. The lower limit of detection for virus in the upper and lower respiratory tract was 1.7 log10 PFU/g of tissue. Statistical differences of mean virus titers compared to rPVM were determined on day 3 and day 6 postinfection by analysis of variance and the Tukey-Kramer post-hoc test. Values that share a superscript letter are not statistically different (P < 0.05).
That is, the reduction in mean titer compared to rPVM.
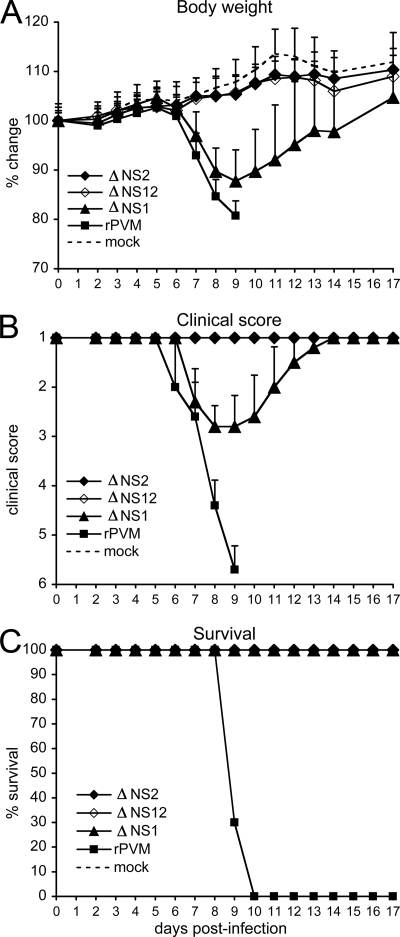
Morbidity and mortality were examined more thoroughly in a second study in which mice in groups of 10 were infected with 200 PFU of the various viruses and evaluated daily for 17 days for body weight (Fig. 5) and for disease assessed using a clinical scoring system (Table 2 and Fig. 5). Consistent with the first experiment, clinical disease signs and a reduction in body weight were first apparent on day 6 in animals infected with wt rPVM, respectively, whereas no disease or weight loss was evident at that time in animals infected with the various gene deletion viruses (Fig. 5). On day 7, most of the rPVM-infected animals showed ruffled fur and deeper or labored breathing. By day 9, seven of the ten rPVM-infected animals had to be euthanized due to severe clinical disease involving reduced mobility, hunching, and severe weight loss; the remaining three rPVM-infected animals had to be euthanized the following day. The ΔNS1 virus induced mild to moderate clinical disease and weight loss beginning on day 7 and peaking on day 9, with the maximum weight loss averaging 15% compared to mock-infected animals. Subsequently, disease signs abated by day 12 in all of the animals inoculated with ΔNS1 recovered, and all of the animals regained body weight. For the animals infected with ΔNS2 and ΔNS12, none developed any clinical signs or showed any reduction in body weight, although they gained weight more slowly than the mock-infected controls.
FIG. 5.
Pathogenicity of the NS gene deletion mutants in mice. Mice (n = 10) were inoculated on day 0 with 200 PFU of rPVM or the indicated gene deletion mutant. (A) The mice were weighed on the indicated days, and the percent body weight change was calculated, compared to the mean weight on day 0. (B) The clinical signs were assessed daily according to a score presented in Table 1. (C) The percent survival rate was calculated for rPVM and the gene-deletion mutants.
Cytokine profiles in mice infected with the gene deletion viruses.
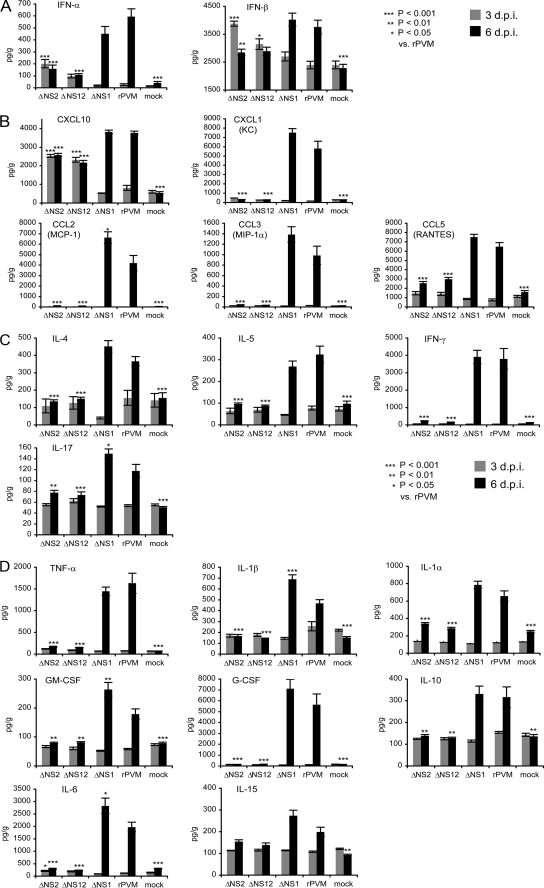
Clarified supernatants were prepared from lung homogenates from the first mouse study described above (harvested 3 and 6 days postinoculation) and analyzed by ELISA to measure the levels of IFN-α and IFN-β (Fig. 6A) and by a bead-based immunoassay to measure 22 additional cyokines and chemokines, namely, interleukin-1α (IL-1α), IL-1β, IL-4, IL-5, IL-6, IL-10, IL-13, IL-15, IL-17, tumor necrosis factor alpha, gamma interferon (IFN-γ), granulocyte-macrophage colony-stimulating factor, CCL2 (MCP-1), CCL3 (MIP-1α), CCL5 (RANTES), granulocyte colony-stimulating factor, CXCL10, CXCL1 (KC), IL-2, IL-7, IL-9, and IL-12 (Fig. 6B to D).
FIG. 6.
Expression of representative cytokines and chemokines in the lungs of mice infected with rPVM or the gene deletion mutants. Mice were infected with 200 PFU of the indicated virus, and clarified supernatants were prepared from lung homogenates obtained on days 3 and 6 postinfection, from the experiment shown in Table 3. The supernatants were analyzed by ELISA to determine the concentrations of IFN-α and IFN-β and by a bead-based immunoassay (Lincoplex mouse cytokine/chemokine kit; LincoResearch, St. Charles, MO) to determine the concentrations for 22 mouse chemokine/cytokine proteins using a Luminex 100 reader (Luminexcorp, Austin, TX). IL-2, IL-7, IL-9, IL-12, and IL-13 were not detected. The panels show the responses of IFN-α and IFN-β (A); chemokines (B); cytokines produced predominantly by T lymphocytes (C); and cytokines produced variously by epithelial cells, macrophages, and other cell types (D). Note that CXCL1 (also known as KC) shares functional properties with IL-8, which has no structural homolog in mice. Statistical significance was calculated by one way analysis of variance tests and Tukey-Kramer post-hoc analyses. Columns marked with asterisks differ significantly (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001) from rPVM at the same time point. The statistical significance compared to mock-inoculated mice is not shown, but note that, on day 3, none of the cytokines/chemokines were significantly upregulated for rPVM or ΔNS1 compared to mock-infected mice, whereas IFN-α and IFN-β, CXCL10, and IL-6 were significantly upregulated for ΔNS2 and ΔNS12.
This analysis segregated the various PVMs into two groups with regard to the pattern of cytokine induction. In the first group, consisting of rPVM and ΔNS1, there was no increase in pulmonary IFN-α and IFN-β compared to mock-inoculated mice on day 3 despite the considerable pulmonary titers of rPVM and ΔNS1 (Fig. 6A). Increases also were not observed on day 3 for any of the 22 cytokines and chemokines analyzed in addition to type I IFN (Fig. 6B to D). The picture was very different on day 6 postinoculation, which was the first day that clinical disease was evident in rPVM-infected mice and was 1 day before disease was observed in ΔNS1-infected mice. High levels of type I IFN were present in the lungs of mice infected with rPVM or the ΔNS1 virus (Fig. 6A). In addition, of the 22 additional cytokines and chemokines that were evaluated, 17 were significantly higher in rPVM- and ΔNS1-infected animals compared to mock-infected controls (representative cytokines are shown in Fig. 6B to D; the other five cytokines in the panel, IL-2, IL-7, IL-9, IL-12, and IL-13, were not detected in the mock- or virus-infected samples. The factors that were increased included a number of proinflammatory chemokines produced by epithelial cells and macrophages, including the CC chemokines CCL2 (MCP-1), CCL3 (MIP-1α), and CCL5 (RANTES) and the CXC chemokines CXCL1 (KC) and CXCL10 (Fig. 6B). The increased factors also included several cytokines that are predominantly produced by T lymphocytes: IL-4 (T helper [Th] 2), IL-5 (Th2), IL-17 (Th17 (1, 13), and IFN-γ that is produced by virus-specific CD4 Th1 and CD8 T cells and NK cells (Fig. 6C). The increased factors also included a number of proinflammatory cytokines variously produced by epithelial cells, macrophages, and other cell types, namely, IL-1β, IL-1α, IL-6, IL-15, and tumor necrosis factor alpha; granulocyte colony-stimulating factor, which is produced primarily by monocytes and macrophages; granulocyte-macrophage colony-stimulating factor, which is produced by a many cell types, including lung epithelial cells, T cells and macrophages; and IL-10, which inhibits inflammation by negatively regulating several inflammatory mediators (9, 37) and is produced by activated T cells, B cells, monocytes/macrophages, and epithelial cells (Fig. 6D). Thus, by day 6 postinfection, rPVM and ΔNS1 induced a strong production of cytokines/chemokines with strong proinflammatory effects by an array of different cell types, as well as T cell-related cytokines.
In contrast, the second group of viruses, ΔNS2 and ΔNS12, induced significant increases in the expression of four factors compared to mock-inoculated animals on day 3 postinoculation, a time when no increases were observed in the first group, as noted above. These were IFN-α (Fig. 6A), IFN-β (Fig. 6A), CXCL10 (Fig. 6B), and IL-6 (Fig. 6D). By day 6, the levels of IFN-α, IFN-β, and IL-6 had largely returned to baseline. Even at their maximum (on day 3), the levels of IFN-α and IL-6 induced by ΔNS2 and ΔNS12 were considerably below the maximum observed (on day 6) for rPVM and ΔNS1. In addition, infection with ΔNS2 and ΔNS12 did not induce increased expression of any of the other 20 cytokines and chemokines that were examined (Fig. 6B to D). As noted, neither ΔNS2 nor ΔNS12 induced clinical disease on any day.
Histopathology in BALB/c mice.
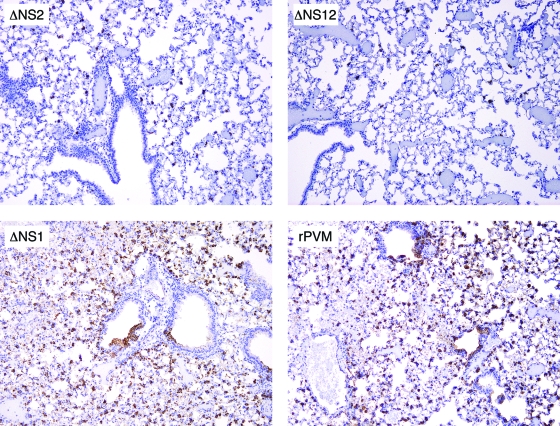
On day 3 postinfection, several small PVM-positive foci were present throughout the lungs in all PVM-infected groups (not shown). Few small perivascular and peribronchial foci of lymphocytes and macrophages were detected. On day 6 postinfection, lung sections of ΔNS2- and ΔNS12-infected mice looked very similar to those from the earlier time point, both with respect to low numbers of antigen-positive cells and few mononuclear infiltrates (Fig. 7). In the lungs of rPVM- and ΔNS1-infected mice, abundant PVM-positive cells were detected on day 6. Bronchiolar epithelial cells and alveolar cells of macrophage and type II pneumocyte morphology were PVM positive. Immunostaining with a macrophage specific antibody (F4/80) confirmed an increase of macrophages in the alveoli of rPVM- and ΔNS1-infected mice on day 6 (data not shown). Histopathological lesions included perivascular and peribronchial inflammation, with perivascular cuffs of lymphocytes (confirmed by staining with a T-cell marker [CD3, not shown]), neutrophils, and macrophages, as well as a few eosinophils. The abundance of antigen-positive cells in rPVM- and ΔNS1-infected mice on day 6 was remarkable. Even though the amount of necrotic cells was relatively low at this time point, direct viral cytopathology and cellular infiltrates might compromise lung function and contribute to the onset of disease evident beginning on this day.
FIG. 7.
Immunohistology for PVM antigens. Lungs of mice infected with rPVM, ΔNS1, ΔNS2, or ΔNS12 were harvested on day 6 postinfection and were formalin fixed. Paraffin sections were stained with hematoxylin-eosin and antibody specific to PVM. Representative sections are shown for each virus. Note the abundant viral antigen (brown color) in NS1 and rPVM lungs and much less viral antigen in the lungs of NS2- and NS12-infected mice (hematoxylin counterstain; magnification, ×100).
DISCUSSION
PVM, the murine counterpart of HRSV, causes fatal respiratory tract disease in BALB/c mice and provides a convenient surrogate model for severe HRSV disease. In the present study, we investigated the effects of deleting the PVM NS1 and NS2 genes on infection in vitro and in vivo. The deletion of NS1 and NS2 individually or together had no effect on the efficiency of PVM replication in IFN-lacking Vero cells, indicating that, at least by this measure, the two proteins did not have discernible direct roles in the viral replicative cycle. However, deletion of the NS2 gene, alone or in combination with the deletion of NS1, was highly attenuating in IFN-competent MEF cells. The ΔNS2 (and ΔNS12) mutant induced high levels of IFN-α/β in MEF cells, whereas IFN-α/β was not detected in cultures infected with wt PVM. In addition, ΔNS2 (and ΔNS12), but not wt PVM, induced increased expression in MEF of two representative IFN-regulated genes, namely, CXCL10, which is an important chemoattractant for monocytes, NK cells, and T lymphocytes (10, 25, 40, 48), and IRF7, which is a key regulator involved in amplifying and broadening the type I IFN response (15). Thus, IFN production and signaling were functional in PVM-infected cells in the absence of NS2, and were inhibited to below the level of detection in the presence of NS2. This identified PVM NS2 as a highly effective IFN antagonist, although the level(s) at which the IFN system is blocked and the mechanism(s) by which this occurs remains to be determined.
In contrast, it is not yet clear whether PVM NS1 plays a role in inhibiting IFN induction or signaling. Deletion of NS1 alone did not affect the ability of PVM to replicate in MEF cells capable of strong IFN responses. Furthermore, deletion of NS1 alone did not result in increased IFN induction or signaling in MEF cells. Third, deletion of NS1 in combination with NS2 did not provide a further increase in IFN production or signaling in addition to that afforded by deletion of NS2 alone. Indeed, IFN induction and signaling were somewhat reduced for ΔNS12 compared to ΔNS2. This might reflect the reduced level of intracellular viral RNA synthesis observed for ΔNS12 compared to ΔNS2, which would provide for reduced activation of the RIG-I-like cytoplasmic pattern recognition receptors that detect viral RNA and initiate signaling through IRF3 and NF-κB. However, when NS1 was deleted in combination with NS2, virus replication in MEFs was further reduced (but only slightly) compared to deletion of NS2 alone. Also, we did not directly evaluate IFN signaling. Therefore, the possibility remains for a minor effect of NS1 in antagonizing the host IFN system, mostly likely in inhibiting type I IFN signaling. This will be investigated in further work.
The identification of NS2 as the predominant IFN antagonist for PVM differs somewhat from the situation with BRSV, for which both NS1 and NS2 appeared to play easily demonstrated roles in blocking IFN induction, with NS2 playing a greater role than NS1 (41). For HRSV, both proteins played a role in blocking IFN induction, but their relative contributions were reversed, and NS1 played the greater role (33). However, in mice, it is the NS2 protein of HRSV that is the major antagonist (20). Thus, there are host and virus species-specific differences in whether both NS proteins have major roles in blocking IFN induction and as to which protein plays the greater role.
Intranasal inoculation of BALB/c mice with a small (200 PFU) dose of wt rPVM resulted in uniformly lethal pneumonia within 9 days. We found that deletion of NS2 alone or in combination with NS1 completely attenuated PVM for mice. No clinical disease signs were observed, and the mice gained weight over the course of the experiment, albeit somewhat more slowly than the mock-infected controls. In comparison, even though deletion of NS1 was not attenuating in vitro, it was significantly attenuating in mice with respect to both disease and virus titer. Specifically, mice infected with ΔNS1 experienced transient pneumonia and weight loss, with a peak on days 8 and 9, from which they appeared to recover fully. Thus, NS2 is a major factor in PVM pathogenesis, presumably due to its ability to suppress the host type I IFN system. NS1 is a second, less-prominent pathogenesis factor whose mechanism of action remains to be clearly defined.
The divergence in outcomes between infections of mice with avirulent ΔNS2 (and ΔNS12) versus highly virulent wt PVM (or the intermediately attenuated ΔNS1 virus) appeared to begin around day 3 postinfection. At that time, the pulmonary virus titer in ΔNS2-infected animals was only 6-fold lower than that of animals infected with wt PVM (4.8 versus 5.6 log10 PFU/g). However, in mice infected with ΔNS2 (or ΔNS12), there was significant pulmonary expression of IFN-α/β, CXCL10, and IL-6 at this time point compared to mock-inoculated animals, whereas no such response was evident on day 3 with wt PVM or ΔNS1. This early cytokine/chemokine response in mice infected with ΔNS2 (and ΔNS12) presumably restricted virus replication through establishment of an IFN-mediated intracellular antiviral state (31), through IFN enhancement of innate and adaptive immunity, and through other effects such as the antiviral activities of CXCL10 (10, 16, 25, 40, 43, 47, 48). As a consequence, by day 6, the titer of ΔNS2 (and ΔNS12) was reduced severalfold, the cytokine/chemokine response was waning, and disease was prevented. This identified a protective, nonpathogenic innate response, namely, one that occurred relatively early (day 3) and consisted of moderate levels of IFN-α/β, as well as a limited number of cytokines and chemokines, namely, 2 (CXCL10 and IL-6) of a total of 22 analyzed.
In contrast, in animals infected with wt PVM (or ΔNS1) the pulmonary virus titers continued to increase following day 3 and were 40-fold higher on day 6, when overt disease became evident. Whereas there had not been a detectable cytokine/chemokine response on day 3, by day 6 there was a heightened, broader response: for example, the level of IFN-α for wt PVM on day 6 was two- to threefold higher than the peak for ΔNS2 on day 3, and 18 of 22 analyzed cytokines/chemokines were induced on day 6 for wt rPVM versus 2 of 22 at any time for ΔNS2. It seems evident that the expression of NS2 in wt rPVM-infected cells was able to efficiently inhibit the innate cytokine/chemokine response during the first several days of infection. However, as infection progressed, substantial virus production and cell damage probably occurred sufficient to activate innate responses that could not be contained by NS2. This might involve signaling through Toll-like receptors, as well as in noninfected dendritic cells, macrophages, and epithelial cells in which NS2 was not expressed. Consistent with Toll-like receptor signaling, promoter analyses showed that most of the cytokines and chemokines detected on day 6 postinfection with rPVM are under the control of NF-κB. Thus, it may have been a case of too much too late; specifically, after several days during which the NS2 protein restrained the innate response, a high level of pulmonary virus and cell damage finally triggered an exaggerated and broadened inflammatory response that occurred too late to prevent disease. Indeed, it likely enhanced disease, since the presence of high levels of viral macromolecules and damaged cells provided for continued exaggerated innate immune stimulation as well as activation of incoming leukocytes, exacerbating pathogenesis in airways already damaged by direct viral cytopathology. It is noteworthy that ΔNS1 closely resembled wt rPVM with regard to the absence of a significant cytokine/chemokine response on day 3 and the presence of a heightened, broad response on day 6: thus, NS1 did not appear to have a significant impact on the expression of the cytokines and chemokines that were analyzed.
We were surprised to find that the peak amount of IFN induced by wt rPVM (and ΔNS1) was substantially greater than for ΔNS2 or ΔNS12, since we had anticipated that deletion of an IFN antagonist would result in higher levels of IFN. Initially, we hypothesized that this late, high expression of IFN-α/β in response to wt PVM (and ΔNS1) might result from plasmacytoid dendritic cells recruited to the infected lung. However, a recent study using transgenic mice with green fluorescent protein as a reporter gene for the IFN-α6 promoter provided evidence that macrophages and conventional dendritic cells, and not plasmacytoid dendritic cells, were the primary IFN-producing cells during viral infection of the respiratory tract (23). Presumably, the source of IFN came from expanded infection of macrophages and conventional dendritic cells, as well as epithelial cells. Although type I IFN is usually considered to be protective, clinical IFN treatment can cause systemic malaise, and there is recent evidence that it also may enhance viral pathogenicity: for example, in IFN-αβR−/− mice, PVM replication was higher than in control mice, but the inflammatory response and pathogenesis was reduced (12). Thus, the heightened IFN response observed on day 6 with wt rPVM and ΔNS1 may contribute to the observed disease.
These results may have relevance for understanding severe HRSV disease in infants and young children. In particular, the profile of proinflammatory cytokines/chemokines detected on day 6 in mice infected with wt rPVM (and ΔNS1) is similar to that present during severe pediatric HRSV infection, and there also has been evidence of extensive HRSV replication in some cases of fatal HRSV disease in infants (reviewed in reference 5). Numerous studies have investigated the basis of pneumovirus pathogenesis in experimental animals (reviewed in reference 5), but individual studies have typically focused on individual factors, such as CD8+ or CD4+ T lymphocytes, or the inflammatory response, or direct virus cytopathology. At least in the present PVM/mouse model, three factors appeared to be associated with the onset of fatal disease: a high (and seemingly poorly controlled) level of virus replication, a heightened proinflammatory response, and a heightened T-cell response. The present results suggest that, in wt PVM infection, the NS2 protein appears to strongly inhibit the host innate response during the early days of infection. This permits extensive viral infection and replication that finally induces a belated, exaggerated innate response and a delayed, robust influx of leukocytes that are strongly activated by high levels of viral antigen in the infected lung. This response probably has the capability to restrict viral replication, but it occurs too late to prevent disease and indeed likely exacerbates disease. It is not unreasonable to speculate that a similar host response of too much too late might account for the severe disease observed in some HRSV-infected infants.
Acknowledgments
We thank Kim Tran and Lijuan Yang for technical assistance during initial experiments and David Stephany of the Flow Cytometry Section, NIAID, for assistance with the Luminex assays. We thank Elizabeth M. Williams, Lawrence J. Faucette, and Lily I. Cheng for technical support for necropsy and immunohistochemistry. We thank the staff of the animal facility, Bldg. 14BS, for care of the mice used in this study.
This research was supported by the Intramural Research Program of NIAID, NIH. The work of C.D.K. was supported by a grant of the Deutsche Forschungsgemeinschaft (Kr 2964/1-2).
Footnotes
Published ahead of print on 3 December 2008.
REFERENCES
- 1.Aujla, S. J., P. J. Dubin, and J. K. Kolls. 2007. Th17 cells and mucosal host defense. Semin. Immunol. 19377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buttmann, M., F. Berberich-Siebelt, E. Serfling, and P. Rieckmann. 2007. Interferon-beta is a potent inducer of interferon regulatory factor-1/2-dependent IP-10/CXCL10 expression in primary human endothelial cells. J. Vasc Res. 4451-60. [DOI] [PubMed] [Google Scholar]
- 4.Cerutti, A., X. Qiao, and B. He. 2005. Plasmacytoid dendritic cells and the regulation of immunoglobulin heavy chain class switching. Immunol. Cell Biol. 83554-562. [DOI] [PubMed] [Google Scholar]
- 5.Collins, P. L., and B. S. Graham. 2008. Viral and host factors in human respiratory syncytial virus pathogenesis. J. Virol. 822040-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, P. L., and B. R. Murphy. 2005. New generation live vaccines against human respiratory syncytial virus designed by reverse genetics. Proc. Am. Thorac Soc. 2166-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, P. L., S. S. Whitehead, A. Bukreyev, R. Fearns, M. N. Teng, K. Juhasz, R. M. Chanock, and B. R. Murphy. 1999. Rational design of live-attenuated recombinant vaccine virus for human respiratory syncytial virus by reverse genetics. Adv. Virus Res. 54423-451. [DOI] [PubMed] [Google Scholar]
- 8.Cook, P. M., R. P. Eglin, and A. J. Easton. 1998. Pathogenesis of pneumovirus infections in mice: detection of pneumonia virus of mice and human respiratory syncytial virus mRNA in lungs of infected mice by in situ hybridization. J. Gen. Virol. 79(Pt. 10)2411-2417. [DOI] [PubMed] [Google Scholar]
- 9.Couper, K. N., D. G. Blount, and E. M. Riley. 2008. IL-10: the master regulator of immunity to infection. J. Immunol. 1805771-5777. [DOI] [PubMed] [Google Scholar]
- 10.Dufour, J. H., M. Dziejman, M. T. Liu, J. H. Leung, T. E. Lane, and A. D. Luster. 2002. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T-cell generation and trafficking. J. Immunol. 1683195-3204. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, J., O. T. Lynch, Y. Suessmuth, P. Qian, C. R. Boyd, J. F. Burrows, R. Buick, N. J. Stevenson, O. Touzelet, M. Gadina, U. F. Power, and J. A. Johnston. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J. Virol. 813428-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garvey, T. L., K. D. Dyer, J. A. Ellis, C. A. Bonville, B. Foster, C. Prussin, A. J. Easton, J. B. Domachowske, and H. F. Rosenberg. 2005. Inflammatory responses to pneumovirus infection in IFN-alpha beta R gene-deleted mice. J. Immunol. 1754735-4744. [DOI] [PubMed] [Google Scholar]
- 13.Harrington, L. E., R. D. Hatton, P. R. Mangan, H. Turner, T. L. Murphy, K. M. Murphy, and C. T. Weaver. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 61123-1132. [DOI] [PubMed] [Google Scholar]
- 14.Hilkens, C. M., J. F. Schlaak, and I. M. Kerr. 2003. Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J. Immunol. 1715255-5263. [DOI] [PubMed] [Google Scholar]
- 15.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434772-777. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh, M. F., S. L. Lai, J. P. Chen, J. M. Sung, Y. L. Lin, B. A. Wu-Hsieh, C. Gerard, A. Luster, and F. Liao. 2006. Both CXCR3 and CXCL10/IFN-inducible protein 10 are required for resistance to primary infection by dengue virus. J. Immunol. 1771855-1863. [DOI] [PubMed] [Google Scholar]
- 17.Indraccolo, S., U. Pfeffer, S. Minuzzo, G. Esposito, V. Roni, S. Mandruzzato, N. Ferrari, L. Anfosso, R. Dell'Eva, D. M. Noonan, L. Chieco-Bianchi, A. Albini, and A. Amadori. 2007. Identification of genes selectively regulated by IFNs in endothelial cells. J. Immunol. 1781122-1135. [DOI] [PubMed] [Google Scholar]
- 18.Jin, H., X. Cheng, V. L. Traina-Dorge, H. J. Park, H. Zhou, K. Soike, and G. Kemble. 2003. Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine 213647-3652. [DOI] [PubMed] [Google Scholar]
- 19.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273210-218. [DOI] [PubMed] [Google Scholar]
- 20.Kotelkin, A., I. M. Belyakov, L. Yang, J. A. Berzofsky, P. L. Collins, and A. Bukreyev. 2006. The NS2 protein of human respiratory syncytial virus suppresses the cytotoxic T-cell response as a consequence of suppressing the type I interferon response. J. Virol. 805958-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krempl, C. D., E. W. Lamirande, and P. L. Collins. 2005. Complete sequence of the RNA genome of pneumonia virus of mice (PVM). Virus Genes. 30237-249. [DOI] [PubMed] [Google Scholar]
- 22.Krempl, C. D., A. Wnekowicz, E. W. Lamirande, G. Nayebagha, P. L. Collins, and U. J. Buchholz. 2007. Identification of a novel virulence factor in recombinant pneumonia virus of mice. J. Virol. 819490-9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumagai, Y., O. Takeuchi, H. Kato, H. Kumar, K. Matsui, E. Morii, K. Aozasa, T. Kawai, and S. Akira. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27240-252. [DOI] [PubMed] [Google Scholar]
- 24.Le Bon, A., C. Thompson, E. Kamphuis, V. Durand, C. Rossmann, U. Kalinke, and D. F. Tough. 2006. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J. Immunol. 1762074-2078. [DOI] [PubMed] [Google Scholar]
- 25.Lindell, D. M., T. E. Lane, and N. W. Lukacs. 2008. CXCL10/CXCR3-mediated responses promote immunity to respiratory syncytial virus infection by augmenting dendritic cell and CD8+ T-cell efficacy. Eur. J. Immunol. 382168-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo, M. S., R. M. Brazas, and M. J. Holtzman. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 799315-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 176660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petry, H., L. Cashion, P. Szymanski, O. Ast, A. Orme, C. Gross, M. Bauzon, A. Brooks, C. Schaefer, H. Gibson, H. Qian, G. M. Rubanyi, and R. N. Harkins. 2006. Mx1 and IP-10: biomarkers to measure IFN-β activity in mice following gene-based delivery. J. Interferon Cytokine Res. 26699-705. [DOI] [PubMed] [Google Scholar]
- 29.Ramaswamy, M., L. Shi, S. M. Varga, S. Barik, M. A. Behlke, and D. C. Look. 2006. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344328-339. [DOI] [PubMed] [Google Scholar]
- 30.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 31.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 748234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 784363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 795353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stetson, D. B., and R. Medzhitov. 2006. Type I interferons in host defense. Immunity 25373-381. [DOI] [PubMed] [Google Scholar]
- 36.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defense. Cell Microbiol. 8907-922. [DOI] [PubMed] [Google Scholar]
- 37.Tebo, J. M., H. S. Kim, J. Gao, D. A. Armstrong, and T. A. Hamilton. 1998. Interleukin-10 suppresses IP-10 gene transcription by inhibiting the production of class I interferon. Blood 924742-4749. [PubMed] [Google Scholar]
- 38.Teng, M. N., and P. L. Collins. 1999. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J. Virol. 73466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng, M. N., S. S. Whitehead, A. Bermingham, M. St Claire, W. R. Elkins, B. R. Murphy, and P. L. Collins. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 749317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trifilo, M. J., C. Montalto-Morrison, L. N. Stiles, K. R. Hurst, J. L. Hardison, J. E. Manning, P. S. Masters, and T. E. Lane. 2004. CXC chemokine ligand 10 controls viral infection in the central nervous system: evidence for a role in innate immune response through recruitment and activation of natural killer cells. J. Virol. 78585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valarcher, J. F., J. Furze, S. Wyld, R. Cook, K. K. Conzelmann, and G. Taylor. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 778426-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Pesch, V., H. Lanaya, J. C. Renauld, and T. Michiels. 2004. Characterization of the murine alpha interferon gene family. J. Virol. 788219-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh, K. B., R. A. Edwards, K. M. Romero, M. V. Kotlajich, S. A. Stohlman, and T. E. Lane. 2007. Expression of CXC chemokine ligand 10 from the mouse hepatitis virus genome results in protection from viral-induced neurological and liver disease. J. Immunol. 1791155-1165. [DOI] [PubMed] [Google Scholar]
- 44.Ward, J. M., C. R. Erexson, L. J. Faucette, J. F. Foley, C. Dijkstra, and G. Cattoretti. 2006. Immunohistochemical markers for the rodent immune system. Toxicol. Pathol. 34616-630. [DOI] [PubMed] [Google Scholar]
- 45.Whitehead, S. S., A. Bukreyev, M. N. Teng, C. Y. Firestone, M. St Claire, W. R. Elkins, P. L. Collins, and B. R. Murphy. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 733438-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright, P. F., R. A. Karron, S. A. Madhi, J. J. Treanor, J. C. King, A. O'Shea, M. R. Ikizler, Y. Zhu, P. L. Collins, C. Cutland, V. B. Randolph, A. M. Deatly, J. G. Hackell, W. C. Gruber, and B. R. Murphy. 2006. The interferon antagonist NS2 protein of respiratory syncytial virus is an important virulence determinant for humans. J. Infect. Dis. 193573-581. [DOI] [PubMed] [Google Scholar]
- 47.Zeng, X., T. A. Moore, M. W. Newstead, J. C. Deng, S. L. Kunkel, A. D. Luster, and T. J. Standiford. 2005. Interferon-inducible protein 10, but not monokine induced by gamma interferon, promotes protective type 1 immunity in murine Klebsiella pneumoniae pneumonia. Infect. Immun. 738226-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng, X., T. A. Moore, M. W. Newstead, J. C. Deng, N. W. Lukacs, and T. J. Standiford. 2005. IP-10 mediates selective mononuclear cell accumulation and activation in response to intrapulmonary transgenic expression and during adenovirus-induced pulmonary inflammation. J. Interferon Cytokine Res. 25103-112. [DOI] [PubMed] [Google Scholar]