Abstract
The multifunctional large (L) polymerase protein of vesicular stomatitis virus (VSV) contains enzymatic activities essential for RNA synthesis, including mRNA cap addition and polyadenylation. We previously mapped amino acid residues G1154, T1157, H1227, and R1228, present within conserved region V (CRV) of L, as essential for mRNA cap addition. Here we show that alanine substitutions to these residues also affect 3′-end formation. Specifically, the cap-defective polymerases produced truncated transcripts that contained A-rich sequences at their 3′ termini and predominantly terminated within the first 500 nucleotides (nt) of the N gene. To examine how the cap-defective polymerases respond to an authentic VSV termination and reinitiation signal present at each gene junction, we reconstituted RNA synthesis using templates that contained genes inserted (I) at the leader-N gene junction. The I genes ranged in size from 382 to 1,098 nt and were typically transcribed into full-length uncapped transcripts. In addition to lacking a cap structure, the full-length I transcripts synthesized by the cap-defective polymerases lacked an authentic polyadenylate tail and instead contained 0 to 24 A residues. Moreover, the cap-defective polymerases were also unable to copy efficiently the downstream gene. Thus, single amino acid substitutions in CRV of L protein that inhibit cap addition also inhibit polyadenylation and sequential transcription of the genome. In contrast, an amino acid substitution, K1651A, in CRVI of L protein that completely inhibits cap methylation results in the hyperpolyadenylation of mRNA. This work reveals that inhibiting cap addition and cap methylation have opposing effects on polyadenylation during VSV mRNA synthesis and provides evidence in support of a link between correct 5′ cap formation and 3′ polyadenylation.
The vesicular stomatitis virus (VSV) genome encodes five genes that are flanked at their termini by a single 3′ leader and a 5′ trailer sequence. The genome is not present as a strand of naked RNA but is instead completely encapsidated by the viral nucleocapsid (N) protein. The structure of the N-RNA template indicates that in order for the polymerase to copy the RNA, the N-RNA must be substantially remodeled (2, 15). The polymerase comprises a large 241-kDa multifunctional subunit (L) that is associated with an accessory phosphoprotein (P) required for the production of full-length viral mRNA (9). Genetic and biochemical evidence demonstrates that L catalyzes ribonucleotide polymerization (34), mRNA cap addition (23, 27), cap methylation (14, 16, 22, 24, 28), and polyadenylation (18). Sequence alignments of all nonsegmented negative-sense (NNS) RNA virus L proteins identified six conserved regions (CR) separated by areas of low sequence similarity (32). Single amino acid substitutions in L have mapped the RNA-dependent RNA polymerase, polyribonucleotidyltransferase, and mRNA cap methyltransferase activities to CRIII, CRV, and CRVI, respectively. Currently, there are no reports of single amino acid substitutions in L that inhibit polyadenylation.
The five genes of the VSV genome are transcribed sequentially (1, 3). Although there is still some debate about precisely how the polymerase accesses the first gene on the template, there is a consensus view of how the genes are sequentially transcribed (42). The polymerase initiates mRNA synthesis at the first gene start (44) and shortly after initiation adds an mRNA cap structure (36, 38, 43). In response to a specific gene end sequence (5, 6, 20), the polymerase terminates synthesis in a process that involves reiterative copying of a U tract to generate the 3′ polyadenylate tail (4). The termination of an upstream gene is critical to permit the polymerase to reinitiate in response to a downstream gene start sequence and produce the next mRNA. The cis-acting signals that govern the polymerase activities at these junctions for termination, polyadenylation, initiation, and mRNA cap addition are defined. An attenuation step, localized to the gene junction, results in the accumulation of less downstream mRNA (21), but our knowledge of how this is accomplished is lacking.
VSV mRNA synthesis can be reconstituted in vitro using purified recombinant polymerase expressed from baculovirus vectors in insect cells (23, 26). Using this system, we defined a set of single amino acid substitutions in CRV of the L protein that prevented mRNA cap addition (23). These residues are highly conserved among the NNS RNA viruses and likely include key catalytic residues of the polyribonucleotidyltransferase activity that is essential for cap addition (27). In reactions reconstituted with L proteins containing alanine substitutions at positions G1154, T1157, H1227, and R1228, the products of RNA synthesis were uncapped and frequently terminated within the N gene, yielding discrete transcripts that were typically <400 nt (23). Although we demonstrated that the polymerases had a specific defect in cap addition, we did not determine where such polymerases were terminating within N. Moreover, because the cap-defective polymerase failed to encounter a gene junction, we were unable to examine their ability to respond to a wild-type termination signal and reinitiate synthesis at a downstream gene.
In the current study, we reconstituted transcription in vitro using a panel of templates that contained additional genes inserted (I) between the leader and N. We found that the cap-defective polymerases efficiently synthesized the full-length I mRNA but that the mRNA lacked both the 5′ cap structure and the 3′ poly(A) tail. The cap-defective polymerases were also unable to efficiently transcribe subsequent genes on the template and instead appear to recycle and copy repeatedly the first gene. Our data suggest that the termination of an upstream gene is insufficient to permit the synthesis of a downstream gene and support a model in which polyadenylation is required for downstream initiation. Moreover, they reveal a previously unappreciated link between 5′ cap addition and 3′ polyadenylation during VSV mRNA synthesis.
MATERIALS AND METHODS
Recombinant viruses.
Recombinant VSV (rVSV) containing genes inserted at the leader-N gene junction were as described previously (4, 41). The sequence of rVSV, which differs from VSV-Indiana (San Juan) as described previously (40), is available on request.
N-RNA template purification.
The N-RNA template was purified from rVSV as described previously (23, 29). Briefly, 4 mg purified virus was disrupted on ice for 1 h in 20 mM Tris-HCl (pH 8.0), 0.1% Triton X-100, 5% glycerol, 5 mM EDTA, 3.5 mM dithioerythritol, 20% dimethyl sulfoxide, and 1.0 M LiCl. The template was recovered by centrifugation (190,000 × g, 3.5 h) through a step gradient of 0.25 ml each of 40, 45, and 50% glycerol in TED buffer (20 mM Tris-Cl [pH 8.0], 1 mM EDTA, 2 mM dithioerythritol) supplemented with 0.1 M NaCl. The pellet was resuspended in 0.3 ml of TED buffer plus 10% glycerol and disrupted on ice, except that the Triton X-100 and EDTA concentrations were reduced to 0.05% and 1 mM, respectively. The N RNA was isolated by banding in a 3.6-ml 20 to 40% (wt/wt) CsCl gradient (150,000 × g, 2.5 h), recovered by side puncture and diluted fourfold with 10 mM Tris-Cl (pH 8.0), 0.1 mM EDTA. The N-RNA was recovered following centrifugation (150,000 × g, 1.5 h) through a 0.5-ml cushion (50% glycerol, TED buffer, 0.1 M NaCl).
Expression and purification of recombinant VSV polymerase.
Polymerase components were expressed from recombinant baculoviruses in Spodoptera frugiperda 21 cells as described previously (23). At 72 h postinfection, the cells were collected, washed twice with ice-cold phosphate-buffered saline, and recovered by centrifugation. The cells were suspended in lysis buffer (50 mM NaH2PO4, 10% glycerol, 0.2% NP-40, 300 mM NaCl, 10 mM imidazole [pH 8.0]) supplemented with EDTA-free protease inhibitor cocktail (Roche) and 1 mM phenylmethylsulfonyl fluoride and disrupted by sonication. The L and P proteins were purified by Ni-nitrilotriacetic acid-agarose (Qiagen), followed by ion-exchange chromatography as described previously (23).
Reconstitution of viral RNA synthesis in vitro.
Reactions were carried out using 1 μg of N-RNA template, 1 to 3 μg of purified L, and 0.5 μg of purified P, nucleoside triphosphates (1 mM ATP and 0.5 mM each CTP, GTP, and UTP), and 30% (vol/vol) rabbit reticulocyte lysate (Promega) as described previously (23). Where indicated, the reaction mixtures were supplemented with 15 μCi of [α-32P]GTP (3,000 Ci mmol−1) (Perkin-Elmer, Wellesley, MA). After 5 h of incubation at 30°C, the RNA was purified by phenol and chloroform extraction and analyzed by electrophoresis on acid-agarose gels as described previously (30, 39).
Primer extension assay.
Minus-sense oligonucleotides, corresponding to nt 27 to 47, 130 to 115, 1439 to 1426, and 2279 to 2260 of the complete rVSV genome sequence, were used in primer extensions to detect VSV leader RNA and N, P, and M mRNA, respectively. Primers annealing to transcripts of the 382-nt chloramphenicol acetyltransferase (CAT) and green fluorescent protein (GFP) genes were designed to yield products of 72, 73, and 81 nt, respectively. Reverse transcriptions were performed using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and the products were analyzed by electrophoresis on 6% polyacrylamide gels and detected by phosphorimager. Where indicated, the products of the in vitro transcription reactions were treated with tobacco acid pyrophosphatase (TAP; Epicenter, WI) to remove the cap structure, prior to reverse transcription, as described previously (38).
Isolation of polyadenylated mRNA.
Polyadenylated mRNA was isolated by oligo(dT) affinity chromatography. Briefly, the total RNA synthesized in vitro was adjusted to 0.5 M NaCl, heated at 65°C for 5 min, and quickly cooled in an ice bath for 3 min. The mixture was added to equilibrated oligo(dT) 17 cellulose beads (New England Biolabs, Beverly, MA). The beads were washed three times with 20 mM Tris-HCl (pH 7.5), 0.5 M NaCl, 1 mM EDTA, followed by one wash in a buffer in which the NaCl concentration was reduced to 0.1 M, and the poly(A) RNA eluted in 70°C 20 mM Tris-HCl (pH 7.5), 1 mM EDTA.
Direct analysis of the mRNA poly(A) tail.
RNA synthesis reactions were performed as previously described, except that cold ATP was reduced to 100 μM and supplemented with 50 μCi of [α-32P] ATP (3,000 Ci mmol−1) (Perkin-Elmer, Wellesley, MA). The purified RNA was digested with 10 units of RNase T1 and the products were resolved on 6% polyacrylamide gels.
3′ RACE and sequencing.
Purified RNA was tailed either with poly(A) or poly(G) using Escherichia coli poly(A) polymerase (NEB, Beverly, MA) in the presence of 0.1 M ATP or GTP. First-strand cDNA synthesis was performed using Superscript III reverse transcriptase with an oligo(dT)17 or oligo(dC)17. The products were amplified by PCR using oligo(dT)17 or oligo(dC)17 and a primer designed to anneal to positions 1 to 17 of the specific mRNA. The DNA products were cloned into pGEM-T (Promega), and individual clones from the independent rapid amplification of cDNA end (RACE) reactions were sequenced at the DF-HCC sequencing facility, Harvard Medical School. Where indicated, the sequences of the 3′-terminal 50 nt were used to generate a pictogram with the program WebLogo 2.8.8 (8), and the data were displayed as a frequency plot.
Quantitative analysis.
Quantitative analysis was performed using a phosphorimager (Typhoon; GE Healthcare) and ImageQuant TL software (GE Healthcare, Piscataway, NJ). Statistical analysis was performed on three to five independent experiments. The significance of the values was determined by a paired Student's t test.
RESULTS
Cap-defective mutants show frequent intragenic termination.
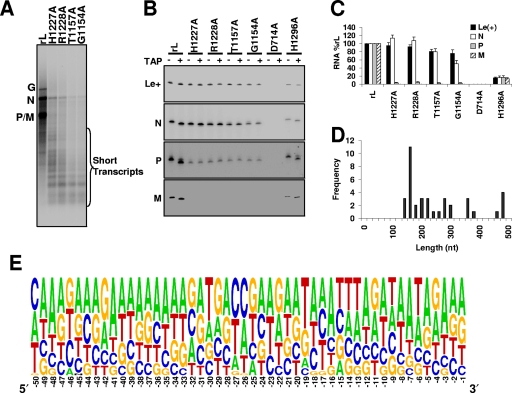
We showed previously that amino acid substitutions G1154A, T1157A, H1227A, and R1228A to highly conserved residues present within region V of the VSV L protein ablate mRNA cap addition. The polymerases defective in mRNA cap addition produced little full-length mRNA in vitro and instead synthesized short transcripts that were typically <400 nt (23). An example of the products from an RNA synthesis reaction reconstituted with highly purified N-RNA template and recombinant P and L proteins demonstrates the accumulation of full-length N, P, M, and G mRNA (Fig. 1A, lane rL). Single amino acid substitutions in CRV that prevent cap addition—G1154A, T1157A, H1227A, and R1228A—were defective in RNA synthesis, producing predominantly truncated transcripts (Fig. 1A). The amount of full-length N mRNA synthesized by the cap-defective polymerases was approximately 1% of that observed for wild-type L protein.
FIG. 1.
Cap-defective mutants show frequent intragenic termination. (A) Transcription reactions performed in vitro in the presence of 1 μg of N-RNA template, 0.5 μg of recombinant P, 1.0 μg of the indicated L, and [α-32P]GTP were analyzed by electrophoresis on acid-agarose gels. The identity of the mRNA is shown at the left, and the short transcripts produced by the cap-defective mutants are indicated by the bracket. Note that half of the products of the wild-type L (rL) reaction were loaded on the gel. (B) A primer extension analysis of the products synthesized by polymerase from the rVSV template is shown. The polymerase used in the reactions is shown at the top, and the products detected by primers designed to anneal within the leader RNA (Le+) and the N, P, and M mRNA are shown in separate panels. The products of primer extension were analyzed by electrophoresis on a 6% polyacrylamide gel and detected by phosphorimage analysis. The relevant sections of the gel are shown. Where indicated (+), the products of in vitro transcription were treated with TAP prior to reverse transcription to show which RNAs contain a cap structure. In addition to the cap-defective polymerases, two controls were included, a catalytically inactive polymerase (D714A) bearing a single amino acid change in CRIII, and a polymerase (H1296A) which contains a single amino acid change in CRV that does not inhibit cap addition. (C) A graph depicting the abundance of each RNA measured in three independent experiments is shown. (D) A graph representing the position within the N gene (within 20 nt) at which transcripts synthesized by H1227A prematurely terminated. (E) A pictogram, assembled as described in Materials and Methods, indicates common attributes of the transcripts shown in panel D produced by frequent intragenic termination within the N gene. The numbering below the pictogram indicates the position from the site of termination, with −1 being the terminating nucleotide.
To determine whether the short transcripts synthesized by the cap-defective L proteins correspond to initiations at the N, P, or M gene starts, we performed a primer extension analysis using primers designed to anneal in the first 81 nt of N, 55 nt of P, and 72 nt of M mRNA. We observed that mutants with defects in cap addition synthesized similar quantities of transcripts that initiated at the N gene start as wild-type L protein, indicating that initiation and elongation to 81 nt were not significantly compromised (Fig. 1B and C). However, the cap-defective polymerases produced few transcripts that initiated at the P gene start, and those that initiated at the M gene start approached the limit of detection (Fig. 1B and C). In contrast, wild-type L protein, or a mutation in CRV (H1296A) that does not affect cap addition, produced transcripts detected by each of the primers. These data indicate that the majority of the truncated transcripts synthesized by the cap-defective polymerases (Fig. 1A) derive from initiation at the N gene start and not from subsequent reinitiation at the P or M gene start. A single amino acid substitution, D714A, to a key catalytic residue of the RNA-dependent RNA polymerase ablated all RNA synthesis (Fig. 1B and C), demonstrating that the observed products were synthesized by the VSV polymerase. As expected, the products of synthesis by wild-type L and H1296A contain an mRNA cap structure, as the products of reverse transcription were 1 nt shorter following treatment of the RNA with TAP. TAP cleaves the cap structure off the RNA while leaving the body of the RNA intact; consequently, reverse transcriptase is unable to extend onto the cap during the primer extension reaction. Consistent with this, the products of primer extension on the uncapped leader RNA, or the N and P transcripts synthesized by the cap-defective polymerases, were unaffected by TAP digestion (Fig. 1B).
The primer extension analysis (Fig. 1B and C) demonstrated that initiation at the N gene start was robust for the cap-defective polymerases but that initiation at the P and M gene starts was dramatically reduced. Coupled with the accumulation of short transcripts (Fig. 1A), this indicated that the cap-defective polymerases likely terminated during the transcription of the N gene. To examine more precisely the transcripts of the N gene, we determined the sequence of transcripts synthesized by H1227A. To do this, we amplified the N gene transcripts by RACE and ligated the resulting cDNA into pGEM-T prior to sequence determination. We obtained 38 sequences and mapped the position within the N gene at which they terminated. Consistent with our previous size analysis and the mobility of the truncated products (Fig. 1A), we found that the majority of the transcripts terminated within the first 500 nt (Fig. 1D). The VSV polymerase normally terminates mRNA synthesis in response to the specific cis-acting signal 3′-AUACUUUUUUUG/C-5′. To determine whether there is a preference for a sequence element that favors the observed premature termination of RNA synthesis, we aligned the last 50 nt of each of the sequenced transcripts of the N gene. These nucleotides were numbered −1 to −50 relative to the last nucleotide of N that was transcribed. A pictogram representation of the alignment of the last 50 nt of 38 sequences revealed a preference for termination on U-rich sequences (Fig. 1E). Although the sequence of the VSV genome is rich in A and U, which will inherently result in a higher preponderance of T's and A's in the sequenced clones, we found that adenylate was the most-frequent nucleotide observed at 32 of the last 50 nt transcribed, and the second-most-favored nucleotide at 12 of the remaining 18 nt. This indicates a bias toward the presence of adenylate residues in close proximity to the point of termination.
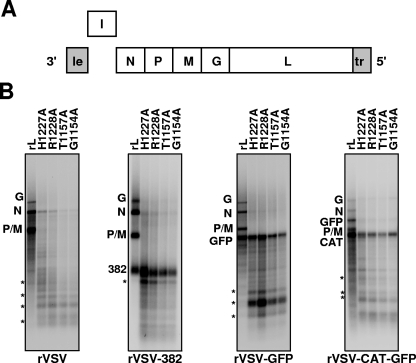
Cap-defective mutants efficiently transcribe a foreign gene inserted at the first position of the viral genome.
Earlier work showed that a polymerase must copy a minimum length of template in order to respond to an authentic gene end sequence (41). This length was >56 nt, which is reached by the cap-defective mutants despite their defect in the synthesis of full-length N mRNA. We therefore examined polymerase activity on templates that contained a single nonessential gene (I) inserted between the leader and the N genes of VSV. As sources of template, we used recombinant viruses, rVSV-382, and rVSV-GFP (Fig. 2A). Briefly, rVSV-382 contains a 382-nt transcriptional unit derived from the Microviridae, Enterobacteria phage φX174, and rVSV-GFP contains a 760-nt transcriptional unit comprising the open reading frame of Aequorea victoria GFP. These inserted (I) transcription units contain the essential VSV gene start and gene end sequences that are necessary for mRNA synthesis. In reactions programmed with template derived from each of these viruses, the cap-defective mutants produced abundant transcripts that corresponded in size to the I mRNA (Fig. 2B, the 382 and GFP products). However, the characteristic truncated products synthesized from the N gene were lacking; instead, alternate truncated transcripts were detected (Fig. 2B, compare transcripts marked by an asterisk). This suggested that the cap-deficient polymerases could efficiently transcribe a single I gene but then recycled to reinitiate at the beginning of the I gene rather than proceed to transcribe the N gene. We therefore examined whether the cap-defective polymerases could efficiently reinitiate and copy a second I gene. To do this, we purified template from rVSV-CAT-GFP, which contains a 701-nt transcription unit encoding CAT, followed by a second 1,098-nt transcription unit comprising a fusion of GFP and the φX174 sequence. The inclusion of the φX174 sequence permitted the CAT and GFP mRNA to be readily distinguished from one another based on their size (Fig. 2B, lanes rL). We found that the cap-defective polymerases efficiently synthesized full-length CAT mRNA, but a product corresponding to full-length 1,098-nt GFP mRNA was not detected (Fig. 2B). In contrast, the wild-type L protein synthesized both CAT and GFP as well as the N, P, M, and G mRNA. These data support the idea that the cap-defective polymerases are unable to transcribe the genome sequentially.
FIG. 2.
Cap-defective mutants efficiently copy a single inserted foreign gene. (A) A schematic of the VSV genome containing an autonomous transcription unit containing all the necessary signals for initiation and termination inserted at the leader-N gene junction. Note that for the template rVSV-CAT-GFP, two I genes containing the conserved VSV gene start and gene end sequences were inserted prior to N. (B) In vitro transcription reactions were reconstituted with 1.0 μg of N-RNA template, 0.5 μg of purified P, and 1.0 μg of the indicated L protein in the presence of [α-32P]GTP as described in Materials and Methods. Purified RNA was analyzed on acid-agarose gels and detected using a phosphorimager. The identity of the template is shown at the bottom of each panel, and the polymerase molecules are shown at the top. The most-abundant products of premature termination synthesized by the cap-defective polymerases are marked on the left-hand side by asterisks. Note that for the wild-type reactions reconstituted with wild-type L, only half of the fraction was loaded on the gel.
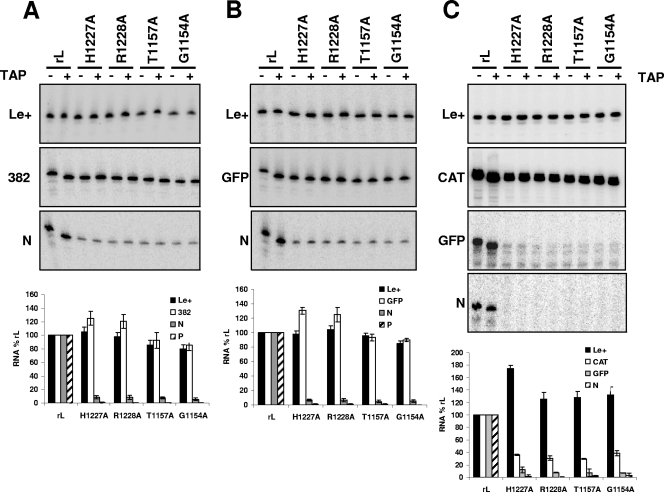
The above analysis showed that the cap-defective polymerases were unable to transcribe a second gene on the template despite the efficient synthesis of full-length transcripts from the first gene. To determine whether the polymerases initiated transcription at the second gene start site, we performed a primer extension assay to detect transcripts of the leader RNA, the I genes, and the VSV N and P genes. For G1154A, T1157A, H1227A, and R1228A, we observed levels of leader RNA that were either elevated or equivalent to those synthesized by wild-type L protein irrespective of the template used. We observed the efficient transcription of the I gene for each cap-defective polymerase (Fig. 3A to C, compare the 382, GFP, and CAT transcripts). The transcripts lacked an mRNA cap structure, as shown by their insensitivity to TAP digestion and the position to which the 5′ end of the transcript mapped (Fig. 3A to C, compare the + and − TAP lanes). In contrast, the transcripts produced by wild-type L protein were capped, as the products of primer extension were 1 nt shorter following TAP cleavage of the RNA. These data confirmed that the cap-defective polymerases fail to cap transcripts produced from the I gene, even though such transcripts appeared to be full length. Despite the efficient transcription of the I gene, full-length N mRNA was not efficiently transcribed (Fig. 2), and initiations at the N gene start were dramatically reduced (Fig. 3). This was not specific to the N gene sequence, as the cap-defective mutants were also unable to efficiently initiate at the GFP gene start when it was located downstream of CAT (Fig. 3C). Although the cap-defective polymerases synthesize some truncated transcripts from the rVSV-CAT-GFP template, abundant full-length CAT mRNA was produced (Fig. 2). Taken together, these data show that despite the efficient synthesis of the first mRNA, the cap-defective polymerases were unable to synthesize full-length transcripts from the second gene and that initiation at the second gene is dramatically reduced.
FIG. 3.
Cap-defective mutants show a defect in sequential transcription. A primer extension analysis of the products synthesized by polymerase from the rVSV-382 (A), rVSV-GFP (B), and rVSV-CAT-GFP (C) templates are shown. The polymerase used in the reactions is shown at the top, and the products detected by the primers designed to anneal within the leader RNA (Le+) and the I and N mRNAs are shown in separate panels. The products of primer extension were analyzed by electrophoresis on a 6% polyacrylamide gel and detected by phosphorimage analysis. The relevant sections of the gel are shown. Where indicated (+), the products of in vitro transcription were treated with TAP prior to reverse transcription to show which RNAs contain a cap structure. The abundance of each RNA was measured in three independent experiments and is represented graphically.
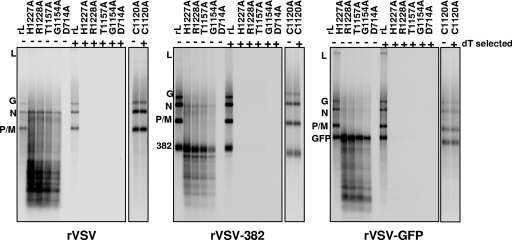
Cap-defective mutants are defective in polyadenylation.
The above observations indicated that the cap-defective polymerases were unable to respond efficiently to the signals encountered at a gene junction. The primer extension analysis revealed that the process of initiation at the 3′ end of the genome to produce leader RNA and at the first gene start was not compromised (Fig. 3). Thus, we suspected that the cap-defective polymerases might have defects in termination and/or polyadenylation. To test this, we examined whether the transcripts of the I genes contained polyadenylate. Using affinity for oligo(dT), we found that mRNAs synthesized by wild-type L were efficiently selected, consistent with the presence of a poly(A) tail. In contrast, the products of the cap-defective polymerases, including the full-length I transcripts, did not bind to oligo(dT), indicating that they lacked polyadenylate (Fig. 4). To demonstrate that the defect in poly(A) tail formation was not an inherent property of all L proteins with amino acid substitutions in CRV, we analyzed transcripts synthesized by C1120A. We previously showed that this substitution, like H1296A, diminished the overall levels of RNA synthesis but was competent to cap mRNA and lacked the truncated transcripts characteristic of the cap-defective polymerases (23). The products of in vitro transcription reactions reconstituted with C1120A were efficiently retained by oligo(dT) chromatography (Fig. 4), indicating that they were polyadenylated.
FIG. 4.
Cap-defective mutants are defective in polyadenylation. In vitro transcription reactions were reconstituted with the indicated N-RNA template and polymerase in the presence of [α-32P]GTP as described in Materials and Methods. Where indicated (+), purified RNA was selected by oligo(dT) chromatography prior to analysis on acid-agarose gels. RNAs were visualized by phosphorimage analysis. The identity of the template is shown at the bottom of each panel, and the polymerase molecules are shown at the top. Note that for the reactions reconstituted with the rVSV template, only a fraction (5%) of the total wild-type L (rL) RNA was analyzed so that the products of the cap-defective polymerases were clearly visible.
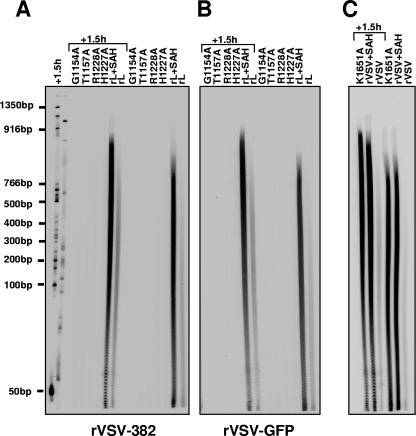
The extent of poly(A) tail formation required for retention by oligo(dT) chromatography is uncertain, and thus, we examined directly the size of the poly(A) present on the RNA. To do this, we took advantage of the ability of RNase T1 to cleave RNA 3′ of guanylate residues. All VSV mRNAs contain a G residue directly preceding the poly(A) tail. Consequently, the cleavage of a VSV mRNA by RNase T1 will produce short fragments from the body of the mRNA but leave the polyadenylate tail intact. Using mRNA synthesized in vitro from templates VSV-382 or VSV-GFP in the presence of [α-32P]ATP, we show that the RNase T1 cleavage yields a range of sizes of polyadenylate synthesized by wild-type L protein (Fig. 5). To size the polyadenylate, we loaded the products of RNase T1 cleavage on a 6% polyacrylamide gel and compared their mobility to a DNA marker (lanes 1 and 2). For wild-type L protein, we observed a poly(A) tail that extended from the 50- to 766-bp markers (Fig. 5). As expected (33), this length increased in the presence of the chemical inhibitor of methylation S-adenosyl homocysteine (SAH), extending beyond the 916-bp marker, and the majority of the RNAs contained polyadenylate that migrated between the 100- and 916-bp markers. In an attempt to define both a lower and an upper limit to the length of the poly(A), we loaded the same samples and DNA marker onto the polyacrylamide gel a second time, 1.5 h later. This shows that the smear of polyadenylate extends beyond the 766-bp marker for wild-type L protein (Fig. 5), and some extends to almost the 1,350-bp marker in the presence of SAH. In marked contrast to the level of polyadenylate observed for wild-type L protein, we did not observe a signal for poly(A) that reached the 50-bp marker for any of the cap-defective polymerases (Fig. 5).
FIG. 5.
Analysis of the polyadenylate tails. (A, B) In vitro transcription reactions were reconstituted with the indicated N-RNA template and polymerase in the presence of [α-32P]ATP. (C) Detergent-activated transcriptions were performed with the indicated recombinant virus as described previously (22). Purified RNA was digested with RNase T1 prior to analysis on 6% polyacrylamide gels and detection by phosphorimage analysis. The identity of the polymerase is shown at the top of the image, and DNA size markers are shown on the left. Lanes that were loaded 1.5 h later are highlighted and are shown as an aid in sizing the polyadenylate.
Although the transcripts synthesized by the cap-defective mutants appeared to be full length by analysis on agarose-urea gels, their exact size is uncertain, and it remained formally possible that such transcripts were not polyadenylated because the polymerase terminated just prior to reaching the gene end sequence. Our polyadenylate tail analysis could also not discriminate whether the cap-defective polymerases synthesized a short poly(A) tail. To confirm the site of termination of transcripts synthesized by these cap-defective polymerases, we cloned the RNAs synthesized in vitro by H1227A from templates containing either GFP or the 382-nt transcription unit. In approximately 60% of our sequences, we found that the polymerase had reached the authentic termination signal 3′-AUACUUUUUUUGA-5′. However, the extent to which the polymerase added adenylate residues varied from 0 to 24, with the majority of the sequences showing no evidence of the reiterative transcription of the U7 tract to generate polyadenylate (Table 1). These data thus confirm that the cap-defective polymerases are able to reach gene end sequences but fail to efficiently polyadenylate in response to an authentic gene end sequence.
TABLE 1.
Sequences of the 3′ ends of full-length I gene transcripts
| Gene | Transcript 3′-end sequencea |
|---|---|
| 382 | ATAAGGCCATATG |
| ATAAGGCCATATGA | |
| ATAAGGCCATATGAAA | |
| ATAAGGCCATATGAAAAAAA (3) | |
| ATAAGGCCATATGAAAAAAAAAA | |
| ATAAGGCCATATGAAAAAAAAAAAAAA | |
| ATAAGGCCATATGAAAAAAAAAAAAAAAA | |
| ATAAGGCCATATGAAAAAAAAAAAAAAAAAAA | |
| ATAAGGCCATATGAAAAAAACTAACAGTAAT | |
| GFP | ATCCCCCCATATGAAAA (2) |
| ATCCCCCCATATGAAAAAAA | |
| ATCCCCCCATATGAAAAAAAAAA | |
| ATCCCCCCATATGAAAAAAAAAAA | |
| ATCCCCCCATATGAAAAAAAAAAAAAAAAA | |
| ATCCCCCCATATGAAAAAAAAAAAAAAAAAAAA | |
| ATCCCCCCATATGAAAAAAAAAAAAAAAAAAAAAAAA |
The sequences at the 3′ ends of the full-length transcripts obtained from RNA transcribed in vitro with polymerase H1227A are shown. The numbers in parentheses indicate multiple independent isolates of the same sequence. The underlined A residues are presumably derived from polymerase slippage on the U tract.
Cap methylase mutants hyperpolyadenylate.
Previously, it was reported that VSV mRNAs synthesized in the presence of an inhibitor of mRNA cap methylation, SAH, possessed giant polyadenylate tails (33). Our data indicated that the failure to cap was linked to a failure to polyadenylate; therefore, we tested directly whether amino acid substitutions in L that inhibit mRNA cap methylation (22) were altered in their polyadenylation capacity. We previously reported that single amino acid substitutions in CRVI of the VSV L protein inhibit mRNA cap methylation at both the guanine-N-7 and ribose-2′-O positions. Specifically, we systematically altered residues K1651, D1762, K1795, and E1833 in CRVI that were predicted to catalyze 2′-O methylation as well as residues G1670, G1672, G1674, and D1735 that were predicted to bind the methyl donor S-adenosyl-l-methionine (SAM). We found that the same SAM binding site was used for both guanine-N-7 and ribose-2′-O methylation and that substitutions to the predicted catalytic residues ablated methylation (22, 24). To test whether substitutions in L that inhibit methylation have the same effect on polyadenylation as SAH, we analyzed the polyadenylate tail of transcripts synthesized by K1651A. We found that the polyadenylate tails were similar in length to those produced by wild-type L in the presence of SAH (Fig. 5), extending beyond the 916-bp marker. These data thus confirm the earlier work that the inhibition of cap methylation results in large polyadenylate. In addition, this finding provides further support of a link between 5′ cap addition and 3′ end formation.
DISCUSSION
In the present study, we show that single amino acid changes in CRV of the VSV L protein that inhibit mRNA cap addition render the polymerase defective in traversing a gene junction. Specifically, we show that substitutions G1154A, T1157A, H1227A, and R1228A in L protein inhibit polyadenylation and dramatically reduce the transcription of the downstream gene. This suggests a link between authentic 5′-end formation and 3′-end formation during VSV mRNA synthesis, and we obtained further evidence in support of this idea by showing that a viral mutant defective in mRNA cap methylation produces large polyadenylate. Our data thus show that single amino acid substitutions in CRV of L protein that affect mRNA cap addition have additional defects in mRNA synthesis, including intragenic termination, polyadenylation, and attenuation. This study provides strong support for a model of mRNA synthesis in which cap addition plays a central role in governing polymerase activity.
Frequent intragenic termination.
We reported previously that polymerases containing single amino acid substitutions in CRV of L protein were defective in mRNA cap addition and produced transcripts that terminate prematurely (23). The size of these transcripts indicated that termination was not completely random but appeared to occur at several distinct sites. Here by sequence analysis, we show that there was a bias toward A residues in the last 50 nt of the transcripts. Consistent with the idea of distinct sites within the N gene favoring termination, the polymerases produced alternately sized products from templates that contained a nonessential gene inserted between the leader and the N genes. The I genes were less U rich than the N gene, perhaps accounting for their efficient transcription.
Our data are consistent with prior work that links the failure to cap the viral mRNAs with the premature termination of RNA synthesis. Specifically, a mutational analysis of the conserved gene start sequence of VSV defined a sequence, 3′-UyG-5′, for the first 3 nt as required for initiation (35). A subsequent in vitro analysis demonstrated that in fact, the requirement for initiation was substantially relaxed but that a cis-acting signal was required to produce a fully capped and methylated 5′ end (36). Failure to modify the 5′ end correctly resulted in the abundant synthesis of short transcripts that ranged in size from 100 to 400 nt. Independent work on the polymerase of respiratory syncytial virus identified a small molecule inhibitor that prevented mRNA capping and polyadenylation (25). RNA synthesized in the presence of this inhibitor terminates prematurely within a gene. Furthermore, viral mutants resistant to this inhibitor contained single amino acid substitutions in CRV of L, consistent with its role in cap formation. Taken together, these earlier studies provide independent corroboration that a defect in mRNA cap addition results in the premature termination of mRNA synthesis. Remarkably, in all cases, transcripts ranged in size from 100 to 400 nt, consistent with the sequence data reported in the present study. We interpret these data as indicating that correct 5′ cap addition results in a change to the elongating polymerase complex that renders the enzyme resistant to intragenic termination. This strategy would ensure that only the authentic transcripts are correctly elongated and terminated and therefore serves as a quality control mechanism for the polymerase during transcription. However, the mechanism by which the cap structure could affect such quality control is currently unknown. Current data cannot exclude the alternate possibility that the uncapped triphosphate RNA serves as a signal to promote termination during RNA synthesis.
In the present study, we also found that the defect in termination can be partially overcome using templates in which nonessential genes are inserted between the leader and the N genes. Although these I genes were typically shorter than the N gene, they were further distinct in that they contained a lower A and U content than that found in the VSV N gene. The authentic VSV termination sequence, 3′-AUACUUUUUUU-5′, is AU rich, and prior work demonstrated that its AU-rich nature is critical for controlling reiterative transcription by the polymerase (4). During authentic termination, the polymerase must realign the nascent strand on the template during reiterative copying of the U tract to generate the polyadenylate. This slippage, which is a prerequisite for mRNA termination, can be overcome by increasing the GC content upstream of the U7 tract (4). Here we found that the transcripts that terminate prematurely within the N gene typically terminated close to U-rich sequences. We suggest that this also reflects the importance of a hybrid between the nascent RNA strand and the RNA template in controlling elongation and that the cap-defective polymerases are sensitized to terminate when this hybrid is weak. This may explain why the cap-defective polymerases are able to synthesize full-length inserted mRNAs that have a higher intrinsic GC content which should result in a stronger hybrid between the nascent strand and the template.
Polyadenylation.
Although the cap-defective polymerase generated full-length I mRNAs, they lacked authentic polyadenylate tails. Mutations in the L gene that inhibit polyadenylation have not been described previously. Analysis of the function of the conserved cis-acting signals present at the gene end demonstrates that the polymerase reiteratively copies the U7 tract to generate the poly(A) tail (4, 5). Polyadenylation therefore likely reflects the polymerase active site reiteratively copying the template, and thus, a separate poly(A) polymerase activity is not required. While we did not determine why the cap-defective polymerases are unable to generate a significant poly(A) tail in response to a wild-type gene end sequence, we interpret our data as reflecting the intrinsic termination sensitivity of the cap-defective polymerases rather than specifically affecting poly(A) polymerase activity. The finding that the positions at which the cap-defective polymerases terminate within the N gene are largely U rich also supports this idea. The frequent intragenic termination and the failure to polyadenylate may simply therefore reflect an effect of the amino acid substitutions or failure to cap the mRNA on the same property of the VSV polymerase. As mentioned above, available evidence indicates that the strength of the hybrid between the nascent RNA strand and the RNA template is important in controlling polyadenylation. If the cap-defective polymerases are prone to termination whenever the hybrid is destabilized, they should be hypersensitive to termination at the gene end sequence.
In contrast to the lack of evidence for polymerases defective in polyadenylation, previous work had demonstrated that a single amino acid change in L protein could result in the production of mRNA with large polyadenylate (19). The amino acid change responsible for this, F1488S, is located in the variable region of L between CRV and CRVI. Other single amino acid substitutions in this region inhibit mRNA cap methylation (13), most likely by indirectly affecting the function of CRVI of L protein. The effect of the F1488S substitution on polyadenylation is consistent with the observation of giant heterogenous polyadenylate on VSV mRNA synthesized in the presence of the methylation inhibitor SAH (33). Previously, we demonstrated that substitutions of residues in CRVI of L predicted to catalyze mRNA cap methylation prevented both 2′-O and G-N-7 methylation (22). In the present study, we show that one of these, K1651A, produces large polyadenylate (Fig. 5), providing further support that failing to methylate the mRNA cap structure can result in the synthesis of large polyadenylate. However, the mRNA cap structures synthesized by viruses containing the F1488 substitutions are in fact methylated (12, 13), illustrating that there is no clear-cut relationship between cap methylation and hyperpolyadenylation.
In addition to SAH, other SAM analogues result in the hyperpolyadenylation of VSV mRNA (17). This led to the suggestion that SAH binding either positively or negatively affects a conformational change in L that results in this hyperpolyadenylation (17). The effect of SAH on polyadenylation was recently examined using a panel of recombinant viruses with defects in cap methylation. Substitutions in CRVI of L that retained the ability to methylate mRNA responded to SAH in the same way as the wild-type L protein by producing large polyadenylate (12). This led to the conclusion that the SAH-induced hyperpolyadenylation of VSV mRNA requires the methyltransferase activity of the L protein. However, the picture for methylation-incompetent mutants was not clear. Similar to the findings for K1651A reported here, substitution D1762E that inhibits mRNA cap methylation produces large polyadenylate (12). In contrast, several other substitutions that inhibit cap methylation, including D1762G, D1762N, G1672P, and G1675P, were reported not to hyperpolyadenylate (12). It was suggested that the D1762E substitution might favor the binding of SAH at the SAM binding site in CRVI, resulting in hyperpolyadenylation without the need for supplemental SAH (12). Although structural models of CRVI of the L proteins of VSV with other NNS RNA viruses and methyltransferases suggest that K1651 may play a role in SAM binding (7, 10, 11, 22), direct evidence for this, and for a role for D1762 in SAM binding, is lacking. Additional experiments will therefore be required to understand the mechanism by which SAH or the failure to methylate the cap structure results in hyperpolyadenylation.
Transcriptional attenuation.
Using ultraviolet irradiation of purified virus particles, experiments 30 years ago established that the VSV polymerase sequentially synthesizes the five mRNAs (1, 3). The mRNAs are not produced in equimolar amounts; rather, their abundance decreases with distance from a single polymerase entry site such that N > P > M > G > L (37). This polarity gradient reflects a localized transcriptional attenuation at each gene junction, where 30% of the polymerase molecules fail to transcribe the downstream gene (21). The mechanism by which the VSV polymerase, or indeed any NNS RNA virus polymerase, synthesizes less downstream mRNA is not understood. Prior to the current study, polymerase mutants that are defective in sequential transcription have not been described. Using templates containing I genes, we show that polymerase proteins that encounter a gene end sequence must have modified authentically the upstream mRNA in order to transcribe the downstream mRNA. What aspect of upstream mRNA synthesis is required to permit downstream mRNA synthesis is uncertain. Our data cannot distinguish whether it is the presence of a 5′ cap structure and/or 3′ polyadenylate on the upstream mRNA that are required to allow copying of the downstream mRNA or whether it is the act of modification that signals to the polymerase to continue copying the genome as mRNA. However, we favor the notion that the polymerase itself must undertake these modifications, as we have thus far been unable to rescue these defects by the addition of exogenous capping enzymes or cap analog (Li and Whelan, unpublished data).
What is the mechanism by which capping regulates polymerase activity?
The above observations beg the question of how the RNA-modifying activities of L influence polymerase function. Previously, we and others speculated that the 5′ cap in the nascent RNA chain serves as a regulatory element to the polymerase that is required for productive elongation and authentic termination (36, 41). This model was initially based upon the finding that substitutions to conserved residues of the gene start site led to the synthesis of prematurely terminated products that failed to react with an antibody against 2,2,7-trimethylguanosine (36, 41). This indicated that the transcripts terminated because they were not capped. However, it remained formally possible that such transcripts terminate prematurely because they lacked the correct sequence at the 5′ terminus. Subsequent work showed that the polymerase could not respond to an authentic termination and polyadenylation signal that was positioned within 56 nt of an initiation signal, and instead, the polymerase read through the wild-type junction to generate a read-through transcript (36, 41). The read-through transcripts appeared to contain direct copies of the gene junction, indicating that the polymerase failed to stutter on the U tract to generate a polyadenylate tail, an essential requirement for termination. Although the stage of synthesis of a transcript at which the mRNA acquires a cap structure is unknown, transcripts less than 37 nt were reportedly not capped (31). This raises the possibility that the polymerase failed to respond to a gene end sequence positioned in close proximity to a gene start because the nascent RNA transcript has not been accurately 5′ processed. In the current study, we now show that a polymerase that cannot cap its mRNA also fails to respond accurately to a gene junction. Thus, premature termination and failure to respond accurately to a gene junction sequence can be achieved by altering the key cis-acting signals for mRNA cap addition or by inhibiting the ability of the polymerase itself to cap the mRNA. The underlying mechanism remains unclear. For example, we do not yet know whether it is the cap structure of the nascent RNA strand or the act of capping by the polymerase that are required for correct elongation. We also do not know whether the cap structure remains associated with the elongating polymerase, somehow rendering the enzyme resistant to intragenic termination but sensitive to termination at an authentic site. Simplistic tests of a model in which the cap itself plays a role in regulating polymerase activity have not yet met with success, as we have been unable to rescue the transcription defects exhibited by our cap-defective polymerases by the addition of exogenous capping enzymes or supplementation with cap analog (Li and Whelan, unpublished). While such results imply that the polymerase itself must cap the RNA in order to correctly elongate and subsequently terminate, direct evidence for this is lacking. Understanding how the 5′ end of the RNA influences the downstream activities of the polymerase thus remains an intriguing question.
The present study highlights a major gap in our understanding of NNS RNA virus mRNA synthesis, specifically, our understanding of what happens to polymerase during the steps of termination and the presumed reinitiation. Dissection of these events using genetic and biochemical approaches is complicated by the population-averaging nature of such experiments. Consequently, addressing this question likely will await the application of new approaches, perhaps those involving the study of single polymerase molecules. In addition to an intrinsic biological interest, answering this question may well have utility in the rational design of antiviral therapeutics.
Acknowledgments
This work was supported by research grant AI0159371 from the NIH to S.P.J.W.
We acknowledge Philip J. Kranzusch for a critical review of the manuscript.
Footnotes
Published ahead of print on 10 December 2008.
REFERENCES
- 1.Abraham, G., and A. K. Banerjee. 1976. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 731504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertini, A. A., A. K. Wernimont, T. Muziol, R. B. Ravelli, C. R. Clapier, G. Schoehn, W. Weissenhorn, and R. W. Ruigrok. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313360-363. [DOI] [PubMed] [Google Scholar]
- 3.Ball, L. A., and C. N. White. 1976. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 756901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, J. N., S. P. J. Whelan, and G. W. Wertz. 1997. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 718718-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr, J. N., S. P. Whelan, and G. W. Wertz. 1997. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J. Virol. 711794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bujnicki, J. M., and L. Rychlewski. 2002. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. 15101-108. [DOI] [PubMed] [Google Scholar]
- 8.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson, S. U., and Y. Yu. 1975. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 151348-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferron, F., S. Longhi, B. Henrissat, and B. Canard. 2002. Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 27222-224. [DOI] [PubMed] [Google Scholar]
- 11.Galloway, S. E., P. E. Richardson, and G. W. Wertz. 2008. Analysis of a structural homology model of the 2′-O-ribose methyltransferase domain within the vesicular stomatitis virus L protein. Virology 38269-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galloway, S. E., and G. W. Wertz. 2008. S-Adenosyl homocysteine-induced hyperpolyadenylation of vesicular stomatitis virus mRNA requires the methyltransferase activity of L protein. J. Virol. 8212280-12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grdzelishvili, V. Z., S. Smallwood, D. Tower, R. L. Hall, D. M. Hunt, and S. A. Moyer. 2006. Identification of a new region in the vesicular stomatitis virus L polymerase protein which is essential for mRNA cap methylation. Virology 350394-405. [DOI] [PubMed] [Google Scholar]
- 14.Grdzelishvili, V. Z., S. Smallwood, D. Tower, R. L. Hall, D. M. Hunt, and S. A. Moyer. 2005. A single amino acid change in the l-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 797327-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, T. J., X. Zhang, G. W. Wertz, and M. Luo. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313357-360. [DOI] [PubMed] [Google Scholar]
- 16.Hercyk, N., S. M. Horikami, and S. A. Moyer. 1988. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology 163222-225. [DOI] [PubMed] [Google Scholar]
- 17.Hunt, D. M. 1989. Effect of analogues of S-adenosylmethionine on in vitro polyadenylation by vesicular stomatitis virus. J. Gen. Virol. 70535-542. [DOI] [PubMed] [Google Scholar]
- 18.Hunt, D. M. 1983. Vesicular stomatitis virus mutant with altered polyadenylic acid polymerase activity in vitro. J. Virol. 46788-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt, D. M., and K. L. Hutchinson. 1993. Amino acid changes in the L polymerase protein of vesicular stomatitis virus which confer aberrant polyadenylation and temperature-sensitive phenotypes. Virology 193786-793. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, L. N., N. Englund, and A. K. Pattnaik. 1998. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J. Virol. 721805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23477-484. [DOI] [PubMed] [Google Scholar]
- 22.Li, J., E. C. Fontaine-Rodriguez, and S. P. Whelan. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 7913373-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., A. Rahmeh, M. Morelli, and S. P. Whelan. 2008. A conserved motif in region V of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 82775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J., J. T. Wang, and S. P. Whelan. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 1038493-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liuzzi, M., S. W. Mason, M. Cartier, C. Lawetz, R. S. McCollum, N. Dansereau, G. Bolger, N. Lapeyre, Y. Gaudette, L. Lagace, M. J. Massariol, F. Do, P. Whitehead, L. Lamarre, E. Scouten, J. Bordeleau, S. Landry, J. Rancourt, G. Fazal, and B. Simoneau. 2005. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J. Virol. 7913105-13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathur, M., T. Das, and A. K. Banerjee. 1996. Expression of L protein of vesicular stomatitis virus Indiana serotype from recombinant baculovirus in insect cells: requirement of a host factor(s) for its biological activity in vitro. J. Virol. 702252-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino, T., and A. K. Banerjee. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 2585-97. [DOI] [PubMed] [Google Scholar]
- 28.Ogino, T., M. Kobayashi, M. Iwama, and K. Mizumoto. 2005. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2804429-4435. [DOI] [PubMed] [Google Scholar]
- 29.Ongradi, J., C. Cunningham, and J. F. Szilagyi. 1985. The role of polypeptides L and NS in the transcription process of vesicular stomatitis virus New Jersey using the temperature-sensitive mutant tsE1. J. Gen. Virol. 661011-1023. [DOI] [PubMed] [Google Scholar]
- 30.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 691011-1020. [DOI] [PubMed] [Google Scholar]
- 31.Piwnica-Worms, H., and J. D. Keene. 1983. Sequential synthesis of small capped RNA transcripts in vitro by vesicular stomatitis virus. Virology 125206-218. [DOI] [PubMed] [Google Scholar]
- 32.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 711153-1162. [DOI] [PubMed] [Google Scholar]
- 33.Rose, J. K., H. F. Lodish, and M. L. Brock. 1977. Giant heterogeneous polyadenylic acid on vesicular stomatitis virus mRNA synthesized in vitro in the presence of S-adenosylhomocysteine. J. Virol. 21683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sleat, D. E., and A. K. Banerjee. 1993. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J. Virol. 671334-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stillman, E. A., and M. A. Whitt. 1997. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J. Virol. 712127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stillman, E. A., and M. A. Whitt. 1999. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J. Virol. 737199-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villarreal, L. P., M. Breindl, and J. J. Holland. 1976. Determination of molar ratios of vesicular stomatitis virus induced RNA species in BHK21 cells. Biochemistry 151663-1667. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J. T., L. E. McElvain, and S. P. J. Whelan. 2007. Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J. Virol. 8111499-11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wertz, G. W., S. Whelan, A. LeGrone, and L. A. Ball. 1994. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc. Natl. Acad. Sci. USA 918587-8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whelan, S. P., L. A. Ball, J. N. Barr, and G. T. Wertz. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. USA 928388-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2000. Identification of a minimal size requirement for termination of vesicular stomatitis virus mRNA: implications for the mechanism of transcription. J. Virol. 748268-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 28361-119. [DOI] [PubMed] [Google Scholar]
- 43.Whelan, S. P., and G. W. Wertz. 1999. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J. Virol. 73297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whelan, S. P., and G. W. Wertz. 2002. Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc. Natl. Acad. Sci. USA 999178-9183. [DOI] [PMC free article] [PubMed] [Google Scholar]