Abstract
Porcine endogenous retroviruses (PERV) are members of the gammaretrovirus genus and display integration preferences around transcription start sites, a finding which is similar to the preferences of the murine leukemia virus (MLV). Our new genome-wide analysis of the integration profile of a recombinant PERV (PERV A/C), enabled us to examine more than 1,900 integration sites and identify 224 integration hot spots. Investigation of the possible genome features involved in hot-spot formation revealed that the expression level of the genes did not influence distribution of the integration sites of gammaretroviruses (PERV and MLV) or the formation of integration hot spots. However, PERV integration and the presence of hot spots was found to be greater in areas of the genome with high densities of genes with CpG islands. Surprisingly, this was not true for MLV. Simulation of integration profiles revealed that retrovirus integration studies involving the use of the restriction enzyme MseI (widely used in genome integration studies of MLV and gammaretroviral vector) underestimated integration near CpG islands and in gene-dense areas. These results suggest that the integration of gammaretrovirus or gammaretroviral vectors might occur preferentially in genome areas that are highly enriched in genes under CpG island promoter regulation.
Retrovirus-based gene therapy approaches carry both huge hope and danger (15). One of the most threatening risks is the oncogenic transformation following retroviral vector integration. Oncogenic retroviral transformation can involve the disruption of a gene product or of some regulatory elements of a gene (9). This risk results from well-described processes that have been used to identify proto-oncogenes (19, 32). A recent setback affecting SCID patients in a clinical trial highlighted the risk of vector integration (16) and the importance of the choice of retroviral backbone to deliver and integrate the therapeutic gene into the host genome (8).
The randomness of retroviral integration within the host genome has long been challenged (9) until the first genome-scale analysis of human immunodeficiency virus (HIV) integration by Schröder et al. definitively demonstrated the preferential HIV integration within active transcription units (TU) (28). Since then, the integration profiles of different retroviruses have been described (7, 13, 36). It is apparent from all of these studies that the preferred site of retroviral integration varies between different retrovirus genera but that retroviruses within the same genus share similar profiles. For example, integrations of porcine endogenous retrovirus (PERV) and murine leukemia virus (MLV), two gammaretroviruses, occur preferentially near CpG Islands and transcription start sites (TSS) (23, 39), whereas integrations of HIV and simian immunodeficiency virus (SIV), two lentiviruses, are favored in active TU (10, 28). The major viral determinant for target-site selection has been convincingly attributed to the integrase protein (IN), probably with the help of host partners (20). All of these studies have clearly demonstrated that retroviral integrations are not totally random, and this raises the crucial question of the impact of the vector backbone to be used in retrovirus-based therapy and the associated risks.
In a previous study, we characterized the integration profile of the PERV in human cells in vitro (23). However, some questions still need to be examined, especially the propensity of PERV to integrate or not with higher frequency in some areas of the genome, defining the so-called integration hot spots. PERV consists of different virus subgroups: PERV-A and PERV-B have been shown to infect human cells in culture, albeit with low titers, whereas PERV-C has a more restricted host range and cannot infect human cells in vitro (17). As a result of recent characterization of a natural PERV A/C recombinant (17), 500 times more infectious in vitro, we decided to reinvestigate its integration profile in the human genome.
Discrepancies within our different data sets led us to refine our analyses of the integration profiles of both PERV and MLV. We show here that the choice of restriction enzymes used to identify the integration sites does have an impact on the final result of integration profiles. MseI leads to an underestimation of the integration in high CpG islands areas, and gammaretrovirus integrations occur preferentially in genome areas enriched in genes and CpG islands.
MATERIALS AND METHODS
Virus preparation and cell infection.
The PERV plasmids used throughout the present study have been described previously (3, 4, 17). The accession numbers for clones A14/220 and Ap60 are AY570980 and AY099323, respectively. Infectious PERV particles were produced from plasmids by transfecting HEK-293 cells with Lipofectamine (Gibco). The cells were maintained by serial passage for 4 weeks to amplify virus production before use in further infections. Cell-free supernatants were filtered (0.22-μm pore size) and applied to human cell cultures for 4 h as described previously (23). The C8166 cells were grown in suspension and needed to be centrifuged for replacement of the medium after the 4-h PERV incubation. PERV-infected cells were maintained in culture for either 2 days or 2 weeks.
Integration site identification.
PERV integration sites were cloned by the ligation-mediated PCR (LM-PCR) protocol as described previously (23). After extraction, genomic DNA was cleaved with either MseI or the AvrII/NheI restriction mix and ligated to the double-strand linker used in our earlier study and described by Schröder et al. (28). The products were cleaved with EaeI to prevent internal viral fragment amplification during nested PCR, which was performed by using long terminal repeat and linker-specific primers. Amplicons were purified on membrane (MinElute PCR purification kit; Qiagen) and cloned with a TOPO TA cloning kit (Invitrogen). Sequencing was performed on an ABI Prism 3130-Avant genetic analyzer (Applied Biosystems). Integration sites were mapped to the human genome on the May 2004 freeze (hg17) using the BLAT program. The oligonucleotides used are listed in Table S1 in the supplemental material.
Bioinformatic analysis.
The PERL language was used for data mining. The frequency of integration in the vicinity of the gene was determined by comparing the positions of the integration sites to the genomic positions of the RefSeq genes and CpG Islands. Bias induced by restriction endonucleases MseI and AvrII/NheI in the human genome (hg17 freeze) was determined with restriction maps. Simulations of the integration sites at these positions were then used as random controls.
Transcription profile analysis was done with publicly available Affymetrix HG-U133A and HG-U95A microarrays datasets. The accession numbers for the NCBI GEO data samples used in the present study were GSM21381 (41) and GSM50270 (35) for uninfected HEK-293 and HeLa cells, respectively.
Gene density indexes.
Four complementary gene density indexes were calculated for each RefSeq identification code (ID). The gene density index was the sum of all of the RefSeq IDs present in a ±1-MB window from the TSS of each RefSeq ID. The CpG+ gene density index was the sum of all of the RefSeq ID with at least one CpG island identified within ±5 kb of their TSS present in the same ±1-Mb window, and the CpG− gene density index was the sum of all of the RefSeq ID with no CpG island identified within ±5 kb of their TSS present in the same window. The delta CpG index was the difference between the CpG+ index and the CpG− index. A negative delta CpG index indicated a relative enrichment of CpG− genes in a given area of the genome. A positive delta CpG index indicated a relative enrichment of CpG+ genes in a given area of the genome. Delta indexes close to zero resulted from either a balanced CpG+ or CpG− distribution in the same window or a low gene density index.
GenBank accession numbers.
The sequence data from the present study have been submitted to GenBank under accession numbers FI185198 to FI187345.
RESULTS
PERV integration sites identified in this study.
In the present study, 1,948 unique PERV integration sites were characterized from nine datasets obtained with four different PERV viruses derived from four molecular clones. Clones A14/220 and Ap60 were obtained by a PCR-based method from biological isolates (17). Clones 1 and 2 are chimeric constructions between clones A14/220 and Ap60 (see Fig. S1 in the supplemental material). Isolate A14/220 is a natural recombinant between PERV-A and PERV-C, which was obtained by transmission from activated peripheral blood mononuclear cells from miniature swine to HEK-293 human cells (25). The clone A14/220 genome is mainly related to PERV-C except for the env receptor-binding domain, which is related to PERV-A. Isolate Ap60 is a PERV-A derived from porcine PK15 cells, which naturally produce particles of PERV subtypes A and B (26, 33).
Most infections were performed in the HEK-293 cell line, and one was performed in the C8166 human T-cell line (see Table S2 in the supplemental material). Integration profiles from seven datasets obtained with HEK-293 cells comprising a total of 1,773 integration sites were used for the present study. Some datasets using the high-titer PERV A14/220 were obtained by analyzing integration sites on day 2 postinfection instead of the standard 15 days postinfection. One of our objectives in the present study was to examine the existence of hot spots following PERV integration. It is possible that retrovirus integration profiles differ during the time course of the viruses spreading in the culture. We initially compared results after short (2 days) and long (15 days) cultivation periods and found high reproducibility in the integration profiles (cf. Table 2 and Table S2 in the supplemental material). We therefore analyzed DNA on day 15 postinfection, which gave a better yield of cloned integration sites. As in our previous study, we used a biotin-streptavidin primer tag selection after the ligation-mediated PCR. However, due to the presence of a MseI restriction site in the 3′ long terminal repeat of clones A14/220, we used an AvrII/NheI enzyme mix, instead of MseI, to digest the cellular DNA. The possible introduction of a bias, in favor of the integrations close to restriction sites, has been regularly addressed but considered negligible (22). However, due to the affinity of the PERV integrations for the CpG environment (23), the differences in the G/C composition of the restriction sites recognized by the enzymes MseI (TTAA), AvrII (CCTAGG), NheI (GCTAGC), and SpeI (ACTAGT) remained a matter of concern for us. We estimated this possible bias by first screening the human genome for all of the MseI, AvrII, NheI, and SpeI restriction sites and randomly selected a set of 10,000 restriction sites for each enzyme mix (MseI alone, AvrII/NheI, or AvrII/NheI/SpeI). We then assessed the distribution of the selected sites, with regard to the three genomic features for which we have already reported an enrichment in PERV integration, namely, the TU of RefSeq genes, the proximity to CpG islands (±5 kb), and the proximity of TU to the TSS (±5 kb). The results obtained with the different mixtures of restriction enzymes highlighted some bias in the distribution of the restriction sites in the selected features (Table 1), an underestimation in the CpG islands and TSS being obtained with MseI compared to the AvrII/NheI mix (respectively, 1.77- and 1.44-fold lower with MseI). A small bias was also observed for TU with the different restriction mixes. Compared to our previous study, the replacement of MseI by AvrII/NheI might then theoretically increase the percentage of integration recovery near CpG islands by ±5 kb and TSS by ±5 kb from 39 and 34%, respectively, to 65 and 50%. The results that we obtained with the different PERV clones were in fact very close to our predictions, with values ranging from 52.8% to 68.8% near CpG islands and from 37.5% to 48.7% near TSS (Table 2). To confirm that the observed sway in the integration profiles was induced by the AvrII/NheI mix and did not reflect differences in the integration profile of A14/220-derived PERV, HEK-293 cells were infected with Ap60 (a PERV molecular clone obtained from PK15). The genomic DNA was then extracted and digested with either MseI or the AvrII/NheI mix to compare the integration profiles. The distribution of Ap60 integration with MseI perfectly matched that obtained in our previous study and differed from that obtained with the AvrII/NheI mix (37.5% versus 43% in TU, 42.2% versus 62.8% near CpG islands, and 36.7% versus 43% near TSS).
TABLE 2.
Influence of endonuclease restriction enzyme combinations on the distribution of recombinant and wild-type PERV integrations in the human genome (HEK-293 cells)a
| Site category | Human genome (% distribution) | % Distribution (P)b
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AvrII/NheI LM-PCR
|
MseI LM-PCR
|
|||||||||||
| A14/220c (n = 303) | Clone 1 (n = 242) | Clone 2 (n = 235) | A14/220-1 (n = 309) | A14/220-2 (n = 349) | AP60 (n = 207) | Mean (n = 1,645) | Random AvrII/NheI | PERV PK15d (n = 189) | AP60 (n = 128) | Random MseI | ||
| Within RefSeq genes | 34.3 | 46.5 | 49.2 | 55.7 | 48.4 | 47.6 | 43.0 | 48.4 | 36.5 | 43.9 (0.0054) | 37.5 (0.69) | 35.8 |
| Within ±5 kb of CpG islands | 7.8 | 58.7 | 59.9 | 60.9 | 68.8 | 52.8 | 62.8 | 60.3 | 9.4 | 39.0 | 42.2 | 5.3 |
| Within ±5 kb of RefSeq TSS | 5.9 | 42.2 | 45.0 | 46.0 | 48.7 | 37.5 | 43.0 | 43.8 | 6.7 | 33.9 | 36.7 | 4.6 |
Values for human genome, random AvrII/NheI, and random MseI are for 10,000 sites.
All P values (chi-square) compared to random AvrII/NheI for AvrII/NheI LM-PCR or random MseI for MseI LM-PCR were <0.0001 except for the P values indicated in parentheses.
The dataset at day 2 postinfection (all the other datasets are from day 15 postinfection).
Dataset from Moalic et al. (23).
TABLE 1.
Distribution of restriction sites in the human genome
| Site category | % Distribution (P)a
|
|||
|---|---|---|---|---|
| MseI | AvrII/NheI | AvrII/NheI/SpeI | Random | |
| Within RefSeq genes | 35.8 (<0.001) | 36.5 (<0.001) | 36.1 (<0.001) | 34.3 |
| Within ±5 kb of CpG islands | 5.3 (<1.137E−20) | 9.4 (<2.42E−09) | 8.3 (NS) | 7.8 |
| Within ±5 kb of RefSeq TSS | 4.6 (<3.44E-08) | 6.7 (<0.0006) | 5.9 (NS) | 5.9 |
P values are compared to the random values. NS, not significantly different from random.
We noted that the signature of the integration mechanism, highlighted by a statistically favored sequence of DNA surrounding the integration site (23), was confirmed for our different datasets (see Fig. S2 in the supplemental material). This sequence was even extended from 8 to 12 bases [−4]GCTG(int)GTACCAGC[7], due to the high number of integration sites analyzed.
Description of PERV integration site hot spots.
We did not show evidence for hot spots in the PERV integration profile in our previous study (23) and hypothesized that the underlining of hot spots might be precluded by the low number of integration sites characterized (<200). By using the AvrII/NheI restriction enzyme mix, which focused 60% of the PERV integrations identified in a genome area representing no more than 8% of the human genome (cf. Table 1), we ensure potentially optimal conditions to address the question of the occurrence of hot spots during PERV integration.
The criterion used to define hot spots was the same one applied to define common insertional sites (CIS) (40), with at least two independent insertions contained within <30 kb, three in <50 kb and, four or more in <100 kb. Looking for hot spots within a limited area of the genome (60% of the integrations in 8% of the genome), necessarily generates, with the increase of integration sites identified, the appearance of some hot spots by chance. We therefore simulated different sets of random integration profiles (that matched the profile observed for the PERV integrations) to estimate the number of hot spots that would occur by chance. The simulated integrations had to fulfill the following criteria: (i) they had to be close to AvrII/NheI sites (the mean distance between experimental PERV integration and AvrII/NheI sites was <200 bp and, for convenience, the simulated integration sites were indeed AvrII/NheI sites) and (ii) the profiles for each simulated integration set had to be similar to the mean profile obtained with PERV, i.e., 48.4% in TU of RefSeq genes, 60.3% in CpG ± 5 kb and 43.8 in TSS ± 5 kb.
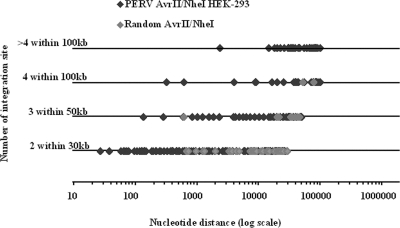
The integration simulations yielded very few hot spots ranging from 1 to 6 hot spots per set of simulations with a sum of 67 hot spots totaling 146 integrations (8.9%) for a total of 1,645 simulated integrations. Conversely, the number of hot spots observed in the PERV integration datasets was significantly higher (P < 0.05, analysis of covariance), ranging from 11 to 37 hot spots per PERV integration set, with a total of 224 hot spots totaling 671 PERV integrations that represented 40.8% of the 1645 PERV integrations identified. Closer examination revealed that the PERV-induced and simulated hot spots displayed significantly different features (Fig. 1). Most of the simulated hot spots were in the 30-kb window (57/67) with two simulated integration sites per hot spot. Only two hot spots were in the 100-kb range, and none had more than four integrations. Conversely, several PERV hot spots were in the 100-kb range, 16 hot spots had more than four integrations, and 1 hot spot culminated with eleven PERV integrations within an 18-kb window. Finally, the window sizes of the hot spots, theoretically defined as 2, 3, or ≥4 integrations in window sizes of 30, 50, or 100 kb, were in fact much narrower with PERV, in which the mean window sizes were, respectively 8, 19.5, and 46 kb versus 11, 29.7, and 64.9 kb for the simulated integrations.
FIG. 1.
Distribution and characteristics of PERV and simulated hot spots. The diagram represents the real windows size of the hot spots for each category of CIS windows. PERV hot-spot (black diamonds) and random hot-spot (gray diamonds) datasets are as defined in the figure. Random hot spots are found mainly in the two integration categories of CIS.
We further investigated the features of hot spots by looking at the effect of the expression level of the genes on the hot-spot distribution.
PERV and MLV integrations are independent of the expression levels of the genes.
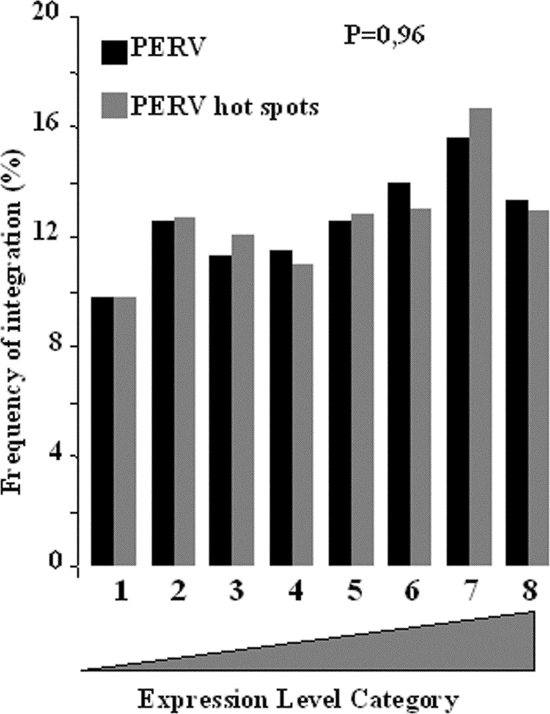
Since a slight preference had previously been described for PERV and MLV integrations close to genes with higher expression levels (23), we inferred that hot spots, which resulted from an increased affinity for some part of the genome, should accentuate this by adding both preferences. Unexpectedly, the distribution of the integrations in the different bins of expression were the same whether these integrations were in hot spots or not (Fig. 2). This indirectly suggested that expression levels might be of little, if any, significance in PERV integration and might possibly be an artifact of the covariation of expression levels with some other features of the genes.
FIG. 2.
Effect of the transcription activity of the gene on PERV hot-spot formation. Expression levels were assayed by using Affymetrix HU133A microarray data sets. All genes on the chips were divided into eight bins according to their relative levels of expression. The frequency distributions of all PERV AvrII/NheI integration sites (black bars) were compared to those of PERV AvrII/NheI integration sites present in hot spots (gray bars). The P value was determined by using the chi-square test.
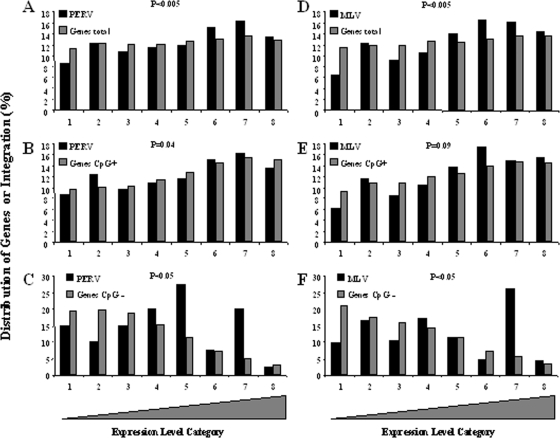
The PERV integration profile was strongly associated with proximity to CpG islands (cf. Table 2), even more than with proximity to a TSS. This suggested that the neighborhood of a CpG island might be more critical for PERV integration than the sole presence of a TSS. Even if CpG islands are strongly associated with TSS, not all genes have CpG islands in close proximity to their TSS (27). We then divided the RefSeq genes into two classes: one with CpG islands contained within ±5 kb from TSS (called CpG+), which represented 72% of the genes, and the other class with the closest CpG island distance being more than 5 kb from TSS (called CpG−), which represented 27% of the genes. Examination of the expression of CpG+ and CpG− genes revealed that the percentage of CpG+ genes regularly increased in the bins with increasing expression levels, with this distribution varying from 10 to 15% from bin 1 (low expression) to bin 8 (high expression) for the CpG+ genes (Fig. 3B and E) and, conversely, from 19 to 3% from bin 1 to bin 8 (Fig. 3C and F) for the CpG− genes (this was consistent for different cell lines [cf. Fig. S3 in the supplemental material]). Classification of the genes according to their expression level indeed resulted in a covariation of the increase in expression level and in the percentage of CpG+ genes. Under these conditions, the slight affinity observed for highly expressed genes cannot be dissociated from the increase in CpG+ genes. We then reanalyzed the distribution of retroviral integrations according to the expression level of the two gene categories (CpG+ or CpG−) (cf. Fig. 3).
FIG. 3.
Influence of CpG island distribution within expression level bins on the distribution of gammaretroviral integration. Expression levels were assayed by using Affymetrix HU133A microarray data sets. All genes on the chips were divided into eight bins according to their relative levels of expression. The distribution of the PERV integration (A, B, and C) or MLV integration (D, E, and F) are given according to the expression level of the genes. Panels A and D represent 100% of the genes present on the chip. Panels B and E represent the distributions of the CpG+ genes present on the chip (∼75% of the genes present in panels A and E). Panels C and F represent the distribution of the CpG− genes present on the chip (∼25% of the genes present in panels A and E). For panels B, C, E, and F, the distribution of the genes is expressed as the percentage of the number of genes that are present in each of the categories (i.e., CpG+ or CpG−). While the distributions of PERV and MLV integration are different from the distributions of the genes in panels A and E (P < 0.005) for the CpG+ genes (B and E), the distributions of the integrations are the same. For the CpG− genes, the distribution of the integration is different from the distribution of the genes (P < 0.005).
The analysis of the distribution of integrations performed on all of the genes (irrespective of the proximity to CpG islands) reproduced the significant slight increase in integration for bins 6 and 7 for PERV (Fig. 3A) and bins 5, 6, and 7 for MLV (Fig. 3D) and a significant strong preference for the same bin for HIV as previously described (see Fig. S4 in the supplemental material). Splitting the genes into the CpG+ and CpG− categories strongly lowered the apparent preference of gammaretrovirus for the highly expressed genes (Fig. 3B and E) in the CpG+ gene population. For HIV, the preference for highly expressed genes was maintained in the CpG+ population (see Fig. S4 in the supplemental material). For the CpG− gene population, the number of integrations was low, and their distributions (both for PERV and MLV) did not match the distribution of the genes (Fig. 3C and F) (P < 0.05): some high expression bins displayed more integration than expected. The apparent slight affinity of gammaretroviruses for highly expressed genes in fact reflected a preferred affinity for (CpG+) genes associated with CpG islands.
Clusters of CpG− genes are cold PERV integration areas in the human genome.
We have shown that the role played by the level of gene expressions in PERV and MLV integration process is probably very marginal but that the CpG environment might be highly relevant. In a recent article, Berry et al. identified CpG islands, gene density, and DNase I hypersensitive sites as genome features that favored gammaretrovirus integration (5). CpG island density and gene density are known to be highly correlated and so we hypothesized that if PERV integration occurred preferentially close to CpG+ genes, then clusters of CpG− genes should be underrepresented, irrespective of the gene density.
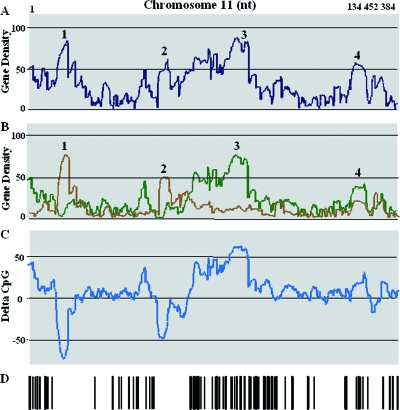
As hypothesized, several areas enriched in CpG+ or CpG− genes have been identified throughout the human genome. Chromosome 11 seems to provide useful information about variation in the gene density index, the CpG+ density index, the CpG− density index, and PERV integration (Fig. 4) and will therefore be used throughout this section for the purpose of our demonstration.
FIG. 4.
Variation of gene density and distribution of PERV integration along chromosome 11. (A) Variation of gene density. For each gene (identified by the RefSeq ID), the gene density was calculated for a 2-Mb window surrounding their TSS. (B) A similar calculation was performed, but the genes were separated into two populations: genes with CpG islands (green line) and genes without CpG islands (brown line). (C) For each gene, a delta index corresponding to the density of the CpG+ genes minus the density of the CpG− genes was calculated. Areas enriched in CpG− genes displayed negative values; areas enriched in CpG+ genes displayed positive values. (D) Bar code representation of PERV integration along chromosome 11. PERV integrations were mainly present in highly positive delta CpG index areas.
Chromosome 11 is 134,452,384-bp long and displayed 119 PERV integrations. Figure 4A shows the variations in gene density index along chromosome 11; Fig. 4B shows the same variations but with the CpG+ and CpG− gene densities calculated separately. Figure 4D represents the distribution of PERV integrations along chromosome 11. The gene density (calculated for a 2-Mb window) varied from 0 to 88 genes. Four areas were of particular interest: area 1 with a maximum full gene density index of 84 containing mainly CpG− genes (CpG− density index of 75), area 2 with a maximum full gene density of 62 (maximum CpG− density index of 49), area 3 with a maximum full gene density index of 88 containing mainly CpG+ genes (CpG+ density index of 75), and area 4 with a maximum gene density of 58 (maximum CpG+ density index of 38). All areas had high gene indexes but showed different PERV integration densities. Areas 3 and 4 (high CpG+ index) were targets for PERV integrations, whereas areas 1 and 2 (high CpG− index) were not. Thus, gene density alone did not appear to be a good predictor of PERV integration. When the genes were categorized as CpG+ or CpG−, correlation with the occurrence of PERV integration was much improved, the rate of integration being high in CpG+ gene areas and low in CpG− gene areas.
To determine whether we could generalize this observation to the entire genome, we then calculated, for each gene, a single index, designated the delta CpG index, which corresponded to the difference between its CpG+ and CpG− gene indexes. This new index clearly defined the area enriched in CpG+ or CpG− genes. Figure 4C illustrates the delta index for chromosome 11. The CpG+ areas are apparent as peaks (positive values), whereas the CpG− enriched areas are apparent as troughs (negative values). The troughs visualized on chromosome 11 did not display any integration (as visualized in Fig. 4C and D), whereas the peaks appeared to be targets for PERV integration. It should be noted that some areas were neither peaks nor troughs and corresponded to either mixed classes of CpG+ and CpG− genes with low delta indexes or to low-gene-density areas. These areas were occasionally targets for PERV integration but with seemingly lower frequency.
Genome areas enriched with CpG+ genes favored PERV integration, were neutral for HIV integration, and disfavored foamy virus (FV) and avian sarcoma-leukosis virus (ASLV) integration.
We have shown for chromosome 11 that the gene density is not a good predictor of PERV integration, that negative delta indexes (i.e., CpG− enriched areas) disfavor PERV integration, and that a delta CpG index provides a more satisfactory prediction of PERV integration. Among the integration profiles of the different retrovirus studied thus far, gene density has been positively correlated with the integration of gammaretroviruses and lentiviruses, but not of alpharetrovirus, deltaretrovirus, or spumavirus (5). We wondered whether nongammaretroviruses might also integrate preferentially in CpG+ areas or areas with a positive delta index. When we considered a 2-Mb window throughout the genome the calculated delta index varied from −75 to +110. and the vast majority of the genes (60%) were between the delta indexes of 1 and 20, 18% of the genes had a delta index below 0, and 22% had a delta index above 20. Comparative analysis of the distribution of the integrations and of the genes, according to the delta index, displayed a significant bias for most of the retrovirus integration profiles observed except for HIV. These biases are represented in Fig. 5, which shows the progressive accumulation (as a percentage) of the genes and of the integrations for MLV, PERV, HIV, and FV, going from the negative delta indexes to the positive delta indexes. The sharp increase between delta indexes 0 and 20 illustrates the fact that 60% of the genes are contained between these two limits. For FV and ASLV, 24 and 18%, respectively, of the integrations were displayed between delta indexes −75 and −1 (whereas only 15.5% of the genes are contained within these indexes) and 94.2 and 91.3% by delta index 20 (for 78.6% of the genes). For MLV integration, 15.1% occurred between delta indexes −75 and 0 and matched the percentage of genes (15.5%), but from delta index 1 to delta index 20, the increase in MLV integration was slightly more rapid than the rise in RefSeq (86.1% MLV versus 78.6% genes). The percent integration in negative delta index classes (from −75 to 0) was lower for HIV (10.8% versus 15.5% for genes), but then the HIV integration curve paralleled the curve of gene distribution. For PERV, the percent integration from delta indexes −75 to 0 was 6.2% and much lower than the gene percentage (15.5%), reaching only 62.7% by delta index 20. Thus, the increase in percent integration occurred more slowly than the increase in the gene percentage. The remaining 37.3% of the PERV integrations were distributed between delta indexes 21 and 111, which (represented only 21.4% of the genes).
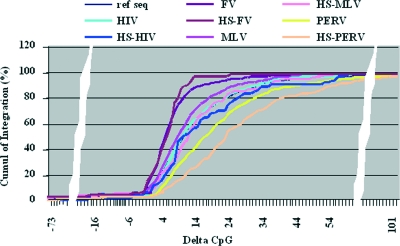
FIG. 5.
Distribution of retroviral integrations according to the delta CpG index. The genes were classified according to their delta CpG index and represented as the sum of the distributions of the gene (in percentages) when moving from the most negative to the most positive delta CpG index. A similar classification was performed for the retroviral integrations of FV, MLV, HIV, and PERV (ASLV [not represented on the figure] displayed the same profile as FV). Integrations present in hot spots were also plotted separately for each data set (hot-spot curves). For all of the viruses except FV, a shift of the curve toward a higher delta CpG index was observed for the integrations present in hot spots (P < 0.005). The curve for the genes (noted RefSeq) is completely overlaid by the HIV integration curve and is not apparent. The retroviral integration datasets used are listed in Table S2 in the supplemental material.
These results show that retrovirus integration, irrespective of the virus, was disfavored for genes localized in the most negative delta index areas. For FV and ASLV, either genes with a low negative delta index or localized in low positive delta index areas were favored for integration, whereas high positive delta indexes were detrimental to integration. For MLV, low positive delta indexes favored integration. For HIV, genes localized in positive delta index area were favored irrespective of the delta index value. For PERV, integration would be favored in genes localized in areas with higher positive delta indexes (χ2 test, P < 0.005).
Assuming then that high positive delta indexes favor PERV integration, such areas should have a higher number of integrations, and we might thus expect PERV hot spots to be over-represented in higher-delta-index areas. This is indeed what we observed, with a significant shift of the integration hot-spot curve toward higher delta index values (χ2 test, P < 0.005). Interestingly, a slight but significant increase was also observed for HIV and MLV (χ2 test, P < 0.05) but not for FV or ASLV.
Choice of restriction enzyme accounted for discrepancies between integration profiles.
In a previous article, we described the features common to PERV and MLV (23). In the present study, with regard to the delta index, the integration profile of MLV appeared to be closer to the FV and ASLV integration profiles than to the PERV integration profile. This behavior of MLV suggested discrepancies between the integration profiles of the two gammaretroviruses MLV and PERV.
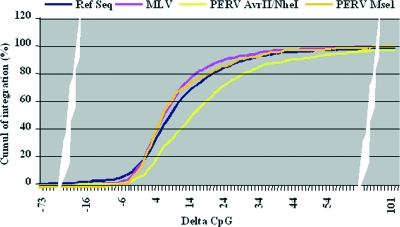
The MLV integration data set described by Wu et al. (39) relied on the use of the restriction enzyme MseI, as did our initial study of PERV integration (23). For the present study, we switched to a different enzyme mix (described at the beginning of Results) and showed that this affected the apparent distribution of integration near CpG islands and TSS (Table 1). We then wondered whether the observed discrepancy between PERV and MLV for the delta index preference was linked to the enzyme mix used. The results, shown in Fig. 6, clearly illustrate that the enzyme mix did result in significant differences in the PERV integration profiles in relation to the delta index. The curve obtained with the MseI enzyme switched PERV integrations toward a lower delta index (P < 0.05) and was identical to the one obtained for MLV integration.
FIG. 6.
Influence of the restriction enzymes used on the distribution of the PERV integration according to the delta CpG index. PERV integrations obtained after either MseI or AvrII/NheI digestion of the DNA were classified according to their delta CpG indexes and are represented as the sum of their distributions (in percentages) when moving from the most negative to the most positive delta CpG index. The distributions of the genes and of the MLV-MseI integrations are also represented. Replacing AvrII/NheI by MseI suppressed the offset toward higher delta CpG indexes for PERV integration (P < 0.05), which then displayed the same profile as the MLV-MseI integration.
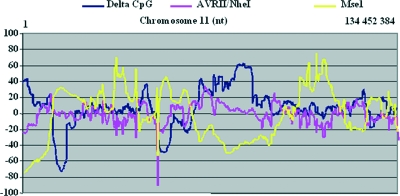
Although the number of MseI restriction sites is enormous (several million throughout the genome), their distribution along the chromosomes is subject to huge variation, and high-delta-index areas displayed the lowest density of MseI sites, as illustrated for chromosome 11 (Fig. 7), with a correlation coefficient between delta index and restriction enzyme sites of r = −0.6 for MseI and r = +0.25 for the AvrII/NheI mix. We then produced random simulations of 10,000 integration sites based on the different restriction enzyme sites to verify the possible influence of enzyme mix on the integration profile with regard to the delta index. The profiles obtained for the different enzymes or the enzyme mix were very similar, with most integrations within the low-delta-index area, which also represents most of the gene-containing area of the genome (Fig. 8). However, a similar random simulation that took into account the integration profile of PERV (i.e., integration close to a CpG island and TSS) according to our two PERV integration studies showed clear differences in the distribution of the simulated integrations according to the restriction enzymes used. With MseI, most of the simulated integrations landed in low-positive-delta-index area, whereas with the AvrII-NheI mix the simulated integration curve was skewed toward the gene distribution curve (P < 0.005), reflecting more simulated integrations in the higher-delta-index range, a curve very close to the one observed for HIV. Interestingly, the Rb2 AvrII/NheI simulation differed from the PERV integration curve (see Fig. 5) and did not shift this curve toward very high delta CpG index values. The shift of the PERV-AvrII/NheI experimental data must be considered a true feature of PERV and not as a bias that might have been introduced by the enzyme mix. In contrast, the integration profiles observed with the MLV-MseI or PERV-MseI datasets are likely, at least in part, to be due to a skewed distribution of the integrations linked to the enzyme used.
FIG. 7.
Variation of MseI and AvrII/NheI restriction site densities along chromosome 11. The densities of the distribution of MseI or AvrII/NheI restriction sites were calculated for a 2-Mb window size along chromosome 11. Density variations were then calculated by reference to a theoretical density (that is, the number of restriction sites present on chromosome 11/the number of bases of chromosome 11). Important variations in the density of restriction sites are observed. MseI displayed the widest variations in site density, and variations were inversely correlated with the delta CpG index (r = −0.6). AvrII/NheI displayed fewer variations, and no correlation with delta CpG index was noted (r = 0.25).
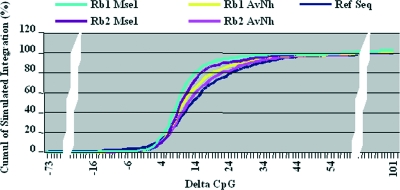
FIG. 8.
Simulation of the delta CpG index distribution of integrations obtained with either MseI or AvrII/NheI. PERV integration profiles were simulated based on the use of either MseI or AvrII/NheI. The profiles obtained had to be similar to the one obtained for the PERV. Rb1 is a profile with a distribution of integrations of 39% CpG, 34% TSS, and 44% RefSeq; Rb2 is a profile with a distribution of integrations of 60% CpG, 40% TSS, and 44% RefSeq. Both MseI profiles (Rb1 MseI and Rb2 MseI) displayed a more rapid accumulation of simulated integration in the low-delta-CpG area than the distribution of the genes (dark blue line) or the accumulation of simulated integration with AvrII/NheI profiles (Rb1 AvrII/NheI and Rb2 AvrII/NheI) (P < 0.005).
DISCUSSION
Hot spots have been regularly observed in the genome wide surveys of retroviral integration but remain poorly explained. Retroviral integration hot spots were initially described with HIV in SupT1 cells, but this phenomenon was not observed in the other cell types studied (28). A recent study compared the hot spots of gammaretroviral and lentiviral vector integration in human CD34+ hematopoietic cells (18). The results showed that a high frequency (>20%) of gammaretroviral integration sites occur in hot spots, whereas they are significantly less frequent in the case of lentiviral vectors, suggesting that a propensity for generating hot spots is a feature of the retrovirus-derived vector. ASLV, for example, which presents an integration pattern close to randomness, displays a very small number of hot spots (<10 hot spots for 500 integrations) that likely occur by chance (22). Our previous study of PERV integration did not reveal significant hot spots. Conversely, in the present study the PERV integrations identified gave rise to a collection of 224 hot spots and some apparent discrepancies with our earlier data set, which led us to reinvestigate the integration profiles of PERV and gammaretroviruses in cell lines.
Integration profiles of retroviruses have been extensively analyzed with regard to numerous features of the genome (5). Each of these features, analyzed separately, participates in retrovirus integration site profiling, but potential covariation of certain features has been insufficiently considered and may have led to misinterpretations. For example, it seemed illogical to us that the slight preference for high-expression genes described for gammaretrovirus integration (23, 39) was not reinforced for those integrations in hot spots. The splitting of the genes into two classes, one with CpG island content and the other without CpG island content, allowed us to pinpoint some covariations and to refine our understanding of gammaretroviral integration. The fact that the apparent slight preference for integration in highly expressed genes can be erased by a subclassification that takes into account both the expression level of genes and the presence (or absence) of CpG islands demonstrates that the gene expression level is not important for gammaretrovirus integration close to CpG-bearing genes (this might not be true for the CpG-deficient genes). This observation must, however, be placed in the context of a genome which, in cultured cell lines, is pervasively transcribed (14). The integration of gammaretrovirus during the correction of X1 or ADA genes in SCID therapy trials has also been associated with the transcribed genes (1, 12, 29), but it must be noted that there is no correlation between the common insertion site localization and the intensity of expression (12). In these studies, the covariation issue was not addressed and, further, the pervasiveness of the chromatin transcription might be significantly different from that observed in cultured cells.
Our results concerning gene expression levels enabled us to highlight the impact of gene density on gammaretroviral integration. Affinity for the CpG area and TSS was clearly demonstrated in previous studies (23, 39). It was interpreted as an affinity for the gene promoter region, and the two parameters (CpG and TSS) were kept in close association. However, even if most gene promoters are in close proximity to CpG islands (∼70%), a significant number are not (27), and this gave us an opportunity to distinguish between the respective influence(s) of CpG islands, TSS, and gene density. Dissociating the genes into two populations (CpG+ and CpG−) allowed the concept of gene density to be refined, and the identification of clusters of genes off CpG (especially on chromosome 11) clearly revealed areas of the genome which, despite a very high density index, were totally devoid of PERV integrations. Gene density is thus of poor prognostic value, with regard to retroviral integration, if no other qualification is applied to the genes. Interestingly, some of the areas that are devoid of PERV integration have also been described as under-represented for HIV integration (38), suggesting that there might be integration cold spots, which are common to gammaretroviruses and lentiviruses, in the human genome. This hypothesis fits with recent data from Shun et al., who showed that, in the absence of LEDGF/P75, the affinity of HIV integration for TU is swayed in favor of promoter and CpG islands and the correlation with gene expression is weaker (31). Some basic requirements for gammaretrovirus and lentivirus integration might thus be shared. Further, Wang et al. have recently shown that HIV integration is favored near the transcription-associated histone modifications, i.e., H3 acetylation, H4 acetylation, and H3 K4 methylations (38), which are mainly present with CpG island-associated genes (14).
Comprehension of the determinants of retroviral integration site preferences in the host genome is fundamental to the use and safety evaluation of retrovirus-based vectors in gene therapy. Since the first genome-scale analysis of HIV integration in human cells by Bushman's group (28), the integration profiles of different genera of retroviruses have been described. All of these studies have made it clear that the different retroviral integration profiles observed are a feature of the different retroviral genera and that integration is essentially driven by the virus-encoded integrase (20) via interactions with specific proteins from the host (37) and/or specific DNA structures. Vectors that rely on retroviral integrases to insert therapeutic genes in the genome of patients will therefore display similar integration profiles. This has been verified experimentally in human cells, in vitro, for lentivirus-derived vector (2) and also recently for cells isolated from different SCID patients treated with a gammaretroviral vector (1, 12, 29). The multiple adverse event that have been observed following either experimental (21, 30) or therapeutic (16) retroviral gene therapy will necessitate a very careful clinical monitoring of vector integrations during therapeutic trials. This monitoring will have to ensure as a prerequisite the full and homogeneous coverage of the patient's genome before granting that no adverse integration events have occurred. Partial recovery of integration events following LM-PCR have already been reported by Nagy et al. (24), and the results presented here provide strong evidence that the digestion of the genome with restriction enzyme mixes will not grant a homogeneous coverage of the genome. Genome coverage has been a matter of concern for sequencing programs, and especially for shotgun sequencing, for several years. Some devices have been developed that ensure homogeneous shearing of DNA (34). More recently, a PCR method, multiple displacement amplification (MDA), has been described for a whole unbiased amplification of genomes (11). The authors of that study claim successful whole-genome amplification from as few as 1 to 10 copies of human genome, which would allow the monitoring of a clinical gene therapy trial. MDA has been recently successfully used for retroviral gene therapy in vitro and in vivo with a NOD/SCID mouse model (6) and might prove to be the most straightforward technique currently available for monitoring retroviral integration. However, parallel LM-PCR experiments from an MDA reaction reveals a partial coverage of the integrations for each LM-PCR, and repeated LM-PCR analyses of the samples will most probably be required to estimate the recovery of integration sites (6).
According to our results, gammaretroviral vectors might be one of the worst choices for use in human gene therapy since they target gene-dense areas. The use of lentiviral vectors might be a safer alternative since these avoid the promoter region of the targeted genes but can efficiently disrupt the genes. Integrating vectors based on an ASLV backbone, with an integration profile close to randomness and an affinity for low-gene density areas, might provide a favorable balance between hope and danger.
Supplementary Material
Acknowledgments
We thank Sophie Roucan for restriction site mapping (GenOuest bioinformatics platform). We thank Birke Bartosch for help with PERV constructions.
Y.M. was supported by a grant from the Foundation for Medical Research (FDT20061208658).
Footnotes
Published ahead of print on 26 November 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aiuti, A., B. Cassani, G. Andolfi, M. Mirolo, L. Biasco, A. Recchia, F. Urbinati, C. Valacca, S. Scaramuzza, M. Aker, S. Slavin, M. Cazzola, D. Sartori, A. Ambrosi, C. Di Serio, M. G. Roncarolo, F. Mavilio, and C. Bordignon. 2007. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J. Clin. Investig. 1172233-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr, S. D., A. Ciuffi, J. Leipzig, P. Shinn, J. R. Ecker, and F. D. Bushman. 2006. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol. Ther. 14218-225. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., D. Stefanidis, R. Myers, R. Weiss, C. Patience, and Y. Takeuchi. 2004. Evidence and consequence of porcine endogenous retrovirus recombination. J. Virol. 7813880-13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., R. A. Weiss, and Y. Takeuchi. 2002. PCR-based cloning and immunocytological titration of infectious porcine endogenous retrovirus subgroup A and B. J. Gen. Virol. 832231-2240. [DOI] [PubMed] [Google Scholar]
- 5.Berry, C., S. Hannenhalli, J. Leipzig, and F. D. Bushman. 2006. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleier, S., P. Maier, H. Allgayer, F. Wenz, W. J. Zeller, S. Fruehauf, and S. Laufs. 2008. Multiple displacement amplification enables large-scale clonal analysis following retroviral gene therapy. J. Virol. 822448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3848-858. [DOI] [PubMed] [Google Scholar]
- 8.Bushman, F. D. 2007. Retroviral integration and human gene therapy. J. Clin. Investig. 1172083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Press, New York, NY. [PubMed]
- 10.Crise, B., Y. Li, C. Yuan, D. R. Morcock, D. Whitby, D. J. Munroe, L. O. Arthur, and X. Wu. 2005. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J. Virol. 7912199-12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean, F. B., S. Hosono, L. Fang, X. Wu, A. F. Faruqi, P. Bray-Ward, Z. Sun, Q. Zong, Y. Du, J. Du, M. Driscoll, W. Song, S. F. Kingsmore, M. Egholm, and R. S. Lasken. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 995261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deichmann, A., S. Hacein-Bey-Abina, M. Schmidt, A. Garrigue, M. H. Brugman, J. Hu, H. Glimm, G. Gyapay, B. Prum, C. C. Fraser, N. Fischer, K. Schwarzwaelder, M. L. Siegler, D. de Ridder, K. Pike-Overzet, S. J. Howe, A. J. Thrasher, G. Wagemaker, U. Abel, F. J. Staal, E. Delabesse, J. L. Villeval, B. Aronow, C. Hue, C. Prinz, M. Wissler, C. Klanke, J. Weissenbach, I. Alexander, A. Fischer, C. von Kalle, and M. Cavazzana-Calvo. 2007. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Investig. 1172225-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derse, D., B. Crise, Y. Li, G. Princler, N. Lum, C. Stewart, C. F. McGrath, S. H. Hughes, D. J. Munroe, and X. Wu. 2007. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 816731-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447799-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, A., and M. Cavazzana-Calvo. 2005. Integration of retroviruses: a fine balance between efficiency and danger. PLoS Med. 2e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T-cell proliferation in two patients after gene therapy for SCID-X1. Science 302415-419. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, I., Y. Takeuchi, B. Bartosch, and J. P. Stoye. 2004. Determinants of high titer in recombinant porcine endogenous retroviruses. J. Virol. 7813871-13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hematti, P., B. K. Hong, C. Ferguson, R. Adler, H. Hanawa, S. Sellers, I. E. Holt, C. E. Eckfeldt, Y. Sharma, M. Schmidt, C. von Kalle, D. A. Persons, E. M. Billings, C. M. Verfaillie, A. W. Nienhuis, T. G. Wolfsberg, C. E. Dunbar, and B. Calmels. 2004. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, R., A. Trubetskoy, T. Suzuki, N. A. Jenkins, N. G. Copeland, and J. Lenz. 2003. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J. Virol. 772056-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewinski, M. K., M. Yamashita, M. Emerman, A. Ciuffi, H. Marshall, G. Crawford, F. Collins, P. Shinn, J. Leipzig, S. Hannenhalli, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2006. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Z., J. Dullmann, B. Schiedlmeier, M. Schmidt, C. von Kalle, J. Meyer, M. Forster, C. Stocking, A. Wahlers, O. Frank, W. Ostertag, K. Kuhlcke, H. G. Eckert, B. Fehse, and C. Baum. 2002. Murine leukemia induced by retroviral gene marking. Science 296497. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, R. S., B. F. Beitzel, A. R. Schröder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moalic, Y., Y. Blanchard, H. Felix, and A. Jestin. 2006. Porcine endogenous retrovirus integration sites in the human genome: features in common with those of murine leukemia virus. J. Virol. 8010980-10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy, K. Z., S. Laufs, B. Gentner, S. Naundorf, K. Kuehlcke, J. Topaly, E. C. Buss, W. J. Zeller, and S. Fruehauf. 2004. Clonal analysis of individual marrow-repopulating cells after experimental peripheral blood progenitor cell transplantation. Stem Cells 22570-579. [DOI] [PubMed] [Google Scholar]
- 25.Oldmixon, B. A., J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H. J. Schuurman, and C. Patience. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 763045-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3282-286. [DOI] [PubMed] [Google Scholar]
- 27.Saxonov, S., P. Berg, and D. L. Brutlag. 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 1031412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schröder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110521-529. [DOI] [PubMed] [Google Scholar]
- 29.Schwarzwaelder, K., S. J. Howe, M. Schmidt, M. H. Brugman, A. Deichmann, H. Glimm, S. Schmidt, C. Prinz, M. Wissler, D. J. King, F. Zhang, K. L. Parsley, K. C. Gilmour, J. Sinclair, J. Bayford, R. Peraj, K. Pike-Overzet, F. J. Staal, D. de Ridder, C. Kinnon, U. Abel, G. Wagemaker, H. B. Gaspar, A. J. Thrasher, and C. von Kalle. 2007. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J. Clin. Investig. 1172241-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seggewiss, R., S. Pittaluga, R. L. Adler, F. J. Guenaga, C. Ferguson, I. H. Pilz, B. Ryu, B. P. Sorrentino, W. S. Young III, R. E. Donahue, C. von Kalle, A. W. Nienhuis, and C. E. Dunbar. 2006. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood 1073865-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shun, M. C., N. K. Raghavendra, N. Vandegraaff, J. E. Daigle, S. Hughes, P. Kellam, P. Cherepanov, and A. Engelman. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 211767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki, T., H. Shen, K. Akagi, H. C. Morse, J. D. Malley, D. Q. Naiman, N. A. Jenkins, and N. G. Copeland. 2002. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32166-174. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 729986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorstenson, Y. R., S. P. Hunicke-Smith, P. J. Oefner, and R. W. Davis. 1998. An automated hydrodynamic process for controlled, unbiased DNA shearing. Genome Res. 8848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian, B., Y. Zhang, B. A. Luxon, R. P. Garofalo, A. Casola, M. Sinha, and A. R. Brasier. 2002. Identification of NF-κB-dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 766800-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trobridge, G. D., D. G. Miller, M. A. Jacobs, J. M. Allen, H. P. Kiem, R. Kaul, and D. W. Russell. 2006. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. USA 1031498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Maele, B., K. Busschots, L. Vandekerckhove, F. Christ, and Z. Debyser. 2006. Cellular cofactors of HIV-1 integration. Trends Biochem. Sci. 3198-105. [DOI] [PubMed] [Google Scholar]
- 38.Wang, G. P., A. Ciuffi, J. Leipzig, C. C. Berry, and F. D. Bushman. 2007. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 171186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 3001749-1751. [DOI] [PubMed] [Google Scholar]
- 40.Wu, X., B. T. Luke, and S. M. Burgess. 2006. Redefining the common insertion site. Virology 344292-295. [DOI] [PubMed] [Google Scholar]
- 41.Zagranichnaya, T. K., X. Wu, A. M. Danos, and M. L. Villereal. 2005. Gene expression profiles in HEK-293 cells with low or high store-operated calcium entry: can regulatory as well as regulated genes be identified? Physiol. Genomics 2114-33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.