Abstract
Western lowland gorillas (Gorilla gorilla gorilla) are infected with a simian immunodeficiency virus (SIVgor) that is closely related to chimpanzee and human immunodeficiency viruses (SIVcpz and HIV-1, respectively) in west central Africa. Although existing data suggest a chimpanzee origin for SIVgor, a paucity of available sequences has precluded definitive conclusions. Here, we report the molecular characterization of one partial (BQ664) and three full-length (CP684, CP2135, and CP2139) SIVgor genomes amplified from fecal RNAs of wild-living gorillas at two field sites in Cameroon. Phylogenetic analyses showed that all SIVgor strains clustered together, forming a monophyletic lineage throughout their genomes. Interestingly, the closest relatives of SIVgor were not SIVcpzPtt strains from west central African chimpanzees (Pan troglodytes troglodytes) but human viruses belonging to HIV-1 group O. In trees derived from most genomic regions, SIVgor and HIV-1 group O formed a sister clade to the SIVcpzPtt lineage. However, in a tree derived from 5′ pol sequences (∼900 bp), SIVgor and HIV-1 group O fell within the SIVcpzPtt radiation. The latter was due to two SIVcpzPtt strains that contained mosaic pol sequences, pointing to the existence of a divergent SIVcpzPtt lineage that gave rise to SIVgor and HIV-1 group O. Gorillas appear to have acquired this lineage at least 100 to 200 years ago. To examine the biological properties of SIVgor, we synthesized a full-length provirus from fecal consensus sequences. Transfection of the resulting clone (CP2139.287) into 293T cells yielded infectious virus that replicated efficiently in both human and chimpanzee CD4+ T cells and used CCR5 as the coreceptor for viral entry. Together, these results provide strong evidence that P. t. troglodytes apes were the source of SIVgor. These same apes may also have spawned the group O epidemic; however, the possibility that gorillas served as an intermediary host cannot be excluded.
Simian immunodeficiency viruses (SIVs) are known to naturally infect at least 40 different species of nonhuman primates in Sub-Saharan Africa (9, 72). Although each of these primate species harbors a genetically distinct lineage of SIV, phylogenetic evidence indicates that SIVs have crossed species boundaries on numerous occasions in the past (5, 6, 30, 57). Until recently, SIV cross-species transmissions could be detected only by analyzing blood samples coincident with the capture or killing of primates (10, 32, 73). However, the development of noninvasive viral detection methods has transformed the way infectious agents, including SIV, can be studied in wild-living primate populations (33, 38, 51-54, 70). For example, systematic testing of fecal samples from wild-living chimpanzees (Pan troglodytes) for viral nucleic acids and antibodies traced the origin of pandemic (group M) and nonpandemic (group N) human immunodeficiency virus type 1 (HIV-1) to distinct chimpanzee communities in south-central and southeastern Cameroon, respectively (33). Noninvasive surveys also uncovered a new SIV lineage in wild-living gorillas (Gorilla gorilla) (71). An analysis of 213 gorilla fecal samples from 11 field sites in Cameroon revealed three individuals that were SIV/HIV antibody positive (71). PCR amplification of viral sequences from fecal RNA confirmed infection by distinct SIV strains which comprised a new lineage, termed SIVgor. In phylogenetic trees of diagnostic pol and env sequences, the new gorilla viruses fell within the SIVcpz radiation and were most closely related to HIV-1 group O (71). These findings suggested that gorillas, like humans, had acquired SIV from chimpanzees; however, the paucity of available SIVgor sequences precluded definitive conclusions regarding the origins of SIVgor or HIV-1 group O.
Gorillas are classified into two species, with habitats in west central (Gorilla gorilla) and east (Gorilla beringei) Africa, respectively (Fig. 1). The western species is further subdivided into the Cross River gorilla (Gorilla gorilla diehli) and the western lowland gorilla (Gorilla gorilla gorilla), while the eastern species includes the mountain gorilla (Gorilla beringei beringei), Grauer's gorilla (Gorilla beringei graueri), and possibly the Bwindi gorilla (a G. beringei subspecies of uncertain classification) (16). Both western subspecies have been screened for SIVgor infection, but only western lowland gorillas were found to harbor this virus (71). Moreover, SIVgor infection appears to be rare. Initially, only three gorillas from field sites in southwestern (CP) and south-central (BQ) Cameroon were found to be SIV positive (Fig. 1). Subsequent analyses have uncovered eight additional SIVgor infections (C. Neel and M. Peeters, unpublished); however, all of these were identified at one of the two field sites (CP) where SIVgor was first discovered. Thus, there are currently only two locations in Cameroon where SIVgor infection has been documented. The fact that these two sites are 400 km apart raises the question of whether gorillas acquire SIV sporadically from local sources or whether the currently known viruses all resulted from a single cross-species transmission event.
FIG. 1.
Locations of gorilla and chimpanzee subspecies in west central and east Africa. The natural ranges of western gorillas (Gorilla gorilla, solid green) and eastern gorillas (Gorilla beringei, solid magenta) are shown in relation to the ranges of sympatric central chimpanzees (P. t. troglodytes, hatched green) and eastern chimpanzees (P. t. schweinfurthii, hatched magenta) (16). The western gorilla species is subdivided into the Cross River gorilla (G. g. diehli) on the Cameroonian/Nigerian border and the western lowland gorilla (G. g. gorilla) in southern Cameroon, Gabon, Equatorial Guinea, the Republic of Congo, and the Central African Republic. The eastern species is classified into Grauer's gorilla (G. b. graueri) in the Democratic Republic of the Congo; the mountain gorilla (G. b. beringei) on the border of the Democratic Republic of the Congo, Rwanda, and Uganda; and the Bwindi gorilla (a G. beringei subspecies) in Uganda (the classification of the last is still uncertain). The locations of two study sites (CP and BQ) are indicated. International borders (black lines) and major rivers (blue lines) are also shown.
The finding of SIV in wild-living gorillas came as a surprise and raised a number of issues. First, unlike for other primate lentiviruses that were first identified in captive primates, there was no prior evidence of SIV infection in captive gorillas. Second, the route by which gorillas could have acquired SIV was not immediately obvious. Gorillas are herbivorous and believed to avoid physical interaction with other primates (41, 60, 67). In contrast, chimpanzees are avid hunters that prey on smaller monkeys (13, 29, 40, 61). Thus, while chimpanzees likely acquired SIV through predation (6, 57), this mode of transmission is improbable for SIVgor. Third, the range of western lowland gorillas (G. g. gorilla) overlaps that of central chimpanzees (Pan troglodytes troglodytes) (Fig. 1), and SIVgor is significantly more closely related to SIVcpzPtt from the central subspecies than it is to SIVcpzPts from eastern chimpanzees (Pan troglodytes schweinfurthii) (71). These data would suggest that central chimpanzees transmitted their virus to sympatric gorillas, albeit by an as-yet-unknown route. However, under such a scenario, gorilla viruses might be expected to fall within the radiation of SIVcpzPtt strains in evolutionary trees, which is not the case (71). Finally, given that gorillas acquired a chimpanzee SIV (71), one could ask whether this has occurred only once and only in west central Africa. As shown in Fig. 1, eastern chimpanzees and gorillas also live in sympatry, thus providing potential transmission opportunities. Given these uncertainties, it is clear that additional chimpanzee and gorilla viruses need to be analyzed to elucidate the ancestry of SIVgor.
In the present study, we sought to gain new insight into the evolutionary origin of SIVgor and to begin to probe its biological properties. To accomplish this, we amplified partial (BQ664) and full-length (CP684, CP2135, and CP2139) genomes from fecal samples previously shown to contain SIVgor-specific viral RNA. We then performed analyses to (i) characterize the phylogenetic relationships of SIVgor over the entire length of its genome, (ii) investigate whether SIVgor has undergone recombination, (iii) identify the primate reservoir that gave rise to SIVgor, (iv) examine the possibility of local chimpanzee-to-gorilla SIV transmission, and (v) generate the first replication-competent molecular clone of SIVgor for biological characterization. Our results strongly suggest that gorillas acquired SIVcpzPtt from P. t. troglodytes apes and that the current SIVgor lineage is the result of a single such cross-species transmission event. Although it is still unclear whether chimpanzees or gorillas were the source of HIV-1 group O, in vitro studies demonstrate that SIVgor has many of the biological properties necessary for establishing a persistent infection in humans.
MATERIALS AND METHODS
Ape fecal samples.
To amplify full-length SIVgor genomes, fecal samples representing four naturally infected (female) western lowland gorillas were selected for analysis. These included samples CP684 (collected on 18 April 2004) and BQ664 (9 August 2004), which were previously reported to contain SIVgor viral RNA (71), as well as samples CP2135 (11 February 2007) and CP2139 (11 February 2007), which were identified to contain SIVgor-specific antibodies during a follow-up study at the CP field site (C. Neel and M. Peeters, unpublished). We also selected two fecal samples from wild chimpanzees at the CP field site for molecular characterization. Both CP1973 (17 December 2006) and CP2680 (28 August 2007) contained SIVcpz-specific antibodies and represented two female P. t. troglodytes apes, as determined by mitochondrial DNA, microsatellite, and sex marker analyses as described previously (33).
Amplification of SIVgor sequences.
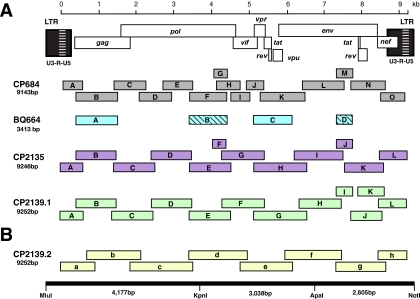
For samples CP2135, CP2139, and CP684, full-length SIVgor sequences were generated by amplifying partially overlapping subgenomic fragments (339 bp to 1,608 bp in length) from fecal virion RNA. For sample BQ664, only gag (1,126 bp) and vif-env (963 bp) sequences were amplified. Reverse transcriptase PCR (RT-PCR) analysis was performed as described previously (33, 51, 52, 65, 71), with some modifications. For CP684, 10 μl of RNA was first incubated with 40 pmol of the outer reverse primer (1 μl) for 10 min at 65°C, rapidly cooled on ice, and then added to the remaining components in a 20-μl reaction volume containing 1× Expand reverse transcriptase buffer, 1 mM deoxynucleoside triphosphate (dNTP), 5 mM dithiothreitol, 1 unit (U)/μl RNase inhibitor (Ambion, Austin, TX), and 2.5 U/μl Expand reverse transcriptase (Roche Diagnostics, Indianapolis, IN). This reaction mixture was incubated for 60 min at 42°C and heat inactivated at 95°C for 2 min, and 10 μl was used for PCR. For CP2135 and CP2139, cDNA was synthesized in a 20-μl reaction volume containing 10 μl fecal RNA, 1× reaction buffer, 20 U of RNase inhibitor (Ambion, Austin, TX), 0.5 mM dNTP, 5 mM dithiothreitol, 2 pmol outer reverse primer, and 200 U of SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). The reaction mixture was incubated at 50°C for 90 min and, after addition of another 200 U of enzyme, for an additional 90 min at 55°C. Following heat inactivation of the enzyme at 70°C for 15 min, 7 μl cDNA was then added to a PCR mixture containing 1× Expand buffer 2, 0.5 mM dNTP, 300 nM of first-round PCR primers, 25 μg bovine serum albumin, and 3.75 U of Expand Long Template enzyme mixture (Roche Diagnostics, Indianapolis, IN). Different combinations of consensus sequence- and strain-specific primers were used to amplify between 8 and 15 subgenomic fragments per proviral genome (Fig. 2); primer sequences and corresponding fragment lengths are listed in Table S1 in the supplemental material. The amplification conditions for first- and second-round PCR included 35 cycles of denaturation (94°C, 0.5 min), annealing (55°C, 0.5 min), and extension (68°C, 1 min). Amplicons were gel purified and sequenced directly using an ABI 3730 DNA analyzer. Chromatograms were examined for positions of base mixtures by using Sequencher version 4.7 (Gene Codes Corporation, Ann Arbor, MI) or Lasergene Seqman Pro version 7.1.0 (DNASTAR, Inc., Madison, WI). In addition, nucleotide differences between adjoining fragments in regions of sequence overlap were recorded (see Table S2 in the supplemental material). The genome of CP2139 was amplified twice (Fig. 2), the first time using a combination of consensus sequence- and strain-specific primers (CP2139.1) and the second time using only strain-specific primers designed to generate larger fragments (CP2139.2). Consensus sequences from these two amplifications were then used to generate a master consensus sequence (see Table S3 in the supplemental material), which served as the template to generate a full-length SIVgor molecular clone.
FIG. 2.
Generation of a replication-competent SIVgor clone. (A) The positions of individual RT-PCR products (open boxes) are shown in relation to the SIVgor provirus. Previously reported BQ664 amplicons are hatched (71). All fragments are drawn to scale, and the length of the assembled population sequence is indicated. Nucleotide sequences are numbered starting at the beginning of the R region in the 5′ LTR (see scale bar). (B) The CP2139 genome was amplified on an independent occasion, using strain-specific primers designed to amplify larger fragments. The resulting second consensus sequence (CP2139.2) differed from the first consensus sequence (CP2139.1) at 54 positions (see Table S3 in the supplemental material). One nucleotide was selected at each of these sites and the resulting master consensus sequence synthesized as three adjoining fragments (schematic on the bottom). Unique restriction enzyme sites allowed directional cloning into a low-copy-number plasmid vector (pBR322-MCS).
Amplification of SIVcpz sequences.
For samples CP1973 and CP2680, a highly conserved SIVcpz pol fragment (220 bp) was amplified, using degenerate HIV-1/SIVcpz/SIVgor consensus primers (F1, 5′-CATGTRGCHAGTGGNTWCMTAGARGCAGARGT-3′; R1, 5′-ACBACYGCNCCTTCHCCTTTC-3′; F2, 5′-AYAAYCCHCAAAGTCAAGGAGTRGT-3′; and R2, 5′-GTCCTTTCCAAATDGGRTCTCTGCTGTC-3′). The amplification conditions for both first- and second-round PCR included 35 cycles of denaturation (94°C, 0.25 min), annealing (55°C, 0.5 min), and extension (68°C, 0.5 min).
Construction of a full-length SIVgor clone.
To obtain a full-length infectious molecular clone of SIVgor, the CP2139 master consensus sequence was used as a template to chemically synthesize three subgenomic fragments (Blue Heron Biotechnology, Bothell, WA), spanning the 5′ long terminal repeat (LTR)-pol (4.2 kb), pol-env (3.0 kb), and env-3′ LTR (2.6 kb) regions (Fig. 2B). To facilitate subsequent cloning, unique MluI and NotI sites were added to the 5′ and 3′ termini of the provirus, respectively (Fig. 2B). These, together with internal KpnI and ApaI sites at positions 4171 and 7208, were then used to assemble the three subgenomic fragments to generate a full-length provirus. To accomplish this, individual fragments were first cloned into in a low-copy-number plasmid vector (pBR-MCS), which was engineered by ligating a synthetic polylinker containing an MluI-KpnI-XhoI-ApaI-NotI multicloning site (consisting of complementary oligonucleotide sequences 5′-AATTCACGCGTGGTACCTCGAGGGCCCGCGGCCGCA-3′ and 5′-AGCTTGCGGCCGCGGGCCCTCGAGGTACCACGCGTG-3′) into pBR322 digested with EcoRI and HindIII. The 4.2-kb fragment was cloned into the MluI and KpnI sites of pBR-MCS. Following digestion of this plasmid with KpnI and NotI, both 3.0-kb KpnI-ApaI and 2.6-kb ApaI-NotI fragments were simultaneously ligated to generate a full-length proviral clone. Transformation of XL2-MRF bacteria (Stratagene, La Jolla, CA) initially yielded only defective clones containing either single-nucleotide substitutions or larger insertions/deletions. One clone contained an ∼150-bp insert at the ApaI site in its env gene. Since the sequence of this clone was otherwise identical to the CP2139 master consensus sequence, it was digested with ApaI and recircularized. Resulting XL2-MRF transformants were screened for appropriately sized inserts, transfected into 293T cells, and tested for infectivity in the JC53-BL assay (21, 77). One functional clone (pBR-CP2139.287) was identified and grown large scale.
Large-scale plasmid preparation.
The pBR-CP2139.287 clone containing the full-length SIVgor genome was propagated in XL2-MRF cells (Stratagene) at 30°C in a Forma orbital benchtop shaker (model 420/2l), using 500 ml LB containing 100 μg/ml ampicillin in 2-liter disposable Erlenmeyer flasks (16-cm diameter, vented caps; Corning) at low levels of agitation (225 rounds per minute). Bacterial cultures were harvested prior to reaching saturating growth density and purified to be endotoxin free (Qiagen, Valencia, CA). Due to the extreme instability of this clone in bacteria, all large-scale plasmid preparations were sequence confirmed. The replication-competent pBR-CP2139.287 clone has been submitted to the National Institutes of Health Research and Reference Program (Rockville, MD).
Phylogenetic methods.
Newly derived full-length SIVgor sequences were translated and compared to previously published full-length HIV-1 and SIVcpz sequences from the database as follows: for HIV-1 group M subtype A, U455A (GenBank accession numbers are listed in Table S4 in the supplemental material); for subtype B, HXB2; for group N, YBF30 and YBF106; for group O, MVP5180 and ANT70; for SIVcpzPtt, MB897, DP943, LB7, MB66, EK505, MT145, CAM3, CAM5, CAM13, GAB1, GAB2, and US; and for SIVcpzPts, ANT, TAN1, TAN2, and TAN3. Amino acid sequences were aligned using CLUSTAL W (66). Sites that could not be aligned unambiguously and sites with a gap in any sequence were discarded. In regions of gene overlap (e.g., Gag/Pol and Pol/Vif genes), the carboxy termini of the overlapping protein sequences were discarded. Trees were inferred by the Bayesian Markov Chain/Monte Carlo method of phylogenetic estimation (79), implemented in MrBayes version 3.1 (50) by using the mixed model of amino acid evolution with gamma-distributed rates at sites and 1 million generations with 25% burn in. Average standard deviations of split frequencies were 0.01 or lower. Four major regions of the proteome were analyzed: Gag, Pol1, Pol2, and Env. The Pol sequence was divided at the position of a recombination breakpoint previously identified in HIV-1 group N (26). Trees of partial pol sequences for SIVcpzPtt strains CP1973 and CP2680 were inferred as described above from an alignment of 67 amino acids, using 10 million generations in MrBayes.
To calculate the time to the most recent common ancestor (MRCA) of the SIVgor clade, regions of an HIV-1/SIVcpz/SIVgor nucleotide sequence alignment corresponding to available BQ664 sequences were concatenated (total length, 2,952 nucleotides). This alignment included new and previously published BQ664 sequences (see Table S4 in the supplemental material), sequences listed above, and additional sequences from HIV-1 group M: for ETH2220 (subtype C), ELI (D) and 02CM.0016BBY (F2); for HIV-1 group N, 04CM-1015-04, 04CM-1131-03, and DJO131; and for HIV-1 group O, SEMP1299, SEMP1300, and VAU. A maximum-likelihood phylogenetic tree was constructed with PAUP* version 4.0b10 (64), using a general time-reversible model with gamma-distributed site-to-site rate variation allowing for invariable sites (GTR+I+G model), with parameters selected using MODELTEST version 3.7 (44, 45) and the Akaike information criterion (2). Relevant branch lengths from the tree were summed to provide the genetic distance of each HIV-1 group O strain from the MRCA of this group. The evolutionary rate for each strain was calculated using its date of isolation and the previously estimated date of the MRCA of group O (35). The average of these rates was then used to estimate the time to the common ancestor of the SIVgor sequences sampled here, again using distances from summed branch lengths and taking account of dates of virus isolation.
For the analysis of SIVgor V3-loop sequences, a phenetic dendrogram was estimated using the unweighted-pair group method with average linkages from a distance matrix of uncorrected sequence differences, using NEIGHBOR from PHYLIP version 3.64 (24). The following additional V3 sequences were included: for HIV-1 group M, A1 92UG037 (see Table S4 in the supplemental material), 97CDKTB48 (subtype A2), BK132 (B), ETH2220 (C), 94UG114 (D), CM240 (01-AE), VI850 (F1), 02CM.0016BBY (F2), DRCBL (G), VI991 (H), and EQTB11C (K); for SIVcpzPtt, BM1034, LB714, LB715, MB801, MB802, MB776, MB803, DP935, and SL995; and for HIV-1 group O, SEMP1300 (O1), 97US08692A (O2), 96CMABB637 (O3), ANT70 (O4), and MVP5180 (O5).
To test for recombination among the SIVcpz ancestors of HIV-1 groups M, N, and O and SIVgor, phylogenetic trees were examined across a concatenated Gag and Pol alignment in windows of 300 residues moved in steps of 5 to 100 amino acids (larger step sizes were used to screen for recombination; smaller step sizes were used to map recombination breakpoints). To test for recombination among the three newly derived SIVgor isolates, full-length nucleotide sequences of the three SIVgor genomes were aligned and tested using GENECONV (http://www.math.wusll.edu/∼sawyer).
Viral infectivity, coreceptor usage, and neutralization phenotype analyses.
Full-length molecular clones of SIVgor, SIVcpz, and HIV-1 were transfected into 293T cells and supernatants equilibrated by particle-associated reverse transcriptase activity as described previously (65). These included the SIVcpzPts clones TAN1, TAN2, and TAN3 (65); the SIVcpzPtt clones GAB2 (11), MT145, MB897, and EK505 (J. Decker and B. H. Hahn, unpublished); and the HIV-1 clones SG3 (28), NL4-3 (1), YU2 (37), and WEAU1.6 (18). In addition, human peripheral blood mononuclear cell (PBMC)-derived viral isolates YBF30 (58) and 97US08692A (78) were also used. Viral infectivity was assessed for JC53BL-13 cells (TZM-bl; NIH AIDS Research and Reference Reagent Program catalogue no. 8129), a HeLa-derived line which has been genetically modified to constitutively express CD4, CCR5, and CXCR4 and to contain integrated luciferase and β-galactosidase reporter genes under the control of an HIV-1 LTR (21, 77). For coreceptor analysis, JC53-BL cells were seeded in 96-well plates at 8,300 cells/well overnight and then treated with the CCR5 antagonist TAK-779 (10 mM) (4), the CXCR4 antagonist AMD3100 (1.2 mM) (20), or a combination of both for 1 hour (NIH AIDS Research and Reference Reagent Program catalogue no. 4983 and 8128, respectively). Virus was added in the presence of 40 μg/ml DEAE-dextran and removed 48 h later. Cells were then lysed and analyzed for luciferase activity (Promega, Madison, WI), using a Tropix luminometer with WinGlow version 1.24 software.
For neutralization assays, 3,000 infectious units of virus were combined in a total volume of 60 μl with or without a 2× concentration of sCD4 in Dulbecco's modified Eagle's medium (DMEM) with 6% fetal calf serum (FCS) and 80 μg/ml DEAE-dextran. After 1 h at 37°C, an equal volume of human plasma (10% [vol/vol] in DMEM plus 6% FCS or fivefold dilutions thereof), monoclonal antibodies, or fusion inhibitor was added. Monoclonal antibodies described in reference 12 were kindly provided by the following individuals: Dennis Burton (b12 and 2G12), Michael Zwick and Dennis Burton (Z13e1), Herman Katinger (2F5 and 4E10), Susan Zolla-Pazner (447-52D), Lisa Cavacini (F425-B4e8), James Robinson (17b, 19e, and 21c), and David Montefiori (HIVIG). The following reagents were obtained commercially: soluble CD4 (514-CD; R&D Systems), T1249 (Triangle Pharmaceuticals), and anti-CD4 monoclonal antibody (catalogue no. 555344, clone RPA-T4; BD Pharmingen). The addition of ligand or antibody brought the final concentration of DEAE dextran to 40 μg/ml. Virus was incubated with test antibodies (with or without sCD4) for 1 h at 37°C and then added to JC53-BL cells. Cells were incubated at 37°C for 2 days and then analyzed for luciferase expression. Controls included cells exposed to no virus or to virus pretreated with normal human plasma. Relative infectivity was calculated by dividing the number of luciferase units at each dilution of test plasma or monoclonal antibodies by values in wells containing normal human plasma. Neutralization was assessed by 50% inhibitory concentration (IC50) determined by linear regression using a least-squares method. All samples were tested in duplicate.
Chimpanzee and human PBMC cultures.
Blood was obtained from normal human volunteers (Research Blood Components, Boston, MA) as well as healthy (HIV-1-uninfected) chimpanzees housed at the Yerkes Regional Primate Center as described previously (65). Briefly, PBMCs were isolated using Ficoll Hypaque Plus (GE Healthcare, Piscataway, NJ). CD4+ T cells were enriched using CD4 microbeads and magnetic cell sorting (Militenyi Biotec, Auburn, CA), stimulated with staphylococcal enterotoxin B (Sigma-Aldridge, St. Louis, MO) for 12 to 15 h (3 μg/ml), and subsequently cocultivated with autologous monocyte-derived macrophages for optimal activation. After 5 to 6 days, CD4+ T cells were removed from the macrophages, placed into DMEM with 10% FCS, and incubated with 30 U/ml interleukin-2 (IL-2). After 24 h, 5 × 105 CD4+ T cells were incubated with transfection-derived viral stocks at a multiplicity of infection of 0.1 (as determined for JC53-BL cells) in 300 μl DMEM containing 10% FCS and 30 U/ml IL-2 for 16 h. CD4+ T cells were washed three times and plated in 24-well plates in DMEM with 10% FCS and 30 U/ml IL-2, and reverse transcriptase activity in culture supernatants was measured every 3 days to monitor viral replication.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences determined in this study are FJ424871 (CP684 consensus sequence), FJ424863 (CP2135 consensus sequence), FJ424864 (CP2139.1 consensus sequence), FJ424865 (CP2139.2 consensus sequence), FJ424866 (pBR-CP2139.287), FJ424867 (BQ664 vif-env), FJ424868 (BQ664 gag), FJ424869 (CP1973 pol), and FJ424870 (CP2680 pol).
RESULTS
SIVgor genome structure.
To amplify complete SIVgor genomes, we selected fecal samples from four infected western lowland gorillas, all of which were sampled in the wild. Two of these (CP684 and BQ664) were previously shown to harbor divergent SIVgor strains based on subgenomic pol and gp41 sequences (71). The other two (CP2135 and CP2139) were identified more recently, in a follow-up study of gorilla communities at the CP field site (C. Neel and M. Peeters, unpublished). All samples were positive for SIVgor antibodies, and their species origins were determined by mitochondrial DNA analysis. All samples were also shown to represent different individuals, based on microsatellite analysis (not shown). Fecal RNA was extracted and subjected to RT-PCR analysis using consensus sequence- as well as strain-specific primers (see Table S1 in the supplemental material). For CP684, CP2135, and CP2139, this approach yielded 12 to 15 partially overlapping fragments, which in each case spanned an entire provirus (Fig. 2A). Amplification of SIVgor sequences from sample BQ664 was more difficult. Despite repeated RT-PCR attempts and multiple primer combinations, only two new fragments in gag and the accessory gene region could be amplified (Fig. 2A). All amplicons were sequenced directly to generate fecal consensus sequences. As expected, these contained a limited number of ambiguous sites, including base mixtures and sequence differences in regions of fragment overlap (see Table S2 in the supplemental material). Although some of the ambiguous sites affected the encoded amino acid, none introduced stop codons or frameshift mutations.
The concatenated genomes of CP2139, CP2135, and CP684 were 9,252, 9,246, and 9,143 nucleotides in length, respectively. All three carried full-length open reading frames for all nine genes found in SIVcpz/HIV-1 (Fig. 2A) as well as all major regulatory sequences. Although the deduced SIVgor, SIVcpz, and HIV-1 protein sequences varied considerably, we found no obvious SIVgor-specific signatures. For example, all three gorilla viruses encoded additional cysteine residues in their fourth variable (V4) envelope domain (two in CP2139 and four in CP2135 and CP684), indicating a more diversified V4 loop structure; however, this was not unique to SIVgor, since additional cysteine residues (usually two) were also observed in the V4 domain of HIV-1 group O and some SIVcpz strains (not shown). Similarly, SIVgor Vpu sequences differed from those of SIVcpz and HIV-1 (including group O) in more than 60% of amino acid residues; however, the predicted secondary structure and hydrophobicity profiles of the deduced SIVgor Vpu proteins suggested a very similar function (not shown). Interestingly, there were some protein domains where SIVgor strains resembled chimpanzee and not human viruses. At position 30 of the Gag matrix protein, the three gorilla viruses (like all strains of SIVcpzPtt) encoded a methionine rather than the arginine found in the ancestors of HIV-1 groups M, N, and O (75). Similarly, in a phenogram of V3 loop sequences, all gorilla viruses clustered with SIVcpz strains (Fig. 3), rather than grouping with HIV-1 group O viruses to which they are much more closely related (see below). Thus, at two sites known to be under host-specific selection pressures in humans and chimpanzees (70, 75), SIVgor resembled SIVcpz rather than HIV-1.
FIG. 3.
V3 loop diversity among different ape lentiviruses. The phenetic clustering of SIVgor (red), SIVcpz (black), and HIV-1 (blue) V3 amino acid sequences is shown. Four major V3 clusters are apparent: (i) HIV-1 group M, (ii) the MB/LB strains of SIVcpz, (iii) all other SIVcpz strains (except GAB2) plus HIV-1 group N and SIVgor, and (iv) HIV-1 group O. Dashes indicate gaps introduced for better alignment.
Origin of SIVgor.
To compare the evolutionary relationships of the newly derived gorilla viruses to each other and to previously characterized SIVcpz and HIV-1 strains, phylogenetic trees were constructed from different regions of the proteome. For the fully sequenced CP2139, CP2135, and CP684 strains, these included Gag, the N-terminal half of Pol (Pol1), the C-terminal half of Pol plus Vif (Pol2), and Env (Fig. 4). For the partially characterized BQ664 strain, individual Gag, Pol, Vif-Env, and gp41 fragments were concatenated prior to analysis (Fig. 5). These studies confirmed and extended previous results from subgenomic SIVgor sequences (71). In all regions of their genome, the gorilla viruses clustered in a monophyletic clade that was most closely related to HIV-1 group O viruses. Within the SIVgor lineage, strains from the CP field site were much more closely related to one another than they were to BQ664. The branching orders of the three CP strains varied among trees from different parts of the genome, but these relationships were not significantly discordant, and closer examination of the nucleotide alignments of the CP strain sequences (using GENECONV) detected no evidence for recombination. Finally, the SIVgor/HIV-1 group O clade was significantly more closely related to SIVcpzPtt from central chimpanzees than to SIVcpzPts from eastern chimpanzees, strongly suggesting a west central African origin for SIVgor.
FIG. 4.
Evolutionary relationships of newly derived SIVgor strains. Trees were inferred for four proteomic regions (Gag, Pol1, Pol2, and Env), where Pol1 ends at a recombination breakpoint previously identified in HIV-1 group N, and Pol2 includes downstream Vif sequences. The SIVgor sequences (red) were compared to representatives of SIVcpz (black) and HIV-1 (blue) groups M (U455 and LAI), N (YBF30 and YBF106), and O (ANT70 and MVP5180). The four lowest SIVcpz strains (ANT, TAN1, TAN2, and TAN3) are from eastern chimpanzees; all others are from central chimpanzees. Numbers on internal branches indicate estimated posterior probabilities (only values above 95% are shown). The scale bars represent 0.05 (or 0.1 for Env) amino acid replacements per site.
FIG. 5.
Evolutionary relationships of newly derived SIVgor strains in partial genome regions. Partial Gag, Pol, Vpr-Vpu, and Env sequences corresponding to BQ664 were concatenated. SIVgor sequences are highlighted in red; HIV-1, SIVcpzPtt, and SIVcpzPts strains are as in Fig. 4. Numbers on internal branches indicate estimated posterior probabilities (only values above 90% are shown). The scale bars represent 0.1 replacements per site.
The phylogenetic relationships in Fig. 4 were derived from large genomic regions, and so, to check for any evidence of recombination involving smaller fragments, we made numerous trees based on shorter (300 amino acids) windows from the alignments. This analysis identified a 900-bp region at the 5′ end of the pol gene that appeared to have a different evolutionary history. While a tree derived from Pol sites 301 to 699 (from a gap-stripped Pol alignment) exhibited the conventional topology with SIVgor and HIV-1 group O viruses clustering exclusive of SIVcpzPtt strains and other HIV-1 groups (Fig. 6, right), in a tree of N-terminal Pol sequences (sites 1 to 300), this was no longer true (Fig. 6, left). Support for this discordant branching was high: the grouping of the SIVcpzPtt strains MT145 and GAB2 together with SIVgor/group O outside the remaining SIVcpzPtt strains had a posterior probability value of 98%. These results indicated that recombination had occurred during the divergence of these sequences.
FIG. 6.
Evidence for an SIVcpzPtt lineage ancestral to SIVgor and HIV-1 group O. Trees were inferred for two parts of the Pol1 region in Fig. 4, separated at a putative recombination breakpoint at position 300. Strains are color coded as in Fig. 4. Numbers on internal branches indicate estimated posterior probabilities (only values above 95% are shown). The scale bars represent 0.05 replacements per site. Green, blue, and red circles indicate nodes that were used to calculate the distances shown in Table 1.
Two different evolutionary scenarios could explain the discordant branching orders in Fig. 6. One possibility is that an ancestor of SIVgor acquired sequences from an ancestral SIVcpzPtt strain; in this case, SIVgor is recombinant. Alternatively, an ancestor of MT145 and GAB2 could have acquired sequences from a divergent SIVcpzPtt lineage that has not yet been sampled or is now extinct; in this case, MT145 and GAB2 are recombinant. To distinguish between these scenarios, we compared the divergences of the various strains in recombinant and nonrecombinant regions. Under the first scenario, the SIVgor/group O lineage would be expected to change its position in the two trees, i.e., this lineage would move inside the SIVcpzPtt clade. As a result, the common ancestor of the entire SIVcpzPtt/SIVgor/HIV-1 clade in the recombinant region (the red node in Fig. 6) would be equivalent, in terms of distance from the root or the tips of the tree, to the ancestor of the SIVcpzPtt/HIV-1 clade in the nonrecombinant region (the blue node in Fig. 6). Alternatively, if MT145 and GAB2 were recombinant, the red node would be equivalent to the ancestor of the entire SIVcpzPtt/SIVgor/HIV-1 clade in the nonrecombinant region (the green node in Fig. 6). To evaluate these alternative hypotheses, we calculated the evolutionary distances from each of these highlighted nodes to the tips of the four strains in the SIVcpzPts clade (Table 1). All distances were measured as average branch lengths, and this was done independently for Gag (Fig. 4) as well as the recombinant (Fig. 6, left) and nonrecombinant (Fig. 6, right) portions of Pol. In order to correct for different rates of evolution in different proteins, each distance was divided by the total length of the tree for that genomic region. Finally, all HIV-1 strains and one SIVcpzPtt recombinant (LB7) were excluded. The results showed that the distance for Pol sites 1 to 300 (for the red node) was similar to those for the green node (and lower than those for the blue node) for Gag and Pol sites 301 to 699 (Table 1), indicating that SIVcpzPtt strains MT145 and GAB2 were recombinant. Although these two SIVcpzPtt strains did not form a statistically supported clade in the recombinant region (Fig. 6, left), the shared mosaicism implied that they were monophyletic. This finding was also supported by results from an analysis of internal branch lengths (not shown) and is consistent with our previous conclusion that GAB2 is recombinant and contains pol gene sequences from a divergent SIVcpzPtt strain (11). Since it is clearly most parsimonious to assume that the recombination event in the ancestry of MT145 and GAB2 occurred during infection of a central chimpanzee, this implies that SIVgor and HIV-1 group O viruses are derived from a divergent SIVcpzPtt lineage that existed (and possibly still exists) in west central Africa.
TABLE 1.
Genetic distances within the SIVcpz/SIVgor/HIV-1 radiation
| Node | Distancea
|
||
|---|---|---|---|
| Gag | Pol sites 1-300 | Pol sites 301-699 | |
| Blue | 0.27 | 0.26 | |
| Green | 0.22 | 0.24 | |
| Red | 0.22 | ||
Distances were calculated as average branch lengths from the tips of each of the four SIVcpzPts strains to the relevant nodes depicted in Fig. 6.
Chimpanzee and gorilla viruses from the same field site.
The finding of divergent SIVgor strains at two field sites located 400 km apart (CP and BQ) (Fig. 1) suggested that wild-living gorillas are endemically infected with SIVgor (71). However, the possibility that local chimpanzee-to-gorilla transmissions had generated the observed SIVgor diversity could not be formally excluded. To examine whether neighboring ape communities harbored epidemiologically linked viruses, we specifically targeted wild-living chimpanzees in one area of high gorilla density (39). Screening 77 chimpanzee fecal samples from the CP field site, we found 9 to be SIVcpz antibody positive. These were identified to represent two naturally infected P. t. troglodytes apes (CP1973 and CP2680) that were sampled 8 months apart. Although the results for initial RT-PCR analyses were negative for diagnostic pol and gp41 regions (33, 70), most likely because of sample degradation, a small, 220-bp pol fragment was eventually amplified. Phylogenetic analysis of the resulting sequences confirmed that the two chimpanzees were infected with distinct SIVcpzPtt strains which were only distantly related to SIVgor strains identified in the same geographic area (Fig. 7). While we have analyzed only two chimpanzees from the CP site, previous surveys have shown that SIVcpzPtt strains exhibit strong phylogeographic clustering (33, 70). The distant relationship between the chimpanzee and gorilla viruses from CP thus indicates that these SIVgor strains were not due to a local transmission from chimpanzees. Furthermore, all SIVgor strains form a single cluster, strongly suggesting that they all descended from the same chimpanzee-to-gorilla transmission. Thus, the diversity within the SIVgor clade most likely reflects viral evolution in gorillas.
FIG. 7.
Evolutionary relationships of SIVgor and SIVcpz strains infecting gorillas and chimpanzees at the CP field site. A tree was inferred for a diagnostic Pol fragment (220 bp). Two SIVcpzPtt strains from the CP field site are boxed. The other strains are color coded as in Fig. 4. Numbers on internal branches indicate estimated posterior probabilities (only values above 95% are shown). The scale bars represent 0.1 replacements per site.
Timing of the MRCA of SIVgor.
The results described above suggest that all currently known SIVgor strains are derived from a single introduction of SIVcpzPtt into western lowland gorillas. To gauge when this introduction might have occurred, we estimated the time to the MRCA of the SIVgor clade. It is not yet possible to obtain a reliable estimate of the rate of SIVgor evolution, because only a few strains have thus far been characterized, and all were isolated within a narrow time period (2004 to 2007). We therefore used an evolutionary rate estimated from closely related HIV-1 group O strains. A phylogeny was obtained from a nucleotide sequence alignment produced by concatenating the four genomic regions available for BQ664, and distances between sequences were calculated by summing branch lengths from that tree. The average estimated distance between BQ664 and the CP strains was 0.295 substitutions per site. The year of the MRCA of HIV-1 group O has been estimated as 1920, with a confidence (95% highest posterior density) interval of 1896 to 1942 (35). With these dates, the rate of nucleotide substitution for HIV-1 group O for the genomic regions used here was estimated to be 1.05 × 10−3 (range, 0.79 × 10−3 to 1.50 × 10−3) per site per year. Then, assuming that SIVgor strains have evolved at the same rate as group O viruses, their MRCA was estimated to have existed around 1864 (range, 1818 to 1906). Thus, the chimpanzee-to-gorilla transmission that spawned the current SIVgor infections seems to have occurred at least 100 to 200 years ago.
Generation and biological characterization of a replication-competent SIVgor clone.
We have previously shown that replication-competent molecular clones of SIVcpz can be derived from fecal viral consensus sequences (65). However, for this strategy to be successful, we found that it was critical to amplify the most predominant viral species in the fecal sample. To ensure that this was the case for SIVgor, we selected one of the three fully sequenced SIVgor strains (CP2139) and reamplified its entire genome by using reextracted fecal RNA as well as strain-specific primers (Fig. 2B). As expected, the second consensus sequence (CP2139.2) was not identical to the first (CP2139.1); there were 54 nucleotide sequence differences, the majority of which were located in the env gene. Table S3 in the supplemental material lists all sequence differences between CP2139.1 and CP2139.2 and indicates which nucleotides were selected for inclusion in the molecular clone.
The CP2139 master sequence was used as the template to chemically synthesize three subgenomic fragments. These were then ligated into a low-copy-number (pBR322-derived) plasmid to generate the proviral clone CP2139.287 (Fig. 2B). To examine its biological activity, CP2139.287 was transfected into 293T cells and the resulting supernatant tested for infectivity in the JC53-BL cell assay. This analysis showed that CP2139.287-derived virus was infectious (600 to 800 IU/ng reverse transcriptase) and entered JC53 cells via the CCR5 coreceptor. As shown in Fig. 8, infectivity of CP2139.287 was completely blocked by the CCR5 antagonist TAK-779 but not by the CXCR4 antagonist AMD3100. This was also true for the primary HIV-1 strains YU-2 (group M), YBF30 (group N), and 97US08692A (group O) but not for NL4-3- and WEAU 1.6-derived viruses, which served as X4 and R5/X4 dual tropic controls, respectively (Fig. 8). Culture experiments also showed that CP2139.287-derived virus replicated efficiently and to high titers in both human and chimpanzee CD4+ T cells, with kinetics very similar to those of previously characterized SIVcpz and HIV-1 strains (Fig. 9). Taken together, these data demonstrated that the reconstructed CP2139.287 clone carried a fully functional, replication-competent SIVgor genome.
FIG. 8.
Coreceptor usage of SIVgor. JC53-BL cells were pretreated with AMD3100 (inhibitor of CXCR4), TAK779 (inhibitor of CCR5), or both prior to addition of the virus preparations indicated. Virus infectivity is plotted on the vertical axis as a percentage of the untreated control. Virus derived from the replication-competent molecular clones NL4.3 (X4-tropic), YU2C (R5-tropic), and WEAU1.6 (dual tropic) as well as the primary isolates 97US08692A (group O) and YBF30 (group N) were included for control.
FIG. 9.
Replication potential of SIVgor in human and chimpanzee CD4+ T cells. The replication kinetics of CP2139.287-derived virus in human (A) and chimpanzee (B) CD4+ T cells are shown in relation to those of HIV-1 (SG3, YU2), SIVcpzPtt (MT145, MB897, EK505, and GAB2), and SIVcpzPts (TAN1, TAN2, and TAN3) reference strains (x axis, number of days postinfection; y axis, nanograms of reverse transcriptase [RT] activity per ml of culture supernatant). Similar replication kinetics were observed for CP2139.287-derived virus in CD4+ T cells from three additional human and two additional chimpanzee donors.
To examine the antigenic properties of SIVgor, we tested CP2139.287-derived virus as well as a panel of HIV-1 group M (NL4-3 and YU-2), N (YBF30), and O (97US08692A) viruses for their sensitivity to anti-HIV-1 monoclonal and polyclonal antibodies as well as receptor and fusion inhibitors (Table 2). These experiments showed that SIVgor was resistant to neutralization by monoclonal antibodies directed against the CD4 binding site (b12), surface glycans (2G12), and membrane-proximal external region (4E10 and Z13e1). SIVgor was also resistant to neutralization by monoclonal antibodies directed against the coreceptor binding site (17b, 21c, and 19e) and the V3 loop (447-52D and F425-B4e8) of HIV-1 in the presence and absence of soluble CD4, highly reactive subtype B and C plasma pools, and two group O plasma samples. In contrast, SIVgor was highly sensitive to the fusion inhibitors T-20 and T-1249 (IC50s of 0.001 μg/ml and 0.004 μg/ml, respectively) and to the membrane-proximal external region monoclonal antibody 2F5 (IC50, 4.6 μg/ml). The infectivity of CP2139.287-derived virus was also blocked by a monoclonal antibody directed against cell surface-expressed CD4 (IC50, 0.23 μg/ml), indicating that access to this receptor was required for SIVgor cell entry. Interestingly, the latter was not the case for the group O virus 97US08692A, which was completely resistant to this antibody (IC50, >2 μg/ml). 97US08692A was also moderately sensitive to the V3 monoclonal antibody 447-52 (despite its divergent V3 crown sequence) (Fig. 3), suggesting a somewhat exposed V3 loop. These data suggest that 97US08692A may be relatively CD4 independent. Future experiments will need to determine whether this property is unique to 97US08692A or general in HIV-1 group O viruses.
TABLE 2.
Neutralization phenotype of SIVgor
| Virusb | Neutralization titera
|
Coreceptor used | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sCD4 (nM) | HIVIGc | Clade B plasma pool | Clade C plasma pool | Group O plasma | Group O plasma | b12 | 2G12 | 2F5 | 4E10 | Z13e1 | T-20 | T-1249 | 447-52D | 447-52D sCD4 | F425-B4e8 | F425-B4e8 sCD4 | 17b | 17b sCD4 | 19e | 19e sCD4 | 21c | 21c sCD4 | Anti-CD4 MAb | TAK-779 (μM) | ||
| SIVgor (CP2139.287) | 154 | >1000 | 20 | <20 | <20 | <20 | >50 | >50 | 4.6 | >50 | >50 | 0.001 | 0.004 | >25 | >25 | >25 | >25 | >10 | >10 | >10 | >10 | >10 | >10 | 0.23 | 0.033 | R5 |
| HIV-1_O (97US08692A) | 21 | >1000 | NAd | NA | NA | NA | >50 | >50 | >50 | >50 | >50 | 0.010 | 0.084 | 12.47 | >25 | >25 | >25 | >10 | >10 | >10 | >10 | >10 | >10 | >2 | 0.201 | R5 |
| HIV-1_N (YBF30) | 498 | >1000 | <20 | <20 | <20 | <20 | >50 | >50 | >50 | 34.63 | >50 | 0.016 | 0.026 | >25 | >25 | >25 | >25 | >10 | >10 | >10 | >10 | >10 | >10 | 0.19 | 0.034 | R5 |
| HIV-1_M (NL4.3) | 2 | 71 | 630 | 358 | 83 | <20 | 0.01 | 0.85 | 0.44 | 1.35 | >50 | 0.102 | 0.001 | 0.02 | 0.01 | >25 | >25 | 0.17 | 0.26 | >10 | >10 | 0.55 | 1.1 | 1.89 | >10 | X4 |
| HIV-1_M (YU2) | 14 | 891 | 178 | 267 | <20 | <20 | 1.63 | >50 | >50 | >50 | >50 | 0.143 | 0.044 | 10.24 | 0.02 | >25 | 1.68 | >10 | >10 | >10 | >10 | >10 | >10 | 0.17 | 0.007 | R5 |
Neutralization titers are expressed as the reciprocal 50% inhibitory plasma dilutions or the IC50s for the respective Env ligands in one representative experiment (performed in duplicate). All titers are given in μg/ml unless otherwise stated.
CP2139.287, NL4.3, and YU2 are molecular clones of HIV-1 strains. 97US08692A and YBF30 are primary HIV-1 isolates.
HIVIG, pooled human immunoglobulin purified from plasma samples of healthy HIV-1-infected individuals.
NA, not available.
DISCUSSION
A primary objective of this study was to determine the origin of SIVgor and to estimate where and when this virus might have been introduced into wild-living gorillas. By sequencing three new full-length SIVgor genomes and conducting detailed recombination analyses, we found evidence for an ancestral SIVcpzPtt lineage from which SIVgor and HIV-1 group O viruses evolved (Fig. 6). Although full-length representatives of this lineage were not identified, we documented its existence in the form of mosaic pol fragments in present-day SIVcpzPtt recombinants. This finding strongly suggests that P. t. troglodytes apes were the original source of SIVgor and that the cross-species transmission took place in the Cameroon/Equatorial Guinea/Gabon area of west central Africa (Fig. 1). To estimate when this event might have occurred, we calculated the time to the MRCA of the current SIVgor clade. This analysis yielded an estimate of 100 to 200 years for the divergence of BQ664 from the three other SIVgor strains (Fig. 5). The transmission that gave rise to SIVgor likely occurred much earlier than that since (i) currently available SIVgor sequences may not represent the entire diversity of extant gorilla viruses, (ii) SIVgor strains that diverged earlier may have gone extinct, and (iii) phylogenetic methods are notorious for underestimating the deeper divergence times of rapidly evolving RNA viruses (31, 56). Once introduced, SIVgor spread within its new host, as evidenced by the presence of viruses at field sites located 400 km apart (Fig. 1). Whether this occurred primarily by sexual or other (e.g., exposure to infectious saliva) routes is not known; however, it is likely that gorilla behavior and social structure facilitated virus dispersal. Western lowland gorillas live in family units of 2 to 30 individuals which typically comprise one dominant male (silverback), three or more sexually active adult females, their offspring, and possibly a few nondominant males (27, 42, 62, 67). The silverback mates with all adult females in the group (27, 48). Adolescent males typically leave their natal groups and become solitary (and highly mobile) until they form their own harems (23), while females also transfer between groups (63). Individual groups have home ranges of about 15 to 20 km2 (8, 22, 47), which often overlap, resulting in frequent encounters of neighboring groups (8, 17, 22, 67). Thus, gorilla mating and social networks provide ample opportunity for a newly introduced SIV to spread, both within and between neighboring communities.
As illustrated in Fig. 4, chimpanzees have transmitted SIVcpz to gorillas and humans on multiple occasions. For humans, exposure to infected chimpanzee blood or mucosal secretions in the context of bushmeat hunting is considered the most plausible scenario for cross-species transmission (30). For gorillas, the route and circumstances of transmission are much less clear. Gorillas are herbivores and do not hunt other mammals (41, 49, 68). However, recent studies have shown that gorillas and chimpanzees commonly feed in the same forest areas and sometimes even in the same fruiting trees (76). Thus, there are focal points for gorilla/chimpanzee encounters that may facilitate virus transmission. Moreover, SIV is not the only virus requiring physical contact for infection that has crossed the species barrier from chimpanzees to gorillas: screening ape fecal samples for hepatitis B virus DNA, we found that this pathogen has been transmitted from wild-living chimpanzees to gorillas on at least two occasions (W. Liu and B. H. Hahn, unpublished). Since hepatitis B virus, like HIV/SIV, is transmitted by direct contact with infectious blood or mucosal secretions (25), encounters between chimpanzees and gorillas that are conducive to cross-species infection must occur. Whether these are physical in nature (i.e., fighting or biting), involve exposure to infectious feces or urine in cofeeding areas, or occur through saliva in partially eaten, discarded fruit (76) will need to be determined. Whatever the circumstances, it appears that successful transmission events are rare.
While it seems clear that chimpanzees were the source of SIVgor, it is not known whether humans acquired HIV-1 group O from naturally infected chimpanzees or gorillas. Thus far, none of the chimpanzee and gorilla communities tested represent likely reservoirs. The known SIVgor strains are too divergent to have been the immediate source of HIV-1 group O (Fig. 4). In addition, the prevalence of SIVgor infection in Cameroon is very low (C. Neel and M. Peeters, unpublished). Finally, none of several hundred P. t. troglodytes apes tested in southern Cameroon, including those at the CP field site (Fig. 8), harbor SIVgor-like viruses (33, 70). Together, these data strongly suggest that the ape reservoir that gave rise to HIV-1 group O exists outside Cameroon. It will be important to determine whether this reservoir still exists and, if so, where it is located. Moreover, it will be important to differentiate between different transmission scenarios (Fig. 10). One possibility is that chimpanzees harboring SIVgor-like viruses infected gorillas and humans independently; identification of SIVcpzPtt strains that join either the branch outside the HIV-1 group O or that outside the SIVgor clade would provide strong support for such a scenario (green lines in Fig. 10A). Alternatively, gorillas may have served as an intermediary host for the human infection; evidence for this would come from the finding of SIVgor strains that join either the branch outside group O or that outside the entire HIV-1 group O/SIVgor clade (Fig. 10B). The viruses invoked in either of these scenarios may exist in as-yet-unsampled apes in Equatorial Guinea, Gabon, or the Republic of the Congo. Indeed, the relative prevalence of group O infections has been reported to be much higher in Equatorial Guinea (9% of HIV-1 infections) than in Cameroon (2% of HIV-1 infections), pointing to this area as a possible starting point of the HIV-1 group O epidemic (3, 15, 43, 74, 80). Formally, there is a third possibility, namely, that chimpanzees harboring SIVgor-like viruses first infected humans, who then passed the virus to gorillas. However, this would imply that HIV-1 group O was far more divergent in the past than is currently the case (Fig. 10C). Moreover, it is hard to imagine how humans could transmit a virus such as SIVgor to wild-living gorillas. Thus, the third scenario is implausible.
FIG. 10.
Possible scenarios for the origin of HIV-1 group O. (A) Chimpanzees harboring SIVgor-like viruses infected gorillas and humans independently. (B) Chimpanzees harboring SIVgor-like viruses infected gorillas, which then passed the virus on to humans. (C) Chimpanzees harboring SIVgor-like viruses infected humans, who then passed the virus to gorillas. Support for each scenario would come from the discovery of one of the hypothetical viruses shown in green. In all three trees, nodes marked by circles indicate ancestral viruses that are assumed to have infected chimpanzees.
A second objective of this study was to generate a replication-competent molecular clone of SIVgor for biological analyses. We accomplished this by synthesizing the complete genome of one of the newly characterized gorilla viruses (CP2139) from fecal consensus sequences. The availability of this SIVgor clone allowed us to compare its baseline biological properties to those of SIVcpz and HIV-1 reference strains. As shown in Fig. 9, SIVgor replicated to high titers in CD4+ T cells from human (n = 4) and chimpanzee (n = 3) donors. We also assessed the sensitivities of SIVgor to a number of Env-specific ligands (Table 2). The results revealed that SIVgor used both CD4 and CCR5 receptors for cell entry (Fig. 8). SIVgor was sensitive to neutralization by the membrane-proximal external region monoclonal antibody 2F5, consistent with the conservation of the epitope recognized (ALLELDKWAD). SIVgor was also highly sensitive to the two fusion inhibitors T20 and T1249 (Table 2), possibly because of a glutamine-to-arginine substitution at position 580 (position 577 in HXB2). The glutamine at position 577 in HXB2 is believed to interact with a tryptophan at position 628 (position 634 in CP2139), and the observed arginine substitution would be expected to destabilize the formation of the gp41 six-helix bundle (Bing Chen, personal communication). In contrast, SIVgor was resistant to neutralization by monoclonal antibodies b12, 2G12, 4E10, and Z13e1, most likely due to epitope variation. SIVgor was also completely resistant to CD4i (17b, 21c, and 19e) antibodies, both in the absence and in the presence of sCD4. Since the coreceptor binding surface is highly conserved between diverse HIV-1 and HIV-2 strains (19), these data strongly suggest that the SIVgor CD4i bridging sheet is shielded as it is in primary strains of HIV-1. SIVgor was also resistant to the V3 monoclonal antibodies 447-52D and F425-B4e8, both of which recognize a highly conserved arginine residue at position 315 in the V3 loop crown (GPGR) (7, 59). Since the crown of the SIVgor V3 loop (GPMT) lacks such an arginine, it is possible that these antibodies failed to neutralize the viruses because of epitope variation. However, the accessibility of V3 to antibodies has been shown to have a major impact on V3-mediated neutralization. The fact that an HIV-1 group O strain (97US08692A) that encodes a similarly divergent V3 crown (GPLA) was resistant to entry inhibition by a monoclonal antibody against surface CD4 and was moderately sensitive to 447-52D (Table 2) suggests that the SIVgor V3 loop, like its CD4i bridging sheet, may be concealed in the functional Env trimer. Finally, we found that SIVgor was resistant to neutralization by heterologous patient antibodies, including two group O plasmas. Altogether, these findings suggest that the sensitivity of SIVgor to Env-specific ligands resembles that of primary HIV-1 and SIVcpz strains in several ways, including those related to CD4 tropism, CCR5 coreceptor preference, and effective concealment of CD4i (and possibly also V3 epitopes) in the functional Env trimer.
Given the biological properties of SIVgor and the extent of ape bushmeat hunting in west central Africa (14, 39, 69), it is clear that wild-living gorillas could represent a reservoir for human infection. Additional field studies are thus needed to determine the prevalence, geographic distribution, species association, and natural history of SIVgor throughout the entire gorilla habitat. Given the current rate of deforestation in central Africa and the extent of bushmeat hunting and consumption, it is critical to collect baseline data on existing sources of human zoonotic diseases now. This approach has been successful for Ebola hemorrhagic fever (36), monkeypox (46), and anthrax (34). Continuation of noninvasive surveys of wild ape populations is thus critical, not only to identify the primate origin of HIV-1 group O but also to ensure that additional reservoirs of HIVs are not overlooked.
Supplementary Material
Acknowledgments
We thank Caroline Tutin, Peter Walsh, Kate Abernethy, and Bing Chen for helpful discussions; the Cameroonian Ministries of Health, Environment and Forestry, and Research for permission to perform this study; the staff from the PRESICA project for logistical support; the Yerkes Primate Center staff for shipping blood samples from uninfected captive chimpanzees; Maria Salazar for technical assistance; and Jamie C. White for artwork and manuscript preparation.
This work was supported in part by the National Institutes of Health (R37 AI50529, R01 AI58715, P30 AI27767, and P30 CA13148), Agence National de Recherches sur le SIDA (ANRS), France (ANRS 12125), the Institut de Recherche pour le Développement (IRD), the Yerkes Regional Primate Research Center (RR-00165), and the Bristol Myers Freedom to Discover Program.
Footnotes
Published ahead of print on 10 December 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akaike, H. 1974. A new look at statistical model identification. IEEE Trans. Automat. Control 19716-723. [Google Scholar]
- 3.Ayouba, A., P. Mauclere, P. M. Martin, P. Cunin, J. Mfoupouendoun, B. Njinku, S. Souquieres, and F. Simon. 2001. HIV-1 group O infection in Cameroon, 1986 to 1998. Emerg. Infect. Dis. 7466-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 965698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailes, E., R. R. Chaudhuri, M. L. Santiago, F. Bibollet-Ruche, B. H. Hahn, and P. M. Sharp. 2002. The evolution of primate lentiviruses and the origins of AIDS, p. 65-96. In T. Leitner (ed.), The molecular epidemiology of human viruses. Kluwer Academic Publishers, Boston, MA.
- 6.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 3001713. [DOI] [PubMed] [Google Scholar]
- 7.Bell, C. H., R. Pantophlet, A. Schiefner, L. A. Cavacini, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2008. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J. Mol. Biol. 375969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bermejo, M. 2004. Home-range use and intergroup encounters in western gorillas (Gorilla gorilla gorilla) at Lossi forest, North Congo. Am. J. Primatol. 64223-232. [DOI] [PubMed] [Google Scholar]
- 9.Bibollet-Ruche, F., E. Bailes, F. Gao, X. Pourrut, K. L. Barlow, J. P. Clewley, J. M. Mwenda, D. K. Langat, G. K. Chege, H. M. McClure, E. Mpoudi-Ngole, E. Delaporte, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2004. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade. J. Virol. 787748-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77773-781. [DOI] [PubMed] [Google Scholar]
- 11.Bibollet-Ruche, F., F. Gao, E. Bailes, S. Saragosti, E. Delaporte, M. Peeters, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 2004. Complete genome analysis of one of the earliest SIVcpzPtt strains from Gabon (SIVcpzGAB2). AIDS Res. Hum. Retrovir. 201377-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boesch, C., and H. Boesch. 1989. Hunting behavior of wild chimpanzees in the Taï National Park. Am. J. Phys. Anthropol. 78547-573. [DOI] [PubMed] [Google Scholar]
- 14.Bowen-Jones, E., and S. Pendry. 1999. The threat to primates and other mammals from the bushmeat trade in Africa, and how this threat could be diminished. Oryx 33233-246. [Google Scholar]
- 15.Brennan, C. A., P. Bodelle, R. Coffey, S. G. Devare, A. Golden, J. Hackett, Jr., B. Harris, V. Holzmayer, K. C. Luk, G. Schochetman, P. Swanson, J. Yamaguchi, A. Vallari, N. Ndembi, C. Ngansop, F. Makamche, D. Mbanya, L. G. Gurtler, L. Zekeng, and L. Kaptue. 2008. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J. Acquir. Immune Defic. Syndr. 49432-439. [DOI] [PubMed] [Google Scholar]
- 16.Butynski, T. M. 2001. Africa's great apes, p. 3-56. In T. S. B. Beck, M. Hutchins, T. L. Maple, B. G. Norton, A. Rowan, E. F. Stevens, and A. Arluke (ed.), Great apes and humans. The ethics of co-existence. Smithsonian Institution Press, Washington, DC.
- 17.Caillaud, D., F. Levrero, S. Gatti, N. Menard, and M. Raymond. 2008. Influence of male morphology on male mating status and behavior during interunit encounters in western lowland gorillas. Am. J. Phys. Anthropol. 135379-388. [DOI] [PubMed] [Google Scholar]
- 18.Clark, S. J., M. S. Saag, W. D. Decker, S. Campbell-Hill, J. L. Roberson, P. J. Veldkamp, J. C. Kappes, B. H. Hahn, and G. M. Shaw. 1991. High titers of cytopathic virus in plasma of patients with symptomatic primary HIV-1 infection. N. Engl. J. Med. 324954-960. [DOI] [PubMed] [Google Scholar]
- 19.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2011407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Clercq, E., N. Yamamoto, R. Pauwels, J. Balzarini, M. Witvrouw, K. De Vreese, Z. Debyser, B. Rosenwirth, P. Peichl, R. Datema, et al. 1994. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob. Agents Chemother. 38668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 748358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doran-Sheehy, D. M., D. Greer, P. Mongo, and D. Schwindt. 2004. Impact of ecological and social factors on ranging in western gorillas. Am. J. Primatol. 64207-222. [DOI] [PubMed] [Google Scholar]
- 23.Douadi, M. I., S. Gatti, F. Levrero, G. Duhamel, M. Bermejo, D. Vallet, N. Menard, and E. J. Petit. 2007. Sex-biased dispersal in western lowland gorillas (Gorilla gorilla gorilla). Mol. Ecol. 162247-2259. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein, J. 2005. PHYLIP (phylogeny inference package) version 3.6. J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle.
- 25.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 3501118-1129. [DOI] [PubMed] [Google Scholar]
- 26.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397436-441. [DOI] [PubMed] [Google Scholar]
- 27.Gatti, S., F. Levrero, N. Menard, and A. Gautier-Hion. 2004. Population and group structure of western lowland gorillas (Gorilla gorilla gorilla) at Lokoue, Republic of Congo. Am. J. Primatol. 63111-123. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh, S. K., P. N. Fultz, E. Keddie, M. S. Saag, P. M. Sharp, B. H. Hahn, and G. M. Shaw. 1993. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology 194858-864. [DOI] [PubMed] [Google Scholar]
- 29.Goodall, J. 1986. The chimpanzees of Gombe: patterns of behavior. Belknap Press, Cambridge, United Kingdom.
- 30.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287607-614. [DOI] [PubMed] [Google Scholar]
- 31.Holmes, E. C. 2003. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 773893-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin, M. J., J. Rogers, J. E. Phillips-Conroy, J. S. Allan, R. C. Desrosiers, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J. Virol. 688454-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keele, B. F., F. Van Heuverswyn, Y. Li, E. Bailes, J. Takehisa, M. L. Santiago, F. Bibollet-Ruche, Y. Chen, L. V. Wain, F. Liegeois, S. Loul, E. M. Ngole, Y. Bienvenue, E. Delaporte, J. F. Brookfield, P. M. Sharp, G. M. Shaw, M. Peeters, and B. H. Hahn. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leendertz, F. H., S. Yumlu, G. Pauli, C. Boesch, E. Couacy-Hymann, L. Vigilant, S. Junglen, S. Schenk, and H. Ellerbrok. 2006. A new Bacillus anthracis found in wild chimpanzees and a gorilla from West and Central Africa. PLoS Pathog. 2e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemey, P., O. G. Pybus, A. Rambaut, A. J. Drummond, D. L. Robertson, P. Roques, M. Worobey, and A. M. Vandamme. 2004. The molecular population genetics of HIV-1 group O. Genetics 1671059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leroy, E. M., P. Rouquet, P. Formenty, S. Souquiere, A. Kilbourne, J. M. Froment, M. Bermejo, S. Smit, W. Karesh, R. Swanepoel, S. R. Zaki, and P. E. Rollin. 2004. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science 303387-390. [DOI] [PubMed] [Google Scholar]
- 37.Li, Y., H. Hui, C. J. Burgess, R. W. Price, P. M. Sharp, B. H. Hahn, and G. M. Shaw. 1992. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 666587-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, W., M. Worobey, Y. Li, B. F. Keele, F. Bibollet-Ruche, Y. Guo, P. A. Goepfert, M. L. Santiago, J. B. Ndjango, C. Neel, S. L. Clifford, C. Sanz, S. Kamenya, M. L. Wilson, A. E. Pusey, N. Gross-Camp, C. Boesch, V. Smith, K. Zamma, M. A. Huffman, J. C. Mitani, D. P. Watts, M. Peeters, G. M. Shaw, W. M. Switzer, P. M. Sharp, and B. H. Hahn. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews, A., and A. Matthews. 2004. Survey of gorillas (Gorilla gorilla gorilla) and chimpanzees (Pan troglodytes troglodytes) in Southwestern Cameroon. Primates 4515-24. [DOI] [PubMed] [Google Scholar]
- 40.Mitani, J. C., and D. P. Watts. 1999. Demographic influences on the hunting behavior of chimpanzees. Am. J. Phys. Anthropol. 109439-454. [DOI] [PubMed] [Google Scholar]
- 41.Nishihara, T. 1995. Feeding ecology of western lowland gorillas in the Nouabale-Ndoki National Park, Congo. Primates 36151-168. [Google Scholar]
- 42.Parnell, R. J. 2002. Group size and structure in western lowland gorillas (Gorilla gorilla gorilla) at Mbeli Bai, Republic of Congo. Am. J. Primatol. 56193-206. [DOI] [PubMed] [Google Scholar]
- 43.Peeters, M., A. Gueye, S. Mboup, F. Bibollet-Ruche, E. Ekaza, C. Mulanga, R. Ouedrago, R. Gandji, P. Mpele, G. Dibanga, B. Koumare, M. Saidou, E. Esu-Williams, J. P. Lombart, W. Badombena, N. Luo, M. Vanden Haesevelde, and E. Delaporte. 1997. Geographical distribution of HIV-1 group O viruses in Africa. AIDS 11493-498. [DOI] [PubMed] [Google Scholar]
- 44.Posada, D., and T. Buckley. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53793-808. [DOI] [PubMed] [Google Scholar]
- 45.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 46.Reed, K. D., J. W. Melski, M. B. Graham, R. L. Regnery, M. J. Sotir, M. V. Wegner, J. J. Kazmierczak, E. J. Stratman, Y. Li, J. A. Fairley, G. R. Swain, V. A. Olson, E. K. Sargent, S. C. Kehl, M. A. Frace, R. Kline, S. L. Foldy, J. P. Davis, and I. K. Damon. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350342-350. [DOI] [PubMed] [Google Scholar]
- 47.Remis, M. J. 1997. Ranging and grouping patterns of a western lowland gorilla group at Bai Hokou, Central African Republic. Am. J. Primatol. 43111-133. [DOI] [PubMed] [Google Scholar]
- 48.Robbins, M. M., M. Bermejo, C. Cipolletta, F. Magliocca, R. J. Parnell, and E. Stokes. 2004. Social structure and life-history patterns in western gorillas (Gorilla gorilla gorilla). Am. J. Primatol. 64145-159. [DOI] [PubMed] [Google Scholar]
- 49.Rogers, M. E., K. Abernethy, M. Bermejo, C. Cipolletta, D. Doran, K. McFarland, T. Nishihara, M. Remis, and C. E. Tutin. 2004. Western gorilla diet: a synthesis from six sites. Am. J. Primatol. 64173-192. [DOI] [PubMed] [Google Scholar]
- 50.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 191572-1574. [DOI] [PubMed] [Google Scholar]
- 51.Santiago, M. L., F. Bibollet-Ruche, N. Gross-Camp, A. C. Majewski, M. Masozera, I. Munanura, B. A. Kaplin, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2003. Noninvasive detection of Simian immunodeficiency virus infection in a wild-living L'Hoest's monkey (Cercopithecus Ihoesti). AIDS Res. Hum. Retrovir. 191163-1166. [DOI] [PubMed] [Google Scholar]
- 52.Santiago, M. L., M. Lukasik, S. Kamenya, Y. Li, F. Bibollet-Ruche, E. Bailes, M. N. Muller, M. Emery, D. A. Goldenberg, J. S. Lwanga, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, D. P. Watts, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, J. F. Brookfield, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 777545-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santiago, M. L., F. Range, B. F. Keele, Y. Li, E. Bailes, F. Bibollet-Ruche, C. Fruteau, R. Noe, M. Peeters, J. F. Brookfield, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2005. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Taï Forest, Cote d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J. Virol. 7912515-12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295465. [DOI] [PubMed] [Google Scholar]
- 55.Reference deleted.
- 56.Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem. Soc. Trans. 28275-282. [DOI] [PubMed] [Google Scholar]
- 57.Sharp, P. M., G. M. Shaw, and B. H. Hahn. 2005. Simian immunodeficiency virus infection of chimpanzees. J. Virol. 793891-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon, F., P. Mauclere, P. Roques, I. Loussert-Ajaka, M. C. Muller-Trutwin, S. Saragosti, M. C. Georges-Courbot, F. Barre-Sinoussi, and F. Brun-Vezinet. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 41032-1037. [DOI] [PubMed] [Google Scholar]
- 59.Stanfield, R. L., M. K. Gorny, C. Williams, S. Zolla-Pazner, and I. A. Wilson. 2004. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure 12193-204. [DOI] [PubMed] [Google Scholar]
- 60.Stanford, C. B., and J. B. Nkurunungi. 2003. Behavioral ecology of sympatric chimpanzees and gorillas in Bwindi Impenetrable National Park, Uganda: diet. Int. J. Primatol. 24901-918. [Google Scholar]
- 61.Stanford, C. B., J. Wallis, H. Matama, and J. Goodall. 1994. Patterns of predation by chimpanzees on red colobus monkeys in Gombe National Park, 1982-1991. Am. J. Phys. Anthropol. 94213-228. [DOI] [PubMed] [Google Scholar]
- 62.Stokes, E. J. 2004. Within-group social relationships among females and adult males in wild western lowland gorillas (Gorilla gorilla gorilla). Am. J. Primatol. 64233-246. [DOI] [PubMed] [Google Scholar]
- 63.Stokes, E. J., R. J. Parnell, and C. Olenjniczak. 2003. Female dispersal and reproductive success in wild western lowland gorillas (Gorilla gorilla gorilla). Behav. Ecol. Sociobiol. 54329-339. [Google Scholar]
- 64.Swofford, D. L. 2003. PAUP* 4.0: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Inc., Sunderland, MA.
- 65.Takehisa, J., M. H. Kraus, J. M. Decker, Y. Li, B. F. Keele, F. Bibollet-Ruche, K. P. Zammit, Z. Weng, M. L. Santiago, S. Kamenya, M. L. Wilson, A. E. Pusey, E. Bailes, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2007. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J. Virol. 817463-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tutin, C. E. 1996. Ranging and social structure of lowland gorilla in the Lope Reserve, Gabon, p. 58. In W. C. McGrew, L. E. Marchant, and T. Nishida (ed.), Great ape societies. Cambridge University Press, New York. NY.
- 68.Tutin, C. E., and M. Fernandez. 1993. Composition of the diet of chimpanzees and comparisons with that of sympatric lowland gorillas in the Lope Reserve, Gabon. Am. J. Primatol. 30195-211. [DOI] [PubMed] [Google Scholar]
- 69.Usongo, L., and R. Ngnegueu. 2001. Great ape hunting and trade in Lobeke, Cameroon. Gorilla J. 2229-31. [Google Scholar]
- 70.Van Heuverswyn, F., Y. Li, E. Bailes, C. Neel, B. Lafay, B. F. Keele, K. S. Shaw, J. Takehisa, M. H. Kraus, S. Loul, C. Butel, F. Liegeois, B. Yangda, P. M. Sharp, E. Mpoudi-Ngole, E. Delaporte, B. H. Hahn, and M. Peeters. 2007. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368155-171. [DOI] [PubMed] [Google Scholar]
- 71.Van Heuverswyn, F., Y. Li, C. Neel, E. Bailes, B. F. Keele, W. Liu, S. Loul, C. Butel, F. Liegeois, Y. Bienvenue, E. M. Ngolle, P. M. Sharp, G. M. Shaw, E. Delaporte, B. H. Hahn, and M. Peeters. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444164. [DOI] [PubMed] [Google Scholar]
- 72.Van Heuverswyn, F., and M. Peeters. 2007. The origins of HIV and implications for the global epidemic. Curr. Infect. Dis. Rep. 9338-346. [DOI] [PubMed] [Google Scholar]
- 73.van Rensburg, E. J., S. Engelbrecht, J. Mwenda, J. D. Laten, B. A. Robson, T. Stander, and G. K. Chege. 1998. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J. Gen. Virol. 791809-1814. [DOI] [PubMed] [Google Scholar]
- 74.Vergne, L., A. Bourgeois, E. Mpoudi-Ngole, R. Mougnutou, J. Mbuagbaw, F. Liegeois, C. Laurent, C. Butel, L. Zekeng, E. Delaporte, and M. Peeters. 2003. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology 310254-266. [DOI] [PubMed] [Google Scholar]
- 75.Wain, L. V., E. Bailes, F. Bibollet-Ruche, J. M. Decker, B. F. Keele, F. Van Heuverswyn, Y. Li, J. Takehisa, E. M. Ngole, G. M. Shaw, M. Peeters, B. H. Hahn, and P. M. Sharp. 2007. Adaptation of HIV-1 to its human host. Mol. Biol. Evol. 241853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walsh, P. D., T. Breuer, C. Sanz, D. Morgan, and D. Doran-Sheehy. 2007. Potential for Ebola transmission between gorilla and chimpanzee social groups. Am. Nat. 169684-689. [DOI] [PubMed] [Google Scholar]
- 77.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 78.Yamaguchi, J., P. Bodelle, L. Kaptue, L. Zekeng, L. G. Gurtler, S. G. Devare, and C. A. Brennan. 2003. Near full-length genomes of 15 HIV type 1 group O isolates. AIDS Res. Hum. Retrovir. 19979-988. [DOI] [PubMed] [Google Scholar]
- 79.Yang, Z., and B. Rannala. 1997. Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol. Biol. Evol. 14717-724. [DOI] [PubMed] [Google Scholar]
- 80.Zekeng, L., J. Obiang Sima, H. Hampl, J. M. Ndemesogo, J. Ntutumu, V. Sima, S. Devare, L. Kaptue, and L. Gurtler. 1997. Update on HIV-1 group O infection in Equatorial Guinea, Central Africa. AIDS 111410-1412. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.