Abstract
Viruses of the order Mononegavirales all encode a large (L) polymerase protein responsible for the replication and transcription of the viral genome as well as all posttranscriptional modifications of viral mRNAs. The L protein is conserved among all members of the Mononegavirales and has six conserved regions (“domains”). Using vesicular stomatitis virus (VSV) (family Rhabdoviridae) experimental system, we and others recently identified several conserved amino acid residues within L protein domain VI which are required for viral mRNA cap methylation. To verify that these critical amino acid residues have a similar function in other members of the Mononegavirales, we examined the Sendai virus (SeV) (family Paramyxoviridae) L protein by targeting homologous amino acid residues important for cap methylation in VSV which are highly conserved among all members of the Mononegavirales and are believed to constitute the L protein catalytic and S-adenosylmethionine-binding sites. In addition, an SeV L protein mutant with a deletion of the entire domain VI was generated. First, L mutants were tested for their abilities to synthesize viral mRNAs. While the domain VI deletion completely inactivated L, most of the amino acid substitutions had minor effects on mRNA synthesis. Using a reverse genetics approach, these mutations were introduced into the SeV genome, and recombinant infectious SeV mutants with single alanine substitutions at L positions 1782, 1804, 1805, and 1806 or a double substitution at positions 1804 and 1806 were generated. The mutant SeV virions were purified, detergent activated, and analyzed for their abilities to synthesize viral mRNAs methylated at their cap structures. In addition, further studies were done to examine these SeV mutants for a possible host range phenotype, which was previously shown for VSV cap methylation-defective mutants. In agreement with a predicted role of the SeV L protein invariant lysine 1782 as a catalytic residue, the recombinant virus with a single K1782A substitution was completely defective in cap methylation and showed a host range phenotype. In addition, the E1805A mutation within the putative S-adenosylmethionine-binding site of L resulted in a 60% reduction in cap methylation. In contrast to the homologous VSV mutants, other recombinant SeV mutants with amino acid substitutions at this site were neither defective in cap methylation nor host range restricted. The results of this initial study using an SeV experimental system demonstrate similarities as well as differences between the L protein cap methylation domains in different members of the Mononegavirales.
The order Mononegavirales includes many medically important pathogens including the lethal rabies, Ebola, Marburg, Nipah, and Hendra viruses. All members of this order share a similar genome organization and common mechanisms of genome replication and gene expression (27, 32, 48). The RNA-dependent RNA polymerase of members of the Mononegavirales is packaged into mature virions and consists of two viral subunits, the phosphoprotein (P) and the large (L) protein. The L polymerase protein, whose large size (more than 2,000 amino acids [aa] in a single polypeptide chain) reflects its multifunctional nature, plays a central role in virus RNA replication and transcription. This protein has six sequence regions (“domains”) with a high degree of homology among all members of the Mononegavirales. Although there is no protein structure data available for any part of the L protein, these domains have been postulated to constitute the specific enzymatic activities of the viral RNA polymerase involved in transcription, mRNA 5′ capping, cap methylation, mRNA 3′ polyadenylation, and replication of viral RNA (27, 32, 48).
The mRNA 5′-cap structures of most members of the Mononegavirales are methylated by the L protein at the guanine-N7 and 2′-O-adenosine positions (1, 3, 18, 37, 42, 44). The methyltransferase (MTase) function of L was originally shown through the characterization of two host range (hr) mutants of vesicular stomatitis virus (VSV) (family Rhabdoviridae). It was shown that these mutants were defective in viral mRNA cap methylation (24, 25) and that purified wild-type (wt) L protein was able to complement their defect during transcription in vitro, demonstrating that the VSV L protein possesses cap MTase activities (21). Later, independent computational analyses (5, 13) proposed that while L proteins share a very low degree of homology with the known S-adenosylmethionine (AdoMet)-dependent MTases at the amino acid level, their domain VI has a prototypical MTase fold, a glycine-rich motif shared by all members of the AdoMet-dependent MTase superfamily and directly involved in AdoMet binding (26, 34), and several potential catalytic residues. Our recent analysis of the VSV hr1 mutant showed that a single-amino-acid substitution (D1671V) in this putative AdoMet-binding glycine-rich motif completely eliminated viral mRNA cap methylation at both the guanine-N7 and 2′-O-adenosine positions (17), thus experimentally supporting the above-described predictions (5, 13). In addition, other authors demonstrated that substitutions at other positions within the VSV L protein domain VI (including an invariant lysine 1651 and aa 1670 and 1672 within the glycine-rich motif) also resulted in various defects in mRNA cap methylation (16, 17, 29, 30). The importance of domain VI in cap methylation was biochemically demonstrated for Sendai virus (SeV) (family Paramyxoviridae) by Ogino et al. (40) using an in vitro assay with a fragment of the L protein that contained domain VI. Intriguingly, only the guanine-N7 but not 2′-O-adenosine MTase activity was detected in that study (40), although SeV normally produces mRNAs methylated at both the guanine-N7 and 2′-O-adenosine positions (44). Although all these studies suggest that a conserved domain VI is the MTase domain of L, many important questions remain, including the MTase specificity of domain VI (guanine-N7, 2′-O-adenosine, or both), and there have been no reports for the L protein in VSV or any other members of the Mononegavirales that directly demonstrate the location of the L protein region physically binding AdoMet.
In addition to defective cap methylation, a link between L protein MTase activities and the phenotype of the VSV mutants was documented. Specifically, VSV mutants defective in cap methylation were temperature-sensitive (ts) and, more interestingly, host range restricted (hr), as manifested by their inability to grow in certain nonpermissive cell lines (e.g., HEp-2 cells) while retaining their ability to grow to high titers in permissive cells (e.g., BHK-21 cells) (16, 17, 24, 25). Previous studies linked the inability of VSV cap methylation-defective mutants to grow in HEp-2 cells to the nontranslatability of primary VSV transcripts (24, 25) and showed that host cells methylate viral mRNA in permissive cell lines through an unknown mechanism (24).
Most previously published studies focused on the cap MTase function of the L protein using VSV experimental systems. While VSV is the prototypic member of the Mononegavirales and serves as a very useful model for understanding the biology of other less studied viruses (e.g., Ebola virus, due to its extreme pathogenicity in humans), we wanted to confirm that previously discovered MTase features of the VSV L protein are shared by other members of the Mononegavirales. To address it, we decided to determine if L protein amino acids that are important for cap methylation in VSV (including the invariant lysine and the residues within the glycine-rich motif comprising the putative catalytic and AdoMet-binding sites) have similar functions in SeV, the prototypic member of the family Paramyxoviridae. Recombinant SeV viruses with amino acid substitutions at several L positions were generated and analyzed for their abilities to synthesize and methylate viral mRNA. In addition, mutant viruses were tested for possible hr and ts phenotypes, which were previously shown for VSV cap methylation mutants. Our study identified important similarities in cap methylation between rhabdoviruses and paramyxoviruses as well as some unexpected differences. The implications for cap MTase function in members of the Mononegavirales are discussed.
MATERIALS AND METHODS
Cell lines and viruses.
African green monkey (Vero) (ATCC CCL-81), human epidermal carcinoma (HEp-2) (ATCC CCL-23), human lung carcinoma (A549) (ATCC CCL-185), and BSR-T7/5 (derived from baby hamster kidney [BHK-21] cells and constitutively expressing bacteriophage T7 polymerase) (4) cells were used for virus infections and plasmid transfections. Monolayer cultures of these cell lines were maintained in Dulbecco's modified Eagle's medium (Cellgro) supplemented with 9% fetal bovine serum (Gibco). Recombinant wt (rWT) SeV (strain Fushimi) (28) and SeV-GFP-Fmut (rWT-GFP) with an enhanced green fluorescent protein (GFP) upstream of the NP gene (49) were kindly provided by Wolfgang J. Neubert (Max Planck Institute of Biochemistry, Germany). To grow and purify wt or mutant SeV, Vero or BSR-T7 cells were infected with wt or mutant viruses at a multiplicity of infection (MOI) of 0.1 cell infectious units (CIU)/ml in MegaVir HyQSFM4 serum-free medium (SFM) (HyClone) and in the presence of 4 μg/ml acetylated trypsin (28) and incubated for 48 to 120 h at 34°C. Cleavage by a cellular protease is necessary for the SeV fusion (F) protein to be biologically active in vivo, making the viral particle infectious and allowing for multiple rounds of virus replication. SeV-GFP viruses were grown similarly but without acetylated trypsin in the medium, as they have a wt monobasic trypsin-dependent cleavage site in the F protein mutated to an oligobasic cleavage site, allowing F activation in any cell type through a ubiquitous furin-like protease (49). The released viruses were purified from the medium as described previously (17); suspended at about 5 mg/ml in a solution containing 1 mM Tris-HCl (pH 7.4), 1 mM EDTA, and 10% dimethyl sulfoxide; and stored at −80°C. Recombinant wt VSV (Indiana serotype) and its derivative, VSV rHR1-1 (referred to as hr1 in this paper), with a single amino acid substitution, D1671V, in the L protein were described previously (17).
Virus growth analysis.
SeV infectivity, expressed as CIU/ml, was measured by virus titration on Vero cells and counting infectious foci visually using light microscopy and/or an immunofluorescence (IF) assay for SeV mutants or by GFP-based fluorescence for SeV-GFP viruses. For IF, SFM from six-well plates was aspirated 2 or 3 days postinfection, and cells were washed with phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde (Sigma) for 10 min, and permeabilized for 2 min on ice with a solution containing 20 mM HEPES (pH 7.5), 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, and 0.5% Triton X-100. Cells were then blocked in PBS with 5% bovine serum albumin for 20 min and incubated with anti-SeV primary antibodies (1:100) for 1 h. Cells were washed, incubated with goat anti-rabbit IgG-fluorescein isothiocyanate antibodies (Santa Cruz) for 1 h in the dark, and viewed under a fluorescent microscope to determine virus titer. For multistep growth analysis, Vero or HEp-2 cells in six-well plates were infected at an MOI of 0.001 CIU/cell in 1.5 ml SFM per well. One hour postinfection (p.i.), medium was aspirated, cells were washed with PBS, and 1.5 ml SFM with 1 μg/ml acetylated trypsin was added to each well. Supernatants were harvested at 12-h (for Vero cells) or 24-h (for HEp-2 cells) intervals and flash-frozen at −80°C. Virus titers were determined using a 96-well plate format by infecting Vero cells with the serial dilutions (1:8) of wt or mutant SeV (collected at various time points) and incubation at 37°C with shaking. At 1 h p.i., viruses were aspirated, and cells were overlaid with 100 μl SFM with 2 μg/ml of acetylated trypsin, and cells were analyzed at 48 h p.i.
Plasmids and mutagenesis.
pGEM plasmids containing wt genes for SeV NP, L, and Pstop (expressing P but not C due to a stop codon in the C open reading frame, referred to here as wt P) under the control of the T7 promoter were described previously (10). Plasmid pGEM-L and mutagenic primers were used for the L protein domain VI deletion and for site-directed mutagenesis (Table 1). Primers also contained silent restriction sites (Table 1) for screening and confirmation purposes. Using an overlapping PCR approach (22), two rounds of PCR were done using wt plasmid pGEM-L as a template, common flanking primers VG19 and VG20, and two specific primers designed for amino acid substitutions (Table 1). The final PCR products were digested with XhoI and MfeI and cloned into XhoI-MfeI-digested plasmid pGEM-Lwt. All L plasmids were tested for the presence of silent sites by digestion with the appropriate silent site enzymes, followed by sequence analysis to confirm the presence of the desired mutations and the absence of any spontaneous secondary mutations. SeV plasmids pTM-NP, pTM-P, and pTM-L; full-length SeV antigenomic plasmid pRS3Gg (28); and SeV plasmid pRSIdeFmut (a full-length SeV antigenomic plasmid with the GFP gene inserted upstream of the NP gene), used for the rescue of recombinant SeVs, were kindly provided by Wolfgang J. Neubert (Max Planck Institute of Biochemistry, Germany).
TABLE 1.
Sequences of primers used in this study to generate SeV mutant L genes
| L mutant or primer | L change | Silent site | Primer sequencea |
|---|---|---|---|
| L-1782A | K1782A | AfeI | CATCAACAGTACTAGCTGCTTGgcAGCGCTTGAACTTACCTACCTATT (+) |
| CAATAGGTAGGTAAGTTCAAGCGCTGCCAAGCAGCTAGTACTGTTGAT (−) | |||
| L-1804A | G1804A | EaeI | GATAGGCTATATTTGGccGAAGGAGCTGGGGCCATG (+) |
| CATGGCCCCAGCTCCTTCGGCCAAATATAGCCTATC (−) | |||
| L-1805A | E1805A | AvrII | GATAAAGATAGGCTATACCTAGGGGcAGGAGCTGGGGCCATG (+) |
| GCATGGCCCCAGCTCCTGCCCCTAGGTATAGCCTATCTTTAT (−) | |||
| L-1805V | E1805V | AvrII | GATAAAGATAGGCTATACCTAGGGGtAGGAGCTGGGGCCATG (+) |
| GCATGGCCCCAGCTCCTACCCCTAGGTATAGCCTATCTTTAT (−) | |||
| L-1806A | G1806A | AvrII | GATAAAGATAGGCTATACCTAGGGGAAGcAGCTGGGGCCATGCTTTC (+) |
| GAAAGCATGGCCCCAGCTTCCCCTAGGTATAGCCTATCTTTATC (−) | |||
| L-1804A/1806A | G1804A/G1806A | PstI | GATAGGCTATATTTAGcGGAAGctGCAGGGGCCATGCTTTC (+) |
| GAAAGCATGGCCCCTGCAGCTTCCGCTAAATATAGCCTATC (−) | |||
| L-ΔVI | Deletion of aa 1777-1976 | None | GGCTCTTTGGCATCAACCTTCTATCGAGGCACCCC (+) |
| GGGGTGCCTCGATAGAAGGTTGATGCCAAAGAGCC (−) | |||
| Upstream primer VG19 for cloning and sequencing | CATACCTATGCAGCTTGGCAGAGA (+) | ||
| Downstream primer VG20 for cloning and sequencing | TAACCCTCAGGTTCCTGATCTCAC (−) |
Lowercase letters show substituted nucleotides resulting in the amino acid change. The underlined nucleotides represent alanine codons generated as a result of site-directed mutagenesis. +, plus sense; −, antisense.
Recovery of recombinant SeV.
Recombinant virus rescue was done using the reverse genetics system for SeV described previously by Leyrer et al. (28) using plasmids with SeV wt NP, P, and L genes and SeV full-length genomic cDNA (wt or mutant L gene) all under the control of the T7 promoter. For this study, we used the BSR-T7 cell line stably expressing the T7 RNA polymerase (4) for initial plasmid transfections and Vero cells for consequent virus passages. The K1782A, G1804A, E1805A, G1806A, and G1804A/G1806A mutations were introduced into full-length genomic SeV plasmid pRS3Gg. To obtain a mutant plasmid, pGEM-Lmut was digested with KpnI and NheI, and the fragment containing the L mutation was cloned into KpnI-NheI-cut pRS3Gg. Similarly, K1782A, E1805A, and G1806A mutations were introduced into plasmid pRSIdeFmut to generate recombinant SeV-GFP viruses. To rescue recombinant viruses, 10 μg of full-length plasmid pRS3Gg or pRSIdeFmut containing wt or a mutant L gene along with 1 μg of plasmid pTM-L, 3 μg of plasmid pTM-P, and 5 μg of plasmid pTM-NP plasmid were transfected into BSR-T7 cells in 35-mm dishes using Opti-MEM medium (Gibco) and Lipofectamine (Invitrogen) in a total of 2 ml according to the manufacturer's protocol. All transfection reaction mixtures were incubated for 24 h at 34°C. After 24 h, the transfection medium was aspirated, and 1.5 ml of SFM and 4 μg/ml acetylated trypsin were added to each well (SeV-GFP viruses were grown without trypsin). The cells were then incubated at 34°C for 2 days. On day 3 posttransfection (p.t.), 500 μl of BSR-T7 supernatant was collected and passed (passage 1) onto a fresh monolayer of Vero cells in 1 ml of fresh SFM medium with 4 μg/ml acetylated trypsin. Between 2 and 5 days following passage 1, there were noticeable cytopathic effects (CPE), cellular debris was pelleted, and the medium was harvested. The titers of recombinant SeV mutants on Vero cells with an agar overlay with 4 μg/ml acetylated trypsin were determined, and individual infectious foci were picked and grown on Vero cells. Recombinant viruses were purified as described previously (17), and all mutations were confirmed by reverse transcription-PCR and digestion with the appropriate silent restriction enzymes and by sequence analysis for the presence of the desired mutations and absence of any spontaneous secondary mutations in the L gene.
In vitro transcription with T7-expressed L proteins.
For the virus-driven expression of bacteriophage T7 RNA polymerase, Vero or A549 cells were infected with T7-expressing vaccinia virus (VV-T7) (15). To express wt SeV P and wt or mutant SeV L proteins, 60-mm dishes of A549 or Vero cells were infected with VV-T7 at an MOI of 2.5 PFU/cell for 1 h at 37°C, washed with Opti-MEM (Gibco), transfected with 1.5 μg of SeV plasmids pGEM-Pstop and 1 μg of pGEM-L (wt L or one of the mutant L genes) using Lipofectamine, and incubated at 34°C in Opti-MEM. At 18 h p.t., cytoplasmic extracts were prepared exactly as described previously (8, 17). To assay for SeV mRNA synthesis, 1 μg of wt SeV polymerase-free RNA-N template and 20 μCi of [α-32P]CTP were added to each extract, and reaction mixtures were incubated for 2 h at 30°C. Total RNA was purified using RNeasy columns (Qiagen) and analyzed by 1.5% agarose-6 M urea gel electrophoresis. The gels were fixed in 7% acetic acid, dried, exposed to Kodak X-OMat film for 18 h at −80°C, and quantitated using a PhosphorImager and ImageQuant software (Molecular Dynamics).
In vitro transcription using purified SeV virions.
SeV in vitro transcription by detergent-activated purified virions was conducted essentially as described previously (35). For [α-32P]UTP-labeled RNA, 10 μg of purified virus was incubated at 30°C for 6 h in a 50-μl reaction mixture containing 30 mM HEPES-KOH (pH 7.9); 75 mM NaCl; 50 mM KCl; 6 mM MgCl2; 2 mM dithiothreitol; 2 mM spermine; 0.1% NP-40; 500 μM each of ATP, CTP, and GTP; 50 μM UTP; 50 U of RNasin (Promega); 12 μg of purified tubulin (>99% pure) from bovine brain (Cytoskeleton Inc.); and 20 μCi of [α-32P]UTP. Total RNA was purified using RNeasy columns (Qiagen) and analyzed by 1.5% agarose-6 M urea gel electrophoresis. The gels were fixed in 7% acetic acid, dried, exposed to Kodak X-OMat film for 4 to 18 h at −80°C, and quantitated using a PhosphorImager. To test for viral mRNA cap methylation, in vitro transcription by detergent-activated purified wt or mutant SeV was conducted as described above, but RNA was synthesized in a 200-μl reaction mixture with cold nucleoside triphosphates (1 mM each) and 11 μCi of [3H]AdoMet (55 Ci/mmol) (final concentration, 1 μM AdoMet) in the presence or absence of 100 μM of the methylation inhibitor S-adenosylhomocysteine (AdoHcy). Total RNA was purified using RNeasy columns (Qiagen), diluted in 25 μl of H2O, and used for measurements of [3H]Met incorporation by scintillation counting (20 μl) or analyzed by Northern blotting to measure mRNA levels (5 μl). For Northern blot analysis, mRNA products of in vitro transcription reactions with [3H]AdoMet were separated in a 1.2% agarose-formaldehyde gel system, transferred onto a Hybond-N+ nylon membrane (GE Healthcare), and incubated with an RNA probe complementary to the SeV NP gene. To make this probe, SeV plasmid pGEM3-NP was digested at the MfeI site and transcribed in vitro with [α-32P]CTP using the MAXIscript SP6 kit (Ambion). Radioactive signals were measured using a PhosphorImager and ImageQuant software.
Western blot analysis.
To compare the amounts of the P and L proteins, total protein samples from transfected cytoplasmic lysates (5 μl of a total of 100 μl of lysate) were separated by 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto a polyvinylidene difluoride membrane (Sigma). The blots were incubated with a mixture of antibodies against the SeV L and P proteins and developed with a horseradish peroxidase-conjugated secondary antibody using the Enhanced Chemiluminescence Plus protein detection system (GE Healthcare) according to the manufacturer's protocol.
RESULTS
Generation of transcriptionally active SeV L protein mutants with amino acid substitutions in the putative AdoMet-binding site.
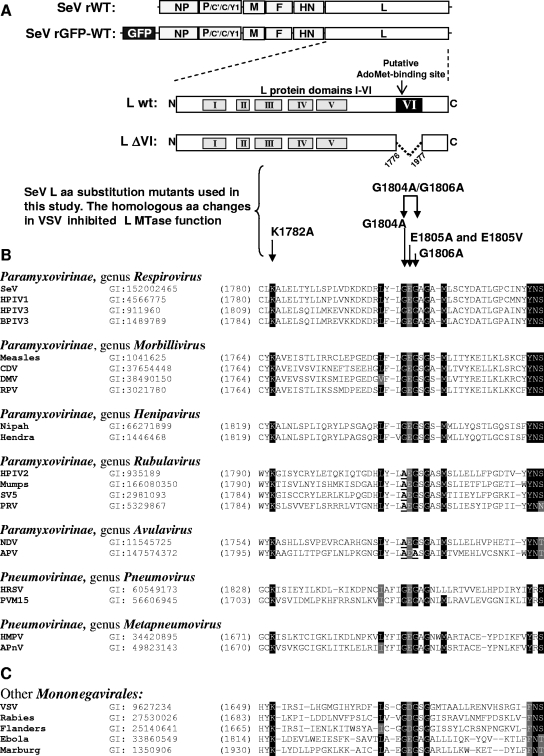
To date, all the domain VI mutagenesis studies have been carried out using the VSV (a rhabdovirus) experimental system (16, 17, 29, 30). Based on those studies and computational predictions (5, 13), it has been postulated that the glycine-rich motif (L amino acid positions 1670 to 1674 in VSV and positions 1804 to 1808 in SeV) constitutes an AdoMet-binding site of L, while an upstream invariant lysine (amino acid positions 1651 in VSV and 1782 in SeV) is a part of the active MTase site (5, 13, 30). Because the L proteins of members of the Mononegavirales are conserved and they all have a glycine-rich motif and lysine at the same positions as those in VSV (Fig. 1), it has been suggested that these amino acid residues are likely to have a similar importance in all members of the Mononegavirales (5, 13, 30). To test this hypothesis experimentally, we targeted the SeV L protein amino acid residues homologous to those that are important for cap methylation in VSV by site-directed mutagenesis. Figure 1 shows sequence alignments comparing a portion of domain VI of the L protein (including the glycine-rich motif and a critical lysine) between various paramyxoviruses (Fig. 1B) and other members of the Mononegavirales (Fig. 1C). Using plasmid SeV pGEM-Lwt (wt SeV L gene under the control of the T7 promoter), single-amino-acid substitutions to alanine were introduced at L amino acid positions K1782, G1804, E1805, and G1806, and a double substitution to alanines was introduced at positions G1804 and G1806 (Fig. 1A and Table 1). Also, two additional mutants were generated: (i) E1805V, based on an analogous D1671V mutation in the VSV hr1 mutant (17), and (ii) L-ΔVI, with a deletion of the entire domain VI to confirm that the presence of this region is critical for L transcriptional activity, as was previously shown for a VSV L protein mutant (7).
FIG. 1.
(A) SeV L protein mutants generated and analyzed in this study. L-ΔVI has a deletion of the entire domain VI. All other mutants have amino acid substitutions that were previously shown to inhibit cap methylation in VSV. (B and C) Multiple alignment of the putative AdoMet-binding region within L protein domain VI of members of the family Paramyxoviridae (B) and its comparison to the putative AdoMet-binding motif of other members of the Mononegavirales (C). Multiple alignment was conducted using the CLUSTAL W program. NCBI database identification numbers and starting amino acid positions are shown for all L proteins. Identical amino acids are highlighted in black, while gray shadows indicate conservative amino acid substitutions. HPIV1, HPIV2, and HPIV3, human parainfluenza virus types 1, 2, and 3, respectively; BPIV3, bovine parainfluenza virus type 3; CDV, canine distemper virus; DMV, dolphin morbillivirus; SV5, simian virus type 5; PRV, porcine rubulavirus; HMPV, human metapneumovirus; APV, avian pneumovirus; HRSV, human respiratory syncytial virus; PVH15, pneumonia virus of mice type 15; APnV, avian pneumovirus.
To test these mutant L proteins for their abilities to transcribe viral RNA and thus to determine if the corresponding recombinant viruses can be rescued, we used a VV-T7-based mammalian expression system described previously (8, 17). Briefly, Vero or A549 cells were infected with VV-T7, transfected with SeV P and L (wt or mutant), and incubated at 34°C. At 18 h p.t., cytoplasmic extracts containing P-L complexes were prepared (8, 17) and supplied with the exogenous wt SeV polymerase-free RNA-N template (isolated from wt SeV virions) and [α-32P]CTP to assay for SeV mRNA synthesis. The transcription products were analyzed by 1.5% agarose-6 M urea gel electrophoresis, visualized by autoradiography, and quantitated using a PhosphorImager. As shown in Fig. 2 for A549 cells (similar results were obtained with Vero cells) (data not shown), most tested proteins were transcriptionally active except for the L-ΔVI deletion mutant and L-E1805V, where mutations completely inactivated L. The latter result is rather unexpected, as a similar substitution in the VSV L protein (D1671V), while abolishing cap methylation, had little effect on VSV transcription (17). Nevertheless, as the other mutations passed the in vitro transcription test (Fig. 2A), they were cloned into the SeV full-length infectious cDNA plasmid for consequent recovery and characterization of recombinant SeV mutant viruses.
FIG. 2.
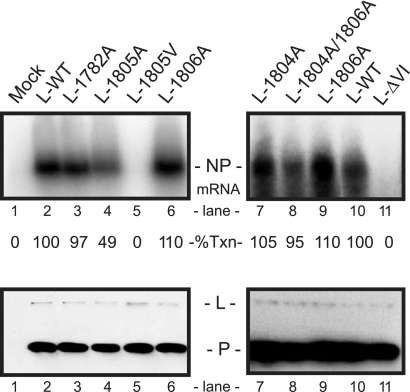
In vitro mRNA synthesis with SeV L mutants in A549 cytoplasmic extracts. To express the SeV P and L proteins, A549 cells were infected with VV-T7 at an MOI of 2.5 PFU/cell and transfected with wt P and wt or mutant L plasmids (the mock sample was VV-T7 infected but had no plasmids). (Top) Cytoplasmic extracts were prepared and incubated at 30°C with polymerase-free SeV RNA-N template in the presence of [α-32P]CTP. Labeled RNA products were purified and analyzed by agarose-urea gel electrophoresis and visualized by autoradiography. The position of the SeV NP mRNA is indicated. “% Txn” shows mRNA levels relative to those of wt SeV L protein (100%) using a PhosphorImager and represents the average of two or three experiments where variation was less than 15%. (Bottom) Immunoblot analysis of a portion of the A549 extracts used for in vitro transcription using a mixture of SeV P and L antibodies. The positions of the P and L proteins are indicated.
Recovery and phenotypic characterization of recombinant SeV with L mutations.
Once it was established that the L genes containing mutations were transcriptionally active (except for L-ΔVI and E1805V), the L mutations were cloned into an SeV full-length genomic cDNA plasmid to generate mutant viruses using the BSR-T7 cell line stably expressing the T7 RNA polymerase (4) for initial plasmid transfections and Vero cells for consequent virus passages. Although BSR-T7 cells are derived from BHK-21 cells (4), which support the replication of cap methylation-defective VSV mutants (17, 24), we wanted to confirm that BSR-T7 cells were suitable for the recovery of SeV mutants that are potentially defective in cap methylation. Similarly, we wanted to verify that potential cap methylation-defective viruses can be passed on Vero cells, which support the robust replication of wt SeV. Therefore, prior to the rescue attempts, we tested wt VSV and the cap methylation-defective VSV hr1 mutant (17) for their abilities to grow on BSR-T7 and Vero cells and compared them to their growth on HEp-2 cells, which do not support the replication of the VSV hr1 mutant or any other tested cap methylation-defective VSV mutant (16, 17). The VSV hr1 mutant was unable to grow in HEp-2 cells, as expected (Table 2), but was only moderately attenuated in Vero cells (2.4 × 109 PFU/ml for wt VSV and 6.0 × 107 PFU/ml for the VSV hr1 mutant) (Table 2) and grew normally in BSR-T7 cells (2.4 × 109 PFU/ml for wt VSV and 1.1 × 109 PFU/ml for the VSV hr1 mutant). Therefore, we concluded that a BSR-T7/Vero recovery system could be successfully used to rescue SeV mutants even if they are defective in cap methylation. Using this approach, we successfully recovered six recombinant viruses, designated rWT, r1782A, r1804A, r1805A, r1806A, and r1804A/1806A. During virus rescue, we noted that while most SeV mutants grew similarly to the rWT in Vero cells (as observed by CPE), CPE development in rK1782A was dramatically delayed (by 48 to 72 h), and rE1805A showed a 24-h delay in CPE development (data not shown). All viruses were confirmed for the presence of the desired mutations and the absence of any spontaneous secondary mutations by virus purification followed by reverse transcription-PCR amplification of the L gene and sequence analysis using primers VG19 and VG20 (Table 1).
TABLE 2.
Comparative titers of recombinant SeVs and VSVs in Vero or HEp2 cells at 34°C or 40°C
| Virus | Virus titer (CIU/ml)
|
Ratio of titers at 34°C/40°C for Vero cells | Ratio of titers of Vero/HEp-2 cells at 34°C | ||
|---|---|---|---|---|---|
| Vero cells
|
HEp-2 cells at 34°C | ||||
| 34°C | 40°C | ||||
| SeV rWT | 2.0 × 108 | 3.4 × 107 | 8.8 × 107 | 5.9 | 2.3 |
| SeV r1782A | 4.0 × 105 | <103 | 4.0 × 103 | >1.2 × 103 | 100 |
| SeV r1804A | 1.5 × 107 | 3.2 × 106 | 5.3 × 106 | 4.7 | 2.8 |
| SeV r1805A | 3.6 × 108 | <103 | 1.6 × 108 | >3.6 × 105 | 2.3 |
| SeV r1806A | 2.4 × 108 | 1.6 × 107 | 1 × 108 | 15 | 2.4 |
| SeV r1804A/1806A | 5.0 × 107 | 8.0 × 106 | 2.5 × 107 | 6.3 | 2.0 |
| SeV rWT-GFP | 1.9 × 108 | 7.0 × 106 | 1.0 × 108 | 27.1 | 1.9 |
| SeV r1805A-GFP | 8.3 × 107 | <103 | 3.3 × 107 | >8.3 × 104 | 2.5 |
| SeV r1806A-GFP | 1.3 × 108 | 4.6 × 106 | 4.4 × 107 | 28.3 | 3.0 |
| rVSV wt | 2.4 × 109 | 1.0 × 107 | 3.2 × 107 | 240 | 75 |
| rVSV hr1 | 6.0 × 107 | <103 | <103 | >6 × 104 | >6 × 104 |
Because previously studied VSV cap methylation mutants were hr and ts (16, 17), we tested all our recombinant SeV mutants for their possible hr and ts phenotypes. The titers of wt and mutant SeVs on Vero and HEp-2 cells at 34°C and 40°C were determined (Table 2). In addition, we used wt rVSV and rVSV hr1 viruses as convenient controls for the conditions used in these studies. The rVSV hr1 mutant grew to high titers on Vero cells (permissive cells) at 34°C (permissive temperature) but, unlike wt rVSV, displayed more than a 60,000-fold reduction in growth in HEp-2 cells at 34°C (nonpermissive cells) and in Vero cells at 40°C (nonpermissive temperature). Therefore, we predicted that SeV mutants defective in cap methylation would be moderately attenuated in Vero cells (as VSV hr1 compared to the wt) but severely attenuated in HEp2 cells. As shown in Table 2 and Fig. 3A, the SeV r1782A mutant showed an hr phenotype with a Vero/HEp-2 titer ratio of 100 (compared to 2.3 for the rWT), which supported a possible role of SeV L protein lysine 1782 at the active MTase site. This ratio was much smaller than that for the VSV hr1 mutant (Table 2) because rK1782A was also attenuated in Vero cells, reaching a maximum titer of only 4.0 × 105 CIU/ml at 120 h p.i., compared to 2.0 × 108 CIU/ml for the rWT at 48 to 72 h p.i. (Table 2). Also, infectious foci counted for r1782A in HEp-2 cells were noticeably smaller than those for the rWT (Fig. 3A).
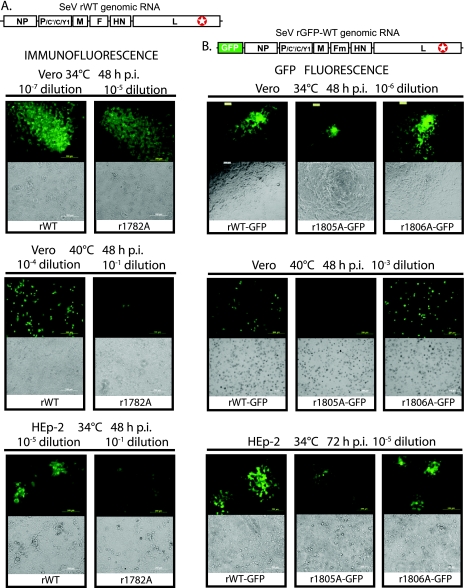
FIG. 3.
Host range and temperature sensitivity analysis of SeV mutants. (A) IF infectious focus assay to analyze SeV r1782A (compared to the SeV rWT) for hr and ts phenotypes. Infectious virus foci were visualized by IF using anti-SeV primary rabbit antibodies and anti-rabbit IgG-fluorescein isothiocyanate antibodies on fixed and permeabilized Vero or HEp-2 cells infected with SeV r1782A or the rWT at 34°C or 40°C. (B) GFP fluorescence focus assay to analyze SeV rWT-GFP, r1805A-GFP, and r1806A-GFP viruses carrying the GFP gene upstream of the NP gene for hr and ts phenotypes. Assays were done using Vero or HEp-2 cells at 34°C or 40°C. Infectious virus foci were visualized by microscopy at 48 or 72 h p.i., as indicated, using fluorescence (top) or bright-field (bottom) channels. Virus dilutions are indicated.
Unexpectedly, all tested SeV mutants with amino acid substitutions in the glycine-rich motif produced similar numbers of infectious foci in Vero and HEp-2 cells, with a Vero/HEp-2 titer ratio of about 2.5. However, r1805A displayed slow growth in both Vero and HEp-2 cells, with about a 24-h delay in infectious focus formation and noticeably smaller foci on both Vero and HEp2 cells.
To independently confirm these observations, we cloned three representative K1782A, E1805A, and G1806A mutations into a plasmid with the full-length SeV genome additionally encoding the GFP gene and successfully rescued two of the three recombinant viruses (r1805A-GFP and r1806A-GFP) containing the appropriate L mutations (Fig. 3). Ten separate attempts were made to rescue r1782A-GFP, but no infectious virus was ever recovered, and no GFP signal was visible during these attempts. We think that the combination of negative factors, the K1782A mutation and GFP insertion, made this virus too attenuated for recovery, at least under our standard rescue conditions. For successfully rescued GFP viruses, virus titrations were conducted using Vero and HEp-2 cells, and virus infection sites for rWT-GFP, r1805A-GFP, and r1806A-GFP were compared using fluorescent microscopy. As shown in Fig. 3B for both Vero and HEp2 cells, rWT-GFP and r1806A-GFP viruses had similarly sized foci, with similar GFP signals at 48 h p.i. The r1805A-GFP virus at 48 h p.i. had smaller sites on both Vero and HEp2 cells. However, we did not observe differences in the relative ability of r1805A-GFP to grow on HEp-2 cells compared to that on Vero cells (by CIU counts). Together, these data using SeV-GFP viruses confirmed that the G1806A mutation had no effect on SeV growth in Vero or HEp-2 cells, while the E1805A mutation similarly attenuated virus replication in Vero and HEp-2 cells.
In addition, virus titration experiments were performed with wt or mutant SeVs (and SeV-GFP) on Vero cells at 34°C and 40°C to determine possible ts phenotypes of these viruses, as previously shown for VSV hr1 and other cap methylation-defective mutants (16, 17). As shown in Table 2, the rVSV hr1 mutant was clearly ts, with a 34°C/40°C titer ratio in Vero cells of more than 60,000, compared to 75 for wt rVSV. However, only two SeV mutants displayed a ts phenotype, r1782A and r1805A. In agreement with this result, the GFP signal was present in Vero cells at 40°C for the rWT-GFP and r1806A-GFP viruses as early as 48 h p.i. (Fig. 3). However, there was no GFP signal in cells infected with r1805A-GFP virus at 40°C at any time point (Fig. 3).
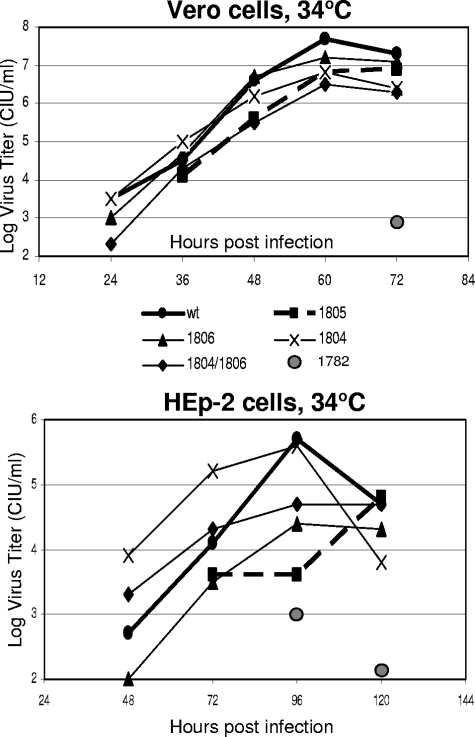
Our titration experiments demonstrated that unlike the r1782A mutant, all recombinant SeVs with amino acid substitutions in the glycine-rich motif did not display hr phenotypes. We wanted to confirm this result using a separate assay to test for the ability of these mutants to generate infectious particles in HEp-2 versus Vero cells (rather than their ability to form infectious foci, as in our titration experiments). Therefore, we conducted a multistep growth kinetic assay for these viruses by infecting Vero cells at a low MOI, harvesting cell supernatants at various time points, and assaying them using Vero cells to determine viral titers for each time point. As shown in Fig. 4, most recombinant viruses, except for SeV r1782A, displayed similar growth kinetics in Vero cells, with all titers peaking at 60 h p.i. The rK1782A mutant grew very slowly in Vero cells, producing about 2.5 × 102 CIU/ml at 72 h p.i. (Fig. 4) and reaching only 4.0 × 105 CIU/ml at 120 h p.i. While the r1804A, r1806A, and r1804A/1806A viruses all behaved similarly to the rWT, r1805A had about a 12-h delay in virus production. In HEp-2 cells, r1782A could be detected only at 96 and 120 h p.i. (maximum titer, 2 × 103 CIU/ml at 96 h p.i.), and the infection of HEp-2 cells with r1805A was clearly delayed, with viral titers beginning to increase after 72 h p.i. In addition, two SeV mutants, r1804A and r1806A, behaved very unusually in HEp-2 cells, with r1804A growing considerably faster than the rWT and r1806A growing considerably slower than the rWT. The presence of both mutations in r1804A/1806A produced an intermediate-growth phenotype, suggesting that these mutations had a reciprocal effect when present together. Despite these differences in growth kinetics, all recombinant viruses, except for r1782, were able to grow in HEp-2 cells to relatively high titers, which was consistent with our titration experiments (Table 2) and suggested that the amino acid substitutions in the glycine-rich motif did not abolish the L protein MTase function.
FIG. 4.
Multistep growth kinetics of wt and recombinant SeV in Vero and HEp-2 cells at 34°C. wt SeV or recombinant viruses were used to infect Vero (top) or HEp2 (bottom) cells at an MOI of 0.001 CIU/cell. Supernatants were harvested at 12-h (top) or 24-h (bottom) intervals and flash-frozen. Supernatants were assayed using Vero cells, and virus titers were determined for each time interval.
Analysis of mRNA cap methylation by recombinant SeV mutants.
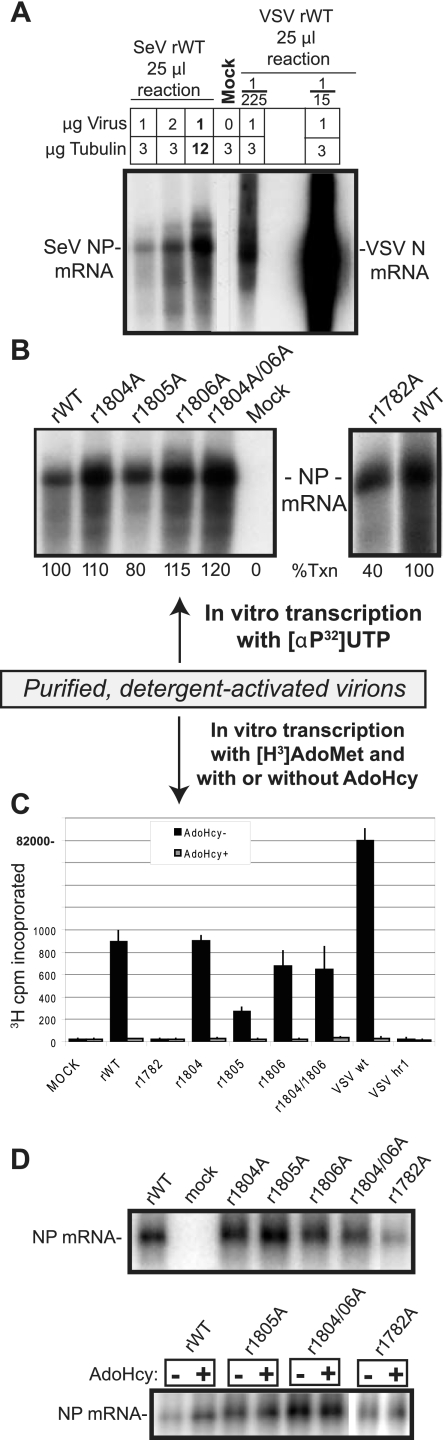
In addition to a phenotypic analysis of SeV mutants, we directly tested mutants for their abilities to methylate viral mRNAs in vitro. The limitation of the above-described VV-T7-based in vitro transcription assay with plasmid-expressed P and L proteins is its dependence on the vaccinia virus vector, which provides trans-active viral MTases (24), thus making these systems unusable for our studies of SeV MTase function. Therefore, the effects of SeV L protein mutations on viral mRNA cap methylation were studied using detergent-activated purified viruses naturally carrying active virion-bound polymerase, as was conducted previously for VSV (16, 17). In addition to recombinant SeV, we used wt rVSV and rVSV hr1 viruses as positive and negative controls for cap methylation throughout all these assays. It is important that in contrast to the VSV system, the reactions with detergent-activated purified SeV (and many other members of the Mononegavirales) virions require the addition of cytoplasmic extracts to each reaction mixture (11, 35, 38, 39). However, such an addition would be very undesirable for our experiments, as these extracts might contain trans-active cellular cap MTases, which could complement L protein defects in cap methylation and thus prevent discrimination between mutants based on their ability to methylate mRNA caps. Therefore, we optimized the SeV in vitro transcription conditions using purified tubulin, which has been shown to stimulate SeV virion transcription even when other cellular components are absent (35, 39). Interestingly, our optimal reaction conditions, producing amounts of viral mRNA similar to those of reactions with cell lysate from Vero cells (data not shown), generated about 200-fold less viral mRNA than did VSV virions transcribed under the same conditions (Fig. 5A). Nevertheless, despite these big differences in the efficiencies of mRNA synthesis, [α-32P]UTP-labeled SeV mRNA was easily detectable (Fig. 5A), and we proceeded to compare all our SeV mutants for their ability to (i) synthesize and (ii) methylate viral mRNAs in vitro.
FIG. 5.
In vitro mRNA synthesis with detergent-activated purified mutant SeV. (A) Optimization of SeV virion transcription. In vitro transcription by detergent-activated purified SeV and VSV virions was performed under the same conditions in the presence of [α-32P]UTP and various amounts of virus and purified tubulin. A 1/225 or a 1/15 dilution of a 25-μl transcription reaction mixture was loaded for wt VSV, while the entire product of a 25-μl transcription reaction mixture was loaded for SeV. The mock sample had all reaction mixture components except for virus. The positions of the SeV NP and VSV N mRNAs are indicated. For all reactions, total RNA was purified and analyzed by urea-agarose gel electrophoresis and visualized by autoradiography. (B) Ten micrograms of purified wt or mutant SeV was detergent activated and used for in vitro mRNA synthesis in a 50-μl transcription reaction mixture in the presence of [α-32P]UTP. The mRNA products were purified, separated by urea-agarose gel electrophoresis, and visualized by autoradiography. The position of the SeV NP mRNA is indicated. “% Txn” shows mRNA levels relative to those of wt SeV (100%) using a PhosphorImager and represents the average of two or three experiments where variation was less than 15%. (C) Eighty micrograms of purified wt SeV (lane 1) or mutant SeV (lanes 2 to 5) was detergent activated and used for in vitro mRNA synthesis in a 200-μl transcription reaction mixture in the presence of [3H]AdoMet and with (gray bars) or without (black bars) the addition of AdoHcy. Under all conditions, RNA was purified, additionally separated from nucleotides using gel filtration columns, and used for the measurement of [3H]Met incorporation into mRNA as assayed by binding to DEAE-cellulose paper and scintillation counting. (D) Northern blot analysis to compare viral mRNA levels produced by SeV mutants in the absence of AdoHcy (top) or to compare mRNA levels produced with and without AdoHcy (bottom). For Northern blotting, 1/10 of the mRNA produced as described (C) was separated in a 1.2% agarose formaldehyde gel system, transferred onto a nylon membrane, and incubated with an RNA probe complementary to the SeV NP gene.
Figure 5B shows a representative gel with [α-32P]UTP-labeled viral mRNA produced by detergent-activated purified wt and mutant SeV virions (all purified viruses were tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, confirming that similar amounts of virus were used for each reaction) (data not shown). We did not observe any dramatic reduction in mRNA synthesis for most mutants, although r1782A produced about 60% less mRNA than did the rWT. Interestingly, the L-1782A protein produced viral mRNA levels similar to those of L-WT in the VV-T7-based in vitro transcription system. The decrease in mRNA synthesis by purified r1782A virions could be a result of a partial loss of virion activity, as r1782A virus was collected for purification 5 days p.i. due to its slow growth (compare to 2 to 3 days p.i. for the rWT and other mutants). Interestingly, r1804A, r1806A, and r1804A/1806A showed a slight increase (10 to 20%) in viral mRNA synthesis compared to that of the rWT (Fig. 5B). Next, we tested all our mutants for their abilities to methylate cap structures using wt rVSV and rVSV hr1 viruses as positive and negative controls for mRNA cap methylation. For this assay, in vitro transcription by detergent-activated purified virions was conducted with cold nucleoside triphosphates and [3H]AdoMet (methyl group donor) in the presence or absence of AdoHcy, a competitive inhibitor of AdoMet-dependent MTases. The total RNA was purified and used for measurement of [3H]Met incorporation into mRNA as assayed by binding to DEAE-cellulose paper and scintillation counting. As shown in Fig. 5C, most SeV virions tested produced similar mRNA methylation patterns, with cap methylation being completely abolished in the presence of AdoHcy, as in the case of wt VSV. The only SeV mutant that showed no detectable methylation in the absence of AdoHcy (as for the VSV hr1 mutant) was SeV r1782A, which is consistent with its hr phenotype. Interestingly, the SeV r1805A mutant always showed an intermediate level of cap methylation (Fig. 5C), with about a 60% reduction in [3H]Met incorporation into viral mRNA. To test whether no mRNA methylation in r1782A and a decrease in cap methylation in r1805A were results of the inhibition of MTase activity rather than decreased mRNA synthesis, a portion of the total mRNA produced with [3H]AdoMet (Fig. 5C) was examined by Northern blot analysis using a specific riboprobe against the SeV NP gene. As shown in Fig. 5D, most SeV mutants produced mRNA levels similar to those of the rWT (with about 40% for r1782A), further indicating that K1782A and E1805A mutations specifically affected mRNA cap methylation rather than viral mRNA synthesis.
DISCUSSION
In this study, we conducted site-directed mutagenesis of the SeV L polymerase protein by targeting several amino acid residues within domain VI homologous to those previously shown to be important for mRNA cap methylation in VSV (16, 17, 29, 30). The present study is the first mutagenic analysis of L protein domain VI conducted for any members of the Mononegavirales other than VSV. In addition to VSV L protein domain VI, our previous study identified a new region between VSV L aa 1450 and 1481 that was critical for mRNA cap methylation (16, 17). However, we did not find any significant homology between rhabdo- and paramyxoviruses in this variable region between conserved domains V and VI, and in that initial study, we targeted only those amino acids that were homologous between VSV and SeV. Therefore, we generated six mutant SeV L genes, K1782A, G1804A, E1805A, E1805V, G1806A, and a double mutant, G1804A/G1806A; in addition, we made the L-ΔVI mutant, with a deletion of the entire domain VI (7).
When these mutant proteins were tested for their abilities to synthesize mRNA using a VV-T7 expression system, we found that while most mutants retained normal RNA polymerase activity, two mutants, L-E1805V (but not L-E1805A) and L-ΔVI, were completely inactive. The loss of activity in L-ΔVI was not surprising, as even a smaller deletion within domain VI abolished RNA synthesis by the VSV L protein as a result of an inability of this mutant to form the P-L complex required for normal L RNA polymerase activity (7). Unlike mRNA capping, which is tightly coupled with mRNA transcription, viral mRNA synthesis proceeds with similar efficiencies in the absence or presence of the methyl group donor AdoMet (2), and therefore, cap methylation is not required for mRNA synthesis in members of the Mononegavirales. Thus, we think that the inactivation of L-ΔVI transcription had no relation to the cap methylation function of this protein but that the deletion negatively affected the overall conformation of the L protein, resulting in a defect in P protein binding (7) or other functions important for normal L protein RNA polymerization activity. The inactivation of the L-E1805V protein was more surprising, as a similar substitution in the VSV hr1 mutant (D1671V), while abolishing cap methylation, had no effect on VSV mRNA synthesis (17). It is likely that the amino acid substitution in the L-E1805V protein negatively affected L protein folding, resulting in a complete inactivation of this protein; however, we were unable to yet reveal the specific defect of this mutant protein.
All mutations that passed the in vitro transcription test were cloned into the SeV full-length infectious cDNA plasmid, and recombinant infectious viruses r1782A, r1804A, r1805A, r1806A, and rG1804A/G1806A were successfully recovered and characterized. We conducted phenotypic and biochemical analyses of these mutants using wt and hr1 recombinant VSV viruses as convenient positive and negative controls, respectively, for the hr and ts virus growth phenotypes and mRNA cap methylation activities.
First, we conducted a phenotypic analysis of SeV mutants by testing their relative growth at 34°C in Vero cells against HEp-2 cells by virus titration or by multistep growth kinetic analysis using these cell lines. Previous studies linked the inability of VSV cap methylation-defective mutants to grow in HEp-2 cells to a viral defect in mRNA cap guanine-N7 methylation and the consequent nontranslatability of primary VSV transcripts (16, 17, 24, 25). It was also suggested that host cells methylate viral mRNA in permissive cell lines through an unknown mechanism (24). It should be noted that the VSV hr1 mutant, while unable to grow in HEp-2 cells, was also attenuated in Vero cells (titer on Vero cells of 2.4 × 109 CIU/ml for wt VSV, compared to 6.0 × 107 CIU/ml for the VSV hr1 mutant). Therefore, while a Vero/HEp-2 titer ratio could serve as a good indicator of a possible defect in cap methylation (e.g., for the VSV hr1 mutant), we expected an attenuation of cap methylation-defective SeV mutants in both Vero and HEp-2 cells. In agreement with a possible role of lysine 1782 as an active site of the L protein MTase domain, the SeV r1782A mutant was attenuated in both Vero and HEp-2 cells and showed an hr phenotype, with a Vero/HEp-2 titer ratio of 100 (compared to 2.3 for the rWT). Given the importance of the glycine-rich motif in VSV and other MTases and the homology of the substituted amino acids in SeV to those shown to be critical for guanine-N7 methylation in VSV (16, 17, 29, 30), we expected that all other tested SeV mutants would also be hr restricted. To our surprise, most of mutant viruses with amino acid substitutions in the glycine-rich motif grew normally not only in Vero but also in HEp-2 cells, indicating that they were not defective in cap methylation. r1805A displayed slow growth in both Vero and HEp-2 cells, with about a 24-h delay in infectious focus formation and noticeably smaller foci on both Vero and HEp2 cells. However, we did not observe differences in the relative ability of r1805A (or r1805A-GFP) to grow on HEp-2 cells versus that on Vero cells by CIU counts, indicating that r1805A retained at least some MTase activity.
We also tested SeV mutants for their temperature sensitivity in Vero cells at 34°C against that in Vero cells at 40°C. Although the ts phenotype alone could not indicate whether SeV mutations affected viral MTase activities, our previously tested VSV mutants defective in cap methylation were also ts (16, 17). Again, most of the tested SeV mutants were not ts, with the exception of r1782A and r1805A, displaying a clear ts phenotype similar to that of the VSV hr1 mutant. Together, our phenotypic analysis of recombinant SeV mutants identified only one mutant, r1782A, that behaved similarly to the VSV hr1 mutant (hr and ts), indicating that all other tested SeV L mutants retain at least some cap methylation function.
To directly test SeV mutants for their MTase functions, we conducted mRNA cap methylation analyses using an in vitro transcription assay with detergent-activated SeV virions and tested viral mRNA products for the presence of methyl groups. Unfortunately, in contrast to our previous VSV studies (16, 17), we were unable to conduct a very detailed analysis of the SeV cap structure because of very low levels of viral mRNA produced in vitro (about 200-fold less viral mRNA than that for the VSV in vitro transcription system). Nevertheless, our assays (supported by the above-described virus growth analysis) allowed us to make general conclusions about cap methylation function in all tested SeV mutants. Thus, consistent with the above-described phenotypic analyses, the r1782A mutant was completely defective in cap methylation, while r1805A displayed about a 60% decrease in cap methylation. Our data are the first to experimentally support previously reported computational predictions (5, 13), suggesting the importance of the invariant lysine (position 1782 in the SeV L protein) and the glycine-rich motif in different members of the Mononegavirales.
The invariant lysine (L positions 1782 in SeV and 1651 in VSV) is conserved in most members of the Mononegavirales and was predicted to be the first lysine within the so-called K-D-K-E tetrad catalyzing an SN2 reaction-mediated 2′-O methyl transfer in 2′-O MTases (12, 19, 23). In West Nile virus (WNV) (a flavivirus), a similar substitution of K61A (K61 is a putative functional analog of K1782 in SeV L) in the NS5 protein, which also carries both guanine-N7 and ribose 2′-O MTase activities, specifically inhibited 2′-O cap methylation (41, 51). In contrast, the K1782A mutation in SeV L (this study) and the previously analyzed VSV K1651A (homologous to SeV K1782A) substitutions abolished both guanine-N7 and 2′-O methylation (29). This discrepancy between WNV and members of the Mononegavirales can be explained by the different order of cap methylation previously shown for flaviviruses (GpppA→m7GpppA→m7GpppAm) (51) and previously proposed for VSV (GpppA→GpppAm→m7GpppAm) (30, 45). While the inactivation of 2′-O methylation by the substitution of the catalytic lysine (K61) could not affect guanine-N7 methylation in WNV due to the order of cap methylation (41, 51), it prevented guanine-N7 methylation in VSV (30) and in SeV (K1782A mutation in this study), suggesting that paramyxoviruses may use the same order of cap methylation as VSV. It is important that that the cap methylation order for members of the Mononegavirales is still controversial, with some evidence pointing to both orders. The in vitro results using detergent-activated VSV (Indiana strain) virions proposed the following order of MTase reactions: GpppA + AdoMet (low concentration)→GpppAm + AdoMet (high concentration)→7mGpppAm (30, 45). However, the previously reported in vivo data for VSV (Indiana strain) mRNA synthesis in the presence of the methylation inhibitor cycloleucine (36) and in vitro transcription data for the VSV New Jersey serotype (20) suggest that the reverse order of VSV mRNA methylation (GpppA→7mGpppA→7mGpppAm) can also occur. Moreover, a previously reported study showed that SeV produces mRNAs methylated at both the guanine-N7 and 2′-O-adenosine positions or at guanine-N7 only (m7GpppA) but did not detect any mRNAs methylated only at the 2′-O-adenosine position (44). Finally, a previous study showed that Newcastle disease virus (NDV), another paramyxovirus, produces viral mRNAs that are not 2′-O methylated at all (9).
While our results show clear similarities between VSV K1651A and SeV K1782A mutants (both completely defective in cap methylation), the amino acid substitutions in the L protein glycine-rich motif had milder (E1805A) or nonsignificant (G1804A and G1806A) effects on viral mRNA cap methylation. Generally, this region, especially the second glycine residue (G1806 in SeV L), is sensitive to amino acid substitutions, as demonstrated for VSV and many other known AdoMet-dependent MTases (34), including cap mRNA MTases of vaccinia virus (33, 43), reovirus (31), and eukaryotic cells (47, 50). The previously characterized VSV mutants with homologous changes in the glycine-rich motif showed the following cap methylation phenotypes: (i) <1 to 20% for guanine-N7 (depending on in vitro conditions) and about 40% overall methylation for the VSV G1670A mutant (homologous to the SeV G1804A mutant) compared to wt VSV (30), (ii) <1% for guanine-N7 and 2′-O methylation for the VSV D1671V mutant (similar to the E1805V [not rescued] and E1805A mutants) compared to wt VSV (17, 30), and (iii) <1 to 20% for guanine-N7 (depending on in vitro conditions) and about 40% overall for the VSV G1672A mutant (similar to the SeV G1806A mutant) compared to wt VSV (16, 30). Although the VSV G1670A and G1672A mutants retained a substantial 2′-O MTase activity, they were severely inhibited in their guanine-N7 MTase activity (30). Importantly, under in vitro conditions similar to those utilized in this study, these VSV mutants showed no detectable guanine-N7 methylation, while the SeV G1804A and G1806A mutants did not significantly affect guanine-N7 or 2′-O cap methylation. The tolerance of the SeV L protein to the amino acid substitutions G1804A and G1806A is also supported by the fact that the double substitution G1804A/G1806A had little effect on virus growth or mRNA methylation in vitro. Interestingly, we found that while both r1804A and r1806A had normal mRNA synthesis and cap methylation, they behaved very unusually during multistep growth in HEp-2 cells, with r1804A growing considerably faster than the rWT and r1806A growing considerably slower than the rWT. The presence of both mutations in r1804A/1806A produced an intermediate-growth phenotype, suggesting that these mutations had a reciprocal effect when present together. Further experiments are needed to elucidate the molecular basis for the differences between r1804A and r1806A in HEp-2 cells.
While we still do not understand the exact mechanism of the hr restriction of cap methylation mutants of VSV, the hr analysis of SeV mutants justifies the future use of this approach as a supporting assay to determine the cap methylation status of SeV mutants in addition to the direct cap methylation analysis. Thus, although none of the tested SeV mutants showed a Vero/HEp-2 titer ratio as dramatic as that for the VSV hr1 mutant (more than 60,000), the only SeV mutant with asymmetric attenuation in HEp-2 cells was the rK1782A virus (with a Vero/HEp-2 ratio of 100), and this mutant was also completely defective in cap methylation. The only other SeV mutant attenuated in HEp-2 cells (although equally in Vero cells) was r1805A, which also showed a 60% reduction in cap methylation. Therefore, while the ability of a mutant (VSV or SeV) to grow in HEp-2 cells may not be sufficient by itself to determine the methylation status of viral mutants, our data for SeV r1782A and r1805A mutants show that this assay can be successfully used to complement an in vitro cap methylation analysis of SeV mutants.
We propose two main theories to explain the tolerance of the SeV L protein to the G1804A, G1806A, and G1804A/1806A substitutions. First, it is possible that, despite a homology between VSV and SeV at the glycine-rich motif, this region is not an AdoMet-binding site in SeV and possibly other paramyxoviruses and that an actual AdoMet-binding site could be located at a different position in the SeV L protein. Importantly, even in VSV, a putative role of this motif as an AdoMet-binding site was postulated based on computational predictions and site-directed mutagenesis studies, but no experimental biochemical data are available to date for the L protein of any member of the Mononegavirales directly demonstrating that this or any other L region actually binds AdoMet. While different AdoMet-binding site locations in SeV and VSV are a possibility, the complete inactivation of L cap methylation by K1782A (the first lysine of the catalytic K-D-K-E tetrad in 2′-O MTases is generally positioned upstream and in a close proximity to the AdoMet-binding site) and about a 60% decrease in methylation by E1805A support another hypothesis, that while the glycine-rich motif is likely to be the SeV L protein AdoMet-binding site, SeV and possibly other paramyxoviruses are far more flexible to the amino acid substitutions in this motif than VSV. Such tolerance may explain why while most members of the Mononegavirales, including VSV and SeV, have the motif G(D/E)G(S/A)G (glycines important for VSV cap methylation are underlined) (Fig. 1), more variation in this motif can be found in the members of the family Paramyxoviridae, especially in the genera Rubulavirus (subfamily Paramyxovirinae) and Avulavirus (subfamily Pneumovirinae), which have the motif AEG(S/A)G, very similar to the sequence for our mutant SeV r1804A (AEGAG). Interestingly, the avian pneumovirus L protein has a motif, AEASG, which is similar to that of our SeV double mutant r1804A/1806A (AEAAG), which had a wt growth phenotype and a normal mRNA cap methylation pattern. Previously, Li et al. (30) speculated that these differences at the glycine-rich motif between NDV (genus Avulavirus) and VSV may account for the differences of these viruses in cap methylation pattern (the NDV caps are not 2′-O methylated at all) (9). However, our data suggest that these sequence variations among paramyxoviruses reflect their tolerance to amino acid substitutions and are not functionally important, as r1804A, r1806A, and r1804A/1806A mutants displayed a normal cap methylation pattern.
Although we did not find dramatic differences between SeV r1804A, r1806A, r1804A/1806A, and the rWT using our experimental conditions, the wt glycine-rich motif sequence may be beneficial during normal viral infection, and we believe that there must be some evolutionary basis for the sequence conservation of this motif among paramyxoviruses and members of the Mononegavirales in general. Hence, we are planning to conduct a comparative pathogenesis study with mice (which are a natural host for SeV) to determine the role of the mutated amino acid residues during normal infection and thus to understand why the glycine-rich motif is conserved in paramyxoviruses (and other members of the Mononegavirales) if it can be mutated without serious consequences to virus fitness. In addition to a better understanding of the biology of these viruses, these experiments would have important practical implications because targeting amino acid residues critical for cap MTase function in VSV, SeV, and other members of the Mononegavirales could be used to rationally attenuate these viruses (or manipulate their hr) for the development of live attenuated viruses and their use as vaccine (6), oncolytic (46), and gene therapy (14) vectors.
Acknowledgments
We are grateful to Sue Moyer (University of Florida College of Medicine) for providing materials and important advice for this project. We thank Wolfgang J. Neubert (Max Planck Institute of Biochemistry, Germany) for a kind gift of plasmid pRSIdeFmut for SeV-GFP virus recovery. We greatly acknowledge the excellent technical assistance of Sherin Smallwood (University of Florida College of Medicine) at the initial stages of this project. We also thank Megan Moerdyk-Schauwecker for critical comments on the manuscript.
This work was supported by NIH grant 1R15GM084422-01 (to V.Z.G.).
Footnotes
Published ahead of print on 3 December 2008.
REFERENCES
- 1.Abraham, G., D. P. Rhodes, and A. K. Banerjee. 1975. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell 551-58. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, G., D. P. Rhodes, and A. K. Banerjee. 1975. Novel initiation of RNA synthesis in vitro by vesicular stomatitis virus. Nature 25537-40. [DOI] [PubMed] [Google Scholar]
- 3.Barik, S. 1993. The structure of the 5′ terminal cap of the respiratory syncytial virus mRNA. J. Gen. Virol. 74485-490. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bujnicki, J. M., and L. Rychlewski. 2002. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. 15101-108. [DOI] [PubMed] [Google Scholar]
- 6.Bukreyev, A., M. H. Skiadopoulos, B. R. Murphy, and P. L. Collins. 2006. Nonsegmented negative-strand viruses as vaccine vectors. J. Virol. 8010293-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canter, D. M., and J. Perrault. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219376-386. [DOI] [PubMed] [Google Scholar]
- 8.Chandrika, R., S. M. Horikami, S. Smallwood, and S. A. Moyer. 1995. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213352-363. [DOI] [PubMed] [Google Scholar]
- 9.Colonno, R. J., and H. O. Stone. 1976. Newcastle disease virus mRNA lacks 2′-O-methylated nucleotides. Nature 261611-614. [DOI] [PubMed] [Google Scholar]
- 10.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 103079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De, B. P., A. Lesoon, and A. K. Banerjee. 1991. Human parainfluenza virus type 3 transcription in vitro: role of cellular actin in mRNA synthesis. J. Virol. 653268-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 212757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferron, F., S. Longhi, B. Henrissat, and B. Canard. 2002. Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 27222-224. [DOI] [PubMed] [Google Scholar]
- 14.Finke, S., and K. K. Conzelmann. 2005. Recombinant rhabdoviruses: vectors for vaccine development and gene therapy. Curr. Top. Microbiol. Immunol. 292165-200. [DOI] [PubMed] [Google Scholar]
- 15.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grdzelishvili, V. Z., S. Smallwood, D. Tower, R. L. Hall, D. M. Hunt, and S. A. Moyer. 2006. Identification of a new region in the vesicular stomatitis virus L polymerase protein which is essential for mRNA cap methylation. Virology 350394-405. [DOI] [PubMed] [Google Scholar]
- 17.Grdzelishvili, V. Z., S. Smallwood, D. Tower, R. L. Hall, D. M. Hunt, and S. A. Moyer. 2005. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 797327-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta, K. C., D. H. Bishop, and P. Roy. 1979. 5′-terminal sequences of spring viremia of carp virus RNA synthesized in vitro. J. Virol. 30735-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hager, J., B. L. Staker, H. Bugl, and U. Jakob. 2002. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 27741978-41986. [DOI] [PubMed] [Google Scholar]
- 20.Hammond, D. C., and J. A. Lesnaw. 1987. The fates of undermethylated mRNA cap structures of vesicular stomatitis virus (New Jersey) during in vitro transcription. Virology 159229-236. [DOI] [PubMed] [Google Scholar]
- 21.Hercyk, N., S. M. Horikami, and S. A. Moyer. 1988. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology 163222-225. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 167351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodel, A. E., P. D. Gershon, and F. A. Quiocho. 1998. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell 1443-447. [DOI] [PubMed] [Google Scholar]
- 24.Horikami, S. M., F. De Ferra, and S. A. Moyer. 1984. Characterization of the infections of permissive and nonpermissive cells by host range mutants of vesicular stomatitis virus defective in RNA methylation. Virology 1381-15. [DOI] [PubMed] [Google Scholar]
- 25.Horikami, S. M., and S. A. Moyer. 1982. Host range mutants of vesicular stomatitis virus defective in in vitro RNA methylation. Proc. Natl. Acad. Sci. USA 797694-7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingrosso, D., A. V. Fowler, J. Bleibaum, and S. Clarke. 1989. Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J. Biol. Chem. 26420131-20139. [PubMed] [Google Scholar]
- 27.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 28.Leyrer, S., W. J. Neubert, and R. Sedlmeier. 1998. Rapid and efficient recovery of Sendai virus from cDNA: factors influencing recombinant virus rescue. J. Virol. Methods 7547-58. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., E. C. Fontaine-Rodriguez, and S. P. Whelan. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 7913373-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, J., J. T. Wang, and S. P. Whelan. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 1038493-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luongo, C. L., C. M. Contreras, D. L. Farsetta, and M. L. Nibert. 1998. Binding site for S-adenosyl-L-methionine in a central region of mammalian reovirus lambda2 protein. Evidence for activities in mRNA cap methylation. J. Biol. Chem. 27323773-23780. [DOI] [PubMed] [Google Scholar]
- 32.Lyles, D. S., and C. E. Rupprecht. 2007. Rhabdoviridae, p. 1363-1408. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 33.Mao, X., and S. Shuman. 1996. Vaccinia virus mRNA (guanine-7-)methyltransferase: mutational effects on cap methylation and AdoHcy-dependent photo-cross-linking of the cap to the methyl acceptor site. Biochemistry 356900-6910. [DOI] [PubMed] [Google Scholar]
- 34.Martin, J. L., and F. M. McMillan. 2002. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12783-793. [DOI] [PubMed] [Google Scholar]
- 35.Mizumoto, K., K. Muroya, T. Takagi, T. Omata-Yamada, H. Shibuta, and K. Iwasaki. 1995. Protein factors required for in vitro transcription of Sendai virus genome. J. Biochem. (Tokyo) 117527-534. [DOI] [PubMed] [Google Scholar]
- 36.Moyer, S. A. 1981. Alteration of the 5′ terminal caps of the mRNAs of vesicular stomatitis virus by cycloleucine in vivo. Virology 112157-168. [DOI] [PubMed] [Google Scholar]
- 37.Moyer, S. A., G. Abraham, R. Adler, and A. K. Banerjee. 1975. Methylated and blocked 5′ termini in vesicular stomatitis virus in vivo mRNAs. Cell 559-67. [DOI] [PubMed] [Google Scholar]
- 38.Moyer, S. A., S. C. Baker, and S. M. Horikami. 1990. Host cell proteins required for measles virus reproduction. J. Gen. Virol. 71775-783. [DOI] [PubMed] [Google Scholar]
- 39.Moyer, S. A., S. C. Baker, and J. L. Lessard. 1986. Tubulin: a factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc. Natl. Acad. Sci. USA 835405-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogino, T., M. Kobayashi, M. Iwama, and K. Mizumoto. 2005. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2804429-4435. [DOI] [PubMed] [Google Scholar]
- 41.Ray, D., A. Shah, M. Tilgner, Y. Guo, Y. Zhao, H. Dong, T. S. Deas, Y. Zhou, H. Li, and P.-Y. Shi. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 808362-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhodes, D. P., and A. K. Banerjee. 1975. 5′-terminal sequence of vesicular stomatitis virus mRNA's synthesized in vitro. J. Virol. 1733-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha, N., S. Shuman, and B. Schwer. 2003. Yeast-based genetic system for functional analysis of poxvirus mRNA cap methyltransferase. J. Virol. 777300-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagi, T., K. Muroya, M. Iwama, T. Shioda, T. Tsukamoto, and K. Mizumoto. 1995. In vitro mRNA synthesis by Sendai virus: isolation and characterization of the transcription initiation complex. J. Biochem. (Tokyo) 118390-396. [DOI] [PubMed] [Google Scholar]
- 45.Testa, D., and A. K. Banerjee. 1977. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 24786-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Messling, V., and R. Cattaneo. 2004. Toward novel vaccines and therapies based on negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283281-312. [DOI] [PubMed] [Google Scholar]
- 47.Wang, S. P., and S. Shuman. 1997. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J. Biol. Chem. 27214683-14689. [DOI] [PubMed] [Google Scholar]
- 48.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 28361-119. [DOI] [PubMed] [Google Scholar]
- 49.Wiegand, M. A., S. Bossow, S. Schlecht, and W. J. Neubert. 2007. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression. J. Virol. 8113835-13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada-Okabe, T., T. Mio, Y. Kashima, M. Matsui, M. Arisawa, and H. Yamada-Okabe. 1999. The Candida albicans gene for mRNA 5-cap methyltransferase: identification of additional residues essential for catalysis. Microbiology 1453023-3033. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, Y., D. Ray, Y. Zhao, H. Dong, S. Ren, Z. Li, Y. Guo, K. A. Bernard, P. Y. Shi, and H. Li. 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 813891-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]