Abstract
The innate immune system guards against virus infection through a variety of mechanisms including mobilization of the host interferon system, which attacks viral products mainly at a posttranscriptional level. The influenza virus NS1 protein is a multifunctional facilitator of virus replication, one of whose actions is to antagonize the interferon response. Since NS1 is required for efficient virus replication, it was reasoned that chemical inhibitors of this protein could be used to further understand virus-host interactions and also serve as potential new antiviral agents. A yeast-based assay was developed to identify compounds that phenotypically suppress NS1 function. Several such compounds exhibited significant activity specifically against influenza A virus in cell culture but had no effect on the replication of another RNA virus, respiratory syncytial virus. Interestingly, cells lacking an interferon response were drug resistant, suggesting that the compounds block interactions between NS1 and the interferon system. Accordingly, the compounds reversed the inhibition of beta interferon mRNA induction during infection, which is known to be caused by NS1. In addition, the compounds blocked the ability of NS1 protein to inhibit double-stranded RNA-dependent activation of a transfected beta interferon promoter construct. The effects of the compounds were specific to NS1, because they had no effect on the ability of the severe acute respiratory syndrome coronavirus papainlike protease protein to block beta interferon promoter activation. These data demonstrate that the function of NS1 can be modulated by chemical inhibitors and that such inhibitors will be useful as probes of biological function and as starting points for clinical drug development.
Influenza is associated with significant morbidity and mortality and is a continuing worldwide public health problem. Seasonal influenza epidemics affect ca. 5 to 15% of the world's population, and estimates of annual mortality range from 250,000 to 500,000 (75), including approximately 30,000 deaths and 200,000 hospitalizations in the United States (68). Groups at high risk include the elderly, the very young, and those suffering from chronic illness. Medical complications include pneumonia and exacerbation of symptoms associated with chronic illness (60).
In the 20th century, three influenza pandemics were recorded—in 1918, 1957, and 1968. The 1918 pandemic was the most severe and was responsible for an estimated 20 to 40 million deaths, including a significant percentage of young adults (58, 67). The epidemiology of transmission and the genetics of the influenza viruses make it likely that additional pandemics will occur due to emergence of new strains, for which the world's healthcare network is not yet prepared (16, 50, 64). In this regard the spread of H5N1 among avian species and sporadic spillage into humans has attracted much attention (48, 51). Whereas this virus has not yet acquired the ability to transmit from person to person, the small number of humans infected by H5N1 due to direct contact with birds has revealed a dangerously high rate of mortality, ca. 60% (1, 16).
Control of seasonal influenza is an ongoing challenge (73). Due to antigenic drift the widely used seasonal vaccine is unevenly effective from year to year, and its use is lower than optimal even in developed countries such as the United States (7, 46). There are currently two classes of anti-influenza virus drugs that have been used effectively in prevention and treatment. These drugs target the viral M2 ion channel (e.g., amantadine) and neuraminidase proteins (e.g., oseltamivir), respectively (25, 44). Despite these successes there remain concerns regarding drug efficacy, resistance, and cost (26).
In light of the continuing threat to public health, the current state of prevention and treatment options, and the likelihood of emergence of a pandemic strain for which the human population is immunologically unprepared, it makes sense to attempt to develop novel antiviral agents that could be used alone or in combination with existing modalities of treatment. Such agents could take advantage of steps in the virus replicative cycle that have not yet been exploited pharmacologically. These agents could also be designed to attack cellular functions that are required to support virus replication or to enhance the host innate or adaptive immune responses. Novel agents that block virus replication could also be used as molecular probes of the biology of the virus, as well as virus-host interactions.
We have explored the use of a novel target for the development of anti-influenza virus compounds, the NS1 protein. NS1 is a nonstructural protein encoded by segment 8 of influenza virus A. Genetic analyses of NS1 have shown that viral replication, spread, and pathogenesis are very dependent on the function of this protein (3, 4, 6, 10, 11, 13, 17, 18, 22, 27, 29, 30, 36, 63, 66, 74). This satisfies an important criterion for an anti-influenza virus target, since drugs that inhibit the action of the target must be able to slow virus production and/or pathogenesis as a consequence. Several interesting functions for NS1 have been described. NS1 is an RNA-binding protein that can interact with a variety of RNA species, including double-stranded RNA (dsRNA) (10, 21, 23, 24, 54-56, 70). Binding of NS1 to dsRNA inhibits the 2-5A oligoadenylase/RNase L pathway for degradation of viral RNAs, blocks the activity of transcription factor pathways that depend on dsRNA, and inhibits activation of cellular PKR, to which NS1 also binds directly (14, 20, 32, 35, 37, 41-43, 52, 65, 71). NS1 also inhibits the induction of RNA interference through its ability to sequester small interfering RNAs (5, 33). NS1 modifies cellular pre-mRNA processing, including 3′-end formation, by binding to the 30-kDa subunit of CPSF (for cleavage and polyadenylation specificity factor) and binding to poly(A)-binding protein II (31). It also inhibits cellular pre-mRNA splicing (69) and blocks nuclear RNA export by associating with several cellular proteins that mediate RNA export (15, 55, 61). Some of the functions of NS1 serve to inhibit the host antiviral response that is mediated by interferon (IFN) (18; reviewed in references 14 and 31). For instance, NS1 blocks induction of IFN gene transcription and mRNA maturation, thereby preventing the cell from mounting an efficient innate defense (47, 65, 71). Other functions of NS1 act to inhibit host cell gene expression so as to favor viral gene expression, such as effects on cellular RNA metabolism and export.
NS1 is a functionally complex protein and is a central player in the virus's response to host defense mechanisms and the establishment of efficient viral gene expression. Because of its importance to virus replication and virus-host interactions and the fact that it is highly conserved across influenza virus A strains (19, 62), NS1 seems a particularly good target for drug discovery. We report here the identification of chemical compounds that inhibit both NS1 function and influenza virus replication.
MATERIALS AND METHODS
Mammalian cells and viruses.
Vero E6, MA104, and 293 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. MDCK cells were maintained in Iscove medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine. All media and sera were from Invitrogen. For infections, viral stocks were diluted in growth medium supplemented with 0.3% bovine serum albumin, 0.22% sodium bicarbonate, and 0.25 U of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (Invitrogen)/ml. Influenza viruses A/PR/8/34 (PR), A/WSN/33 (WSN), and A/Tx/36/91 (TX) were gifts from Adolfo Garcia-Sastre, and A/HK/19/68 (HK) was a gift from Tom Braciale. The viruses were propagated in 10-day-old embryonated chicken eggs at 37°C. Mutant delNS1 (a gift from Adolfo Garcia-Sastre) was propagated in MDCK cells containing a stably transfected NS1PR gene (a gift from Luis Martinez-Sobrido and Adolfo Garcia-Sastre). The titered stock of respiratory syncytial virus (RSV) strain RSΔsh (49) was a gift from Gail Wertz. Titers of influenza virus stocks were determined by 50% tissue culture infective dose (TCID50) analysis on MDCK cells using the hemagglutination assay protocol of Reed and Muench (57).
Plasmids.
A full-length NS1 cDNA from A/WSN/33 (pCAGGS-NS1, a gift from Peter Palese) was used to PCR amplify NS1 for cloning into the galactose-inducible yeast expression vector pYES2 (Invitrogen) to create pYES-NS1. cDNAs encoding NS1 from A/Tx/36/91 and A/PR/8/34 under the control of chicken β-actin promoter [pCAGGS-NS1(Tx/91) and pCAGGS-NS1(PR/34)] and a reporter plasmid encoding firefly luciferase under the control of IFN-β promoter (p125-Luc [76]) were kindly provided by Adolfo Garcia-Sastre. Severe acute respiratory syndrome (SARS) papainlike protease (PLP) was expressed from a CAGGS plasmid.
Yeast strains and growth.
Strain 9526-6-2 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pdr1::KanMX4 pdr3::KanMX4) was a gift from Dan Burke. It was derived by tetrad dissection from two parent strains that had been modified by one step gene replacements. The pdr1::KanMX4 was constructed in BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the pdr3::KanMX4 was constructed in BY4742 (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). PCR-mediated one-step gene replacements, matings and tetrad dissections were performed as described previously (2). Strains 9526-6-2/pYES2 and 9526-6-2/pYES-NS1 were generated by transformation of 9526-6-2 with the plasmids pYES2 and pYES-NS1, respectively, and were maintained on synthetic complete medium (SC) lacking uracil. For growth experiments and library screening, a single transformed colony was grown overnight, and the cell number was determined by using a Coulter counter (Beckman Coulter Corp.). The cells were diluted to 5 × 105 cells/ml in SC lacking uracil and containing raffinose and 2% galactose. A 95-μl portion of this culture was added to 5 μl of preplated test compounds in 96-well plates such that the final drug concentration was 50 μM and the final dimethyl sulfoxide (DMSO) concentration was 1%. The Diversity Set library (National Cancer Institute Developmental Therapeutics Program) was used for the drug screen. It was provided as 10 mM stocks in 100% DMSO. Optical density readings at 600 nm (OD600) were taken every 12 h for 60 h using a Thermomax microplate reader (Molecular Devices).
Virus replication assays.
Confluent cell monolayers were infected at a multiplicity of infection (MOI) of 0.1 for 48 h in the presence or absence of drug. Compounds were added at the beginning of infection and were present throughout the infection. After 48 h virus titers were determined by TCID50 analysis as described previously (57). Growth of RSΔsh was quantified by TCID50 analysis by scoring green fluorescent protein (GFP) fluorescence at 4 days postinfection.
Reverse transcriptase PCR (RT-PCR).
MDCK cells were infected for 6 h with PR or delNS1 at an MOI of 2.0 in the presence or absence of drug, and the total RNA was isolated by using RNeasy (Qiagen). For first-strand cDNA synthesis, 2 μg of total RNA was primed with random nanomers (New England Biolabs) at a final concentration of 2 μM. Reverse transcription was performed with 10 U of Moloney murine leukemia virus RT (New England Biolabs)/μl in the presence of 1 U of RNase inhibitor/μl. Thereafter, 1/20 volume of cDNA was used as a template for PCR (30 cycles). The following primer pairs were used: canine IFN-β (accession no. XM538679), CCAGTTCCAGAAGGAGGACA and CCTGTTGTCCCAGGTGAAGT; NS1 from A/PR/8/34 (accession no. J02150), CTTCGCCGAGATCAGAAATC and TGGACCATTCCCTTGACATT; M2 from A/PR/8/34 (accession no. V01099), ATGATCTTCTTGAAAATTTGC and CTCCAGCTCTATGCTGAC; and canine β-actin (accession no. XM536230), GGCATCCTGACCCTGAAGTA and GGGGTGTTGAAAGTCTCGAA.
Luciferase reporter assay.
293 cells were transfected with 0.2 μg of p125Luc and 0.8 μg of pCAGGS-NS1(Tx/91) or pCAGGS-NS1(PR/34) using Polyfect transfection reagent (Qiagen). At 16 h posttransfection the cells were stimulated with 50 μg of poly[IC] (Sigma-Aldrich)/ml and treated with 50 μM drug for 24 h. The cells were lysed with 1× passive lysis buffer (Promega), and 20 μl of the lysate was used to measure luciferase activity using the luciferase assay system (Promega) according to the manufacturer's protocol. For SARS-CoV PLP experiments, the luciferase plasmid alone or together with a SARS PLP-expressing plasmid (100 ng/well each plasmid) was transfected into 293T cells in triplicate using Fugene6 (Roche). At 6 h postinfection, the cells were treated with each drug and allowed to incubate for an additional 18 h. At 24 h posttransfection, 500 ng of poly[IC]/well was transfected into cells by using Lipofectamine (Invitrogen) to induce IFN-β gene transcription. At 6 h after poly[IC] addition, the cells were lysed in Easy-Glo lysis reagent and quantitated. All transfection experiments were conducted in triplicate.
Protein labeling and Western blot analysis.
Confluent monolayers of MDCK cells in 35-mm plates were infected at an MOI of 0.1 with A/HK/19/68 in the presence or absence of 50 μM compounds. Compounds were added at the beginning of infection and were present throughout the infection. The cells were labeled with [35S]methionine and [35S]cysteine for the final hour of infection. After 24 h the medium was replaced with Dulbecco modified Eagle medium lacking l-cysteine and l-methionine (Sigma-Aldrich) and containing 10% fetal bovine serum. After 30 min of starvation the cells were labeled for 1 h with 25 mCi of Express35S35S protein labeling mix (Perkin-Elmer)/ml. Labeled cells were washed with phosphate-buffered saline and lysed using passive lysis buffer (Promega). The lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography using a Storm Phosphorimager (GE Healthcare Life Sciences).
For analyzing NS1 expression in yeast, cells transformed with pYES-NS1 were grown in SC containing raffinose and lacking uracil to an OD600 of 0.6 to 0.7. The cells were then induced by replacing the medium with the identical medium but containing 2% galactose, in the presence or absence of drug. Lysates were prepared after 8 h. A total of 20 μg of each sample was resolved by SDS-PAGE, followed by Western blotting and probing with a 1:1,000 dilution of rabbit polyclonal antiserum α-NS1 (a gift from Peter Palese). To detect the expression levels of NS1 in transfected cells, 293 cells were lysed by using passive lysis buffer and subjected to SDS-PAGE and Western blotting. The blots were probed with α-NS1 at a dilution of 1:1,000. As a loading control, the blots were also probed with a 1:2,000 dilution of monoclonal α-tubulin antibody (Sigma-Aldrich).
Cytotoxicity assay.
To determine the cell viability, a trypan blue dye exclusion test was used. MDCK-UK, VeroE6, 293, and MA104 cells at 3 × 105 cells/ml were seeded in the presence or absence of increasing concentrations of compounds and incubated for 48 h. Aliquots of trypsin-treated cells were mixed with an equal volume of phosphate-buffered saline containing 0.4% trypan blue. Cells that excluded the dye were counted by using a hemacytometer. The experiment was performed in duplicate.
RESULTS
Screen for compounds that inhibit NS1 function.
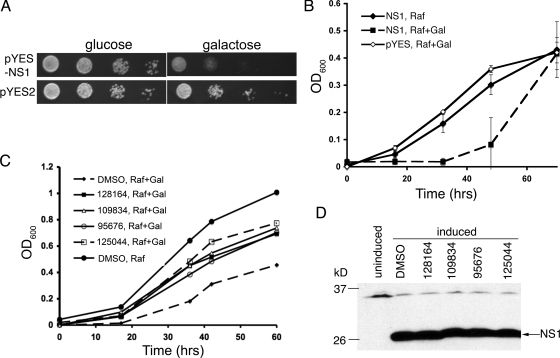
Ward et al. demonstrated that Saccharomyces cerevisiae strains expressing NS1 protein from influenza virus A exhibit a pronounced slow-growth phenotype (72). We investigated whether an NS1- expressing yeast strain could be used to screen for small molecules that suppress the slow-growth phenotype by inhibition of NS1 function. A test strain was generated that carries null alleles for two genes that control drug efflux, PDR1 and PDR3, thus allowing efficient retention of small molecules (59). This in turn was used to create a strain expressing NS1 from A/WSN/33 under the control of the GAL1 promoter, which is inducible by galactose. Shown in Fig. 1A are spot tests of the control and NS1-expressing strains. Growth on medium containing galactose resulted in strong growth inhibition of the NS1-expressing strain but not of the control strain, whereas there was no difference between the two strains on glucose-containing medium.
FIG. 1.
Assay development and screening results. (A) Tenfold serial dilutions (left to right) of strains transformed with the indicated expression plasmids were spotted on plates containing either glucose or galactose as the sole carbon source, and the plates were incubated for 3 days at 28°C. (B) Next, 100 μl cultures of the indicated strains at 5 × 105 cells/ml were incubated at 28°C for the times shown. The OD was measured by using a 96-well plate reader. Values shown are with blank (medium alone) subtracted. NS1, 9526-6-2/pYES-NS1; pYES, 9526-6-2/pYES2; Raf, raffinose; Gal, galactose. (C) Strain 9526-6-2/pYES-NS1 was grown in medium containing raffinose plus 2% galactose in the presence of 50 μM drug or 1% DMSO for the indicated times at 28°C. The final DMSO concentration in the drug-treated cultures was also 1%. (D) Western blot of whole-cell lysates of 9526-6-2/pYES-NS1 grown in the absence (uninduced) or presence (induced) of 2% galactose and either 1% DMSO or the indicated compounds at 50 μM for 8 h. The lysates were separated by SDS-PAGE and blotted for the presence of NS1. The band near the 37-kDa marker is nonspecific.
Time course experiments were performed using a 96-well format to establish conditions for the drug screen. Cells were plated at 5 × 105 cells/ml in either medium containing raffinose as the sole carbon source or medium containing raffinose plus 2% galactose to induce the expression of NS1. Figure 1B shows growth curves for the control and NS1-expressing strains under these conditions over a 68-h period. The NS1-expressing strain grew significantly more slowly than the control strain in the presence of galactose. These data indicated a time period between 36 and 48 h, during which a significant growth differential could be exploited in a screen for small molecules that suppress the slow-growth phenotype.
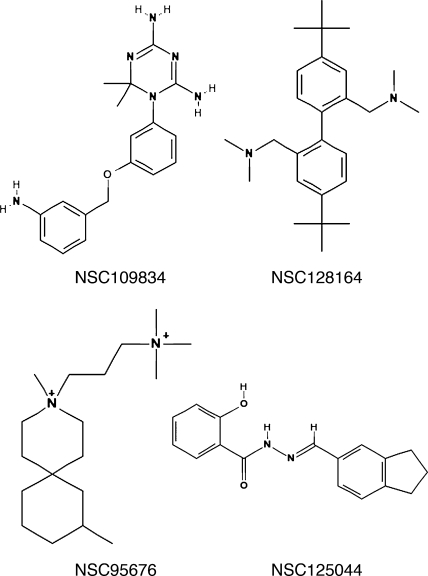
To perform the screen, cells from an overnight culture were plated at 5 × 105 cells/ml in galactose-containing medium in the presence 50 μM drug or 1% DMSO as a control. Approximately 2,000 compounds from the National Cancer Institute Diversity Set library (see http://dtp.nci.nih.gov/branches/dscb/diversity_explanation.html) were screened manually. Optical density readings were taken every 12 h for 60 h. This screen resulted in 15 hits, 9 of which proved to be reproducible when independent samples were obtained from the NCI. The nine hits produced a ≥1.5-fold increase in OD over at least two consecutive OD readings during the time course (data not shown). Of these, four were studied further based on their ability to inhibit influenza virus replication (see below). Of the remaining five, two were toxic to MDCK cells and three showed no activity against influenza virus replication (not shown). Shown in Fig. 1C are individual growth curves for galactose-containing cultures of the NS1-expressing strain treated with each of the positive compounds. Their structures are shown in Fig. 2.
FIG. 2.
Chemical structures of anti-influenza virus compounds.
One possible mechanism for suppression of the NS1-induced growth defect in yeast could be a decrease in NS1 protein expression triggered by addition of the drugs. For example, this could be due to drug effects on transcription from the GAL1 promoter, on plasmid replication or metabolism, or on stability of NS1 RNA or protein. To investigate whether any of the compounds had such an effect, yeast cells expressing NS1 were grown in the presence of drug for 8 h and analyzed for protein expression by Western blotting. As shown in Fig. 1D, none of the compounds caused a change in the level of NS1 protein. These data indicate that in yeast, the positive compounds from the screen act either at the level of NS1 function itself to suppress the slow-growth phenotype, or possibly on cellular functions that specifically modify or bypass NS1 function without altering its expression.
Effects on influenza virus replication.
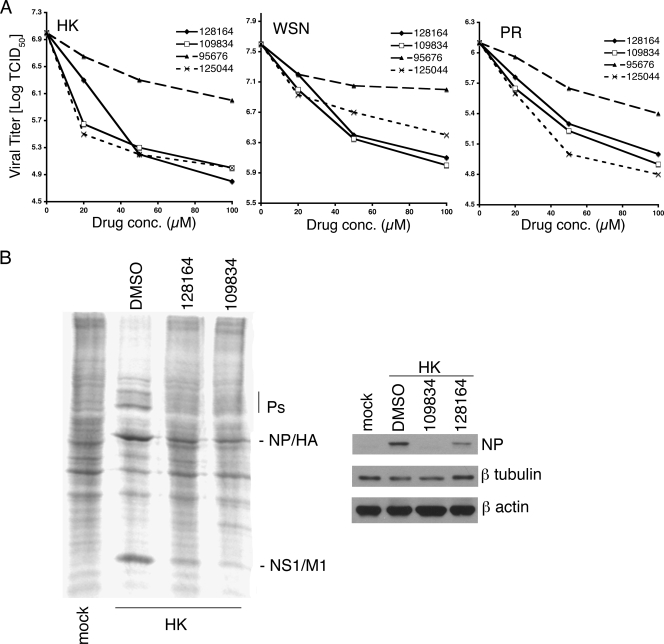
To test the effects of the positive compounds on influenza virus replication, each was used to challenge virus replication assays for A/Hong Kong/19/68 (HK), A/WSN/33 (WSN), and A/PR/8 (PR). MDCK cells were infected at an MOI of 0.1 and incubated for 48 h in the presence of drug or 1% DMSO as control, followed by determination of TCID50 (57). As shown in Fig. 3A, all three viral strains were sensitive to each of the four compounds, with varying overall sensitivity and concentration dependence. The greatest inhibition observed was ∼100-fold for virus HK in cells treated with NSC109834, NSC128164, or NSC125044. A >10-fold inhibition of HK was observed with as little as 10 μM NSC109834 or NSC125044. The 50% inhibitory concentration for inhibition of virus replication was calculated for each compound and is presented in Table 1. Also presented in Table 1 are the selective index (SI) values for each compound.
FIG. 3.
Inhibition of virus replication in MDCK cells. (A) Cells were infected with the indicated viruses at an MOI of 0.1 and treated with drug or 1% DMSO (shown as 0 μM) starting 1 h postinfection. The DMSO concentration for all drug-treated cultures was 1%. After incubation for 48 h, the supernatants were collected and analyzed for TCID50 by the method of Reed and Muench (57). (B) Protein expression in infected cells. In the left panel, MDCK cells were infected with A/HK/19/68 at an MOI of 0.1 for 24 h, in the presence or absence of the indicated compounds at 50 μM. The cells were labeled with [35S]methionine and [35S]cysteine for the final hour of infection. Cell lysates were analyzed by SDS-PAGE and visualized by using a phosphorimager. In the right panel, infected cells were analyzed for total NP protein expression by Western blotting. Blots were also probed for β-tubulin and β-actin as loading controls.
TABLE 1.
50% Inhibitory concentrations and selective indexes of anti-NS1 compounds on different influenza strains cultured in MDCK-UK cellsa
| Anti-NS1 compound | IC50 (μM)/SI
|
||
|---|---|---|---|
| PR | HK | WSN | |
| NSC128164 | 14/16.6 | 5/44.4 | 13/18.5 |
| NSC109834 | 10/32.3 | 2/200.3 | 12/28.2 |
| NSC95676 | 12/104.9 | 20/62.8 | 19/64.5 |
| NSC125044 | 11/12.4 | 7/18.9 | 12/11.8 |
SI = CC50/IC50. The 50% inhibitory concentration (IC50) of the compounds was calculated by interpolation of the dose-response curves shown in Fig. 3A.
To determine the effect of NSC109834 and NSC128164 on viral protein synthesis, cells were infected with HK at an MOI of 0.1 and incubated in the presence of these compounds for 24 h. Protein synthesis was monitored by incorporation of [35S]methionine and [35S]cysteine during the last 45 min of infection, followed by analysis using SDS-PAGE. Figure 3B (left) shows that both compounds significantly affected the level of synthesis of the major viral proteins. Similar assays of infected cells treated with NSC125044 or NSC95676 showed that NSC125044 resulted in a decrease in viral protein synthesis but that NSC96575 produced little or no effect (data not shown). In addition, Western blot analysis was performed on lysates from infected cells. The right panel of Fig. 3B shows that the level of viral NP protein was inhibited in cells treated with NSC109834 and NSC128164. Since NS1 is known to inhibit the cellular IFN response, the observed decrease in viral protein synthesis may be due to an inhibition of viral reinfection and spread during the 24 h experiment. The data in Fig. 3A and B demonstrate that the yeast screen is capable of identifying compounds that have significant anti-influenza virus activity. Of the nine reproducible positives from the screen, four showed significant antiviral activity and are described here. This suggests a hit rate for the screen of ca. 0.2%.
Assays for nonspecific effects.
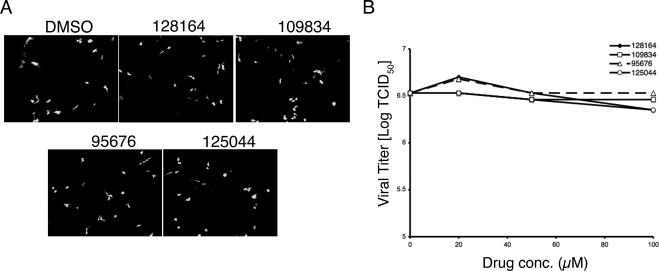
Two types of assays were performed to examine the possibility of nonspecific effects of the compounds that might explain their ability to inhibit influenza virus replication. First, cell growth assays were performed to determine the effects on cell replication. During a 48-h treatment, which is the period used for the virus replication assays shown in Fig. 3, none of the compounds exhibited significant growth toxicity over a dose range of 20 to 100 μM (see Fig. S1 and Table S1 in the supplemental material). Minor effects on cell growth were observed when treatment was extended for up to 6 days (data not shown). A second assay for nonspecific effects was to challenge the replication of another negative-strand RNA virus, RSV. As shown in Fig. 4 none of the compounds had any measurable effect on RSV replication in MA104 cells compared to the DMSO control. Similar results were obtained with RSV infection of MDCK cells (not shown). These data demonstrate that the compounds do not affect the cell's ability to support growth of negative-strand RNA virus replication in general and argue that the effects of the compounds on influenza virus replication are specific for that virus.
FIG. 4.
Effect of anti-influenza virus compounds on RSV replication. (A) MA104 cells were infected at an MOI of 0.1 with the virus RSΔsh (49), in which the SH open reading frame is replaced with that of GFP. The infected cells were treated with the indicated compounds or 1% DMSO (shown as 0 μM) starting at 1 h postinfection. After 48 h the cells were analyzed for GFP expression by fluorescence microscopy and photographed. Representative fields are shown. (B) MA104 cells plated in 96-well plates were infected with serially diluted RSΔsh. After 48 h, quadruplicate wells were scored for the presence or absence of GFP expression by fluorescence microscopy, and the data were analyzed for determination of TCID50.
Interactions with the cellular IFN system.
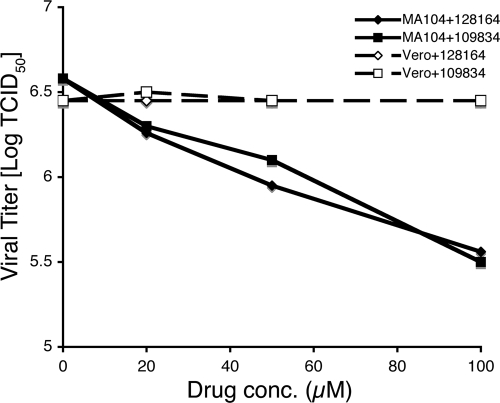
NS1 is a well-characterized inhibitor of the IFN system (14). Blockade of the IFN response by NS1 prevents establishment of the antiviral state, which would otherwise significantly limit virus replication. Accordingly, mutant viruses with altered NS1 function replicate poorly in cells with an intact IFN system but replicate significantly better in IFN-deficient cells (18, 42). Cells with defects in the IFN system might therefore be expected to be resistant to compounds that inhibit NS1 function. To test this, two African green monkey kidney cell lines were compared. Vero E6 cells fail to produce IFN, whereas MA104 cells are IFN competent (8, 12, 40). Each was tested for the ability to support replication of virus PR in the presence of NSC109834 or NSC128164. Figure 5 shows that Vero cells were completely resistant to both compounds, whereas MA104 cells were sensitive, a finding consistent with the idea that the compounds inhibit NS1 function.
FIG. 5.
A/PR/8 replication in drug-treated Vero and MA104 cells. Cells were infected A/PR/8 at an MOI of 0.1 and treated with drug or 1% DMSO (shown as 0 μM) starting 1 h postinfection. After incubation for 48 h, supernatants were collected and analyzed for TCID50 by the method of Reed and Muench (57).
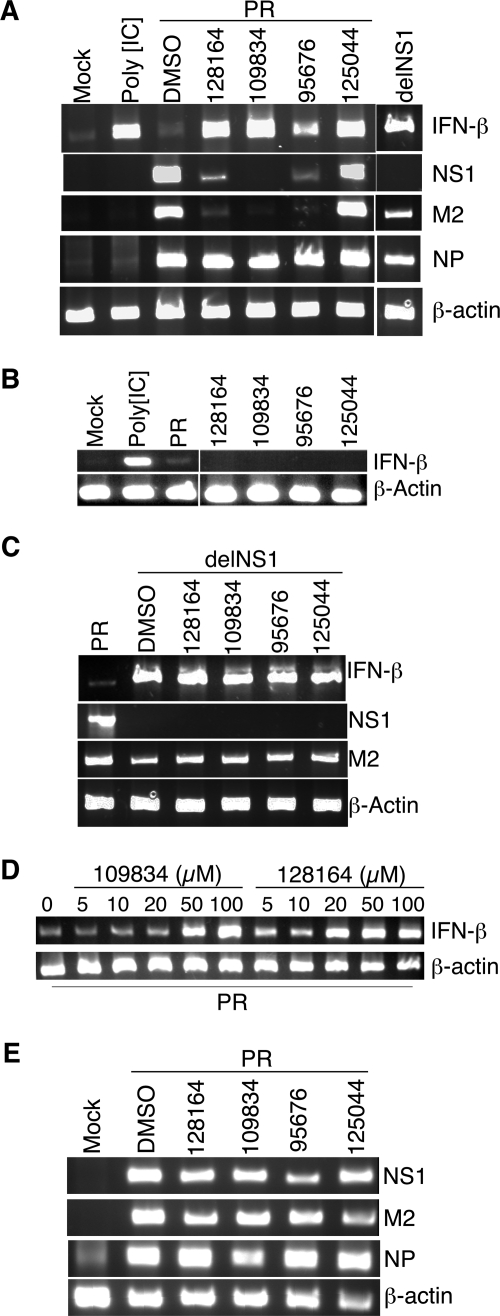
One function of NS1 during infection is inhibition of IFN gene expression through effects on transcriptional activation and mRNA maturation (14, 47, 65, 71). Compounds that block NS1 function would therefore be expected to restore induction of IFN-β mRNA in infected cells. To examine this, MDCK cells were infected with either virus PR or with delNS1, a derivative of PR that is deleted for NS1 coding sequences (18). As expected, delNS1-infected cells showed a pronounced induction of IFN-β as seen by RT-PCR analysis, whereas PR-infected cells contained the same low level of IFN-β mRNA observed in mock-infected cells (Fig. 6A). To test the effects of the drugs, cells infected with PR at an MOI of 2.0 were treated with each compound for 6 h, a period during which an initial round of virus production normally occurs. The cells were harvested for RNA isolation and assayed by RT-PCR for IFN-β mRNA. As shown in Fig. 6A, all four compounds triggered significant activation of IFN-β mRNA. For three of the compounds (NSC128164, NSC109834, and NSC125044) activation of IFN-β was roughly the same as that seen when cells were infected with delNS1 in the absence of drug and was also as strong as for uninfected cells treated with the IFN-β inducer poly[IC]. These data suggest a significant inhibition of NS1 function by these compounds. Importantly, none of the compounds triggered induction of IFN-ββ mRNA in the absence of viral infection (Fig. 6B). Also shown in Fig. 6A are RT-PCR results for three viral RNAs produced during infection: NP, M2, and NS1. Except for NSC125044, a strong reduction for M2 and NS1 RNA was observed, but no effect was seen for NP RNA. These data indicate the possibility of differential effects of some of the compounds on viral RNA production.
FIG. 6.
Drug-dependent restoration of IFN-β mRNA induction in virus-infected cells. (A) MDCK cells were infected with A/PR/8 at an MOI of 2.0 and incubated in the presence of 50 μM concentrations of the indicated compounds for 6 h. Cells were harvested for RT-PCR analysis of cellular IFN-β and β-actin mRNAs, as well as influenza virus RNAs corresponding to NP, M2, and NS1 sequences. Also shown are RT-PCR products for cells treated with poly[IC] for 6 h (second lane) and cells infected with the NS1 deletion virus delNS1 for 6 h (rightmost lane). Mock, uninfected cells. (B) Cells were uninfected, except for the third lane, where cells were infected with PR. Drug treatment and RT-PCR were as described for panel A. (C) Same as panel A except cells were infected with delNS1, as indicated. (D) Cells were treated and analyzed as in panel A, except the drug concentrations are as indicated in the figure. (E) Vero cells were infected with A/PR/8 at an MOI of 2.0 and incubated in the presence of 50 μM concentrations of the indicated compounds for 6 h. Cells were analyzed for the indicated RNAs as in panel A.
Interestingly, we observed that in delNS1-infected cells there was only a moderate decrease in the level of M2 RNA compared to wild-type virus, whereas in cells infected with wild-type virus and treated with NSC128164, NSC109834, or NSC95676, a more severe inhibition of M2 RNA was observed. This result would not be expected if the level of M2 RNA expression were a simple function of NS1 activity. One possibility to explain this result is that the compounds have an NS1-independent activity that results in a decrease in M2 RNA expression. To test this, cells were infected with delNS1 at an MOI of 2.0 and treated with each compound during a 6-h infection. Again, RNA was recovered for RT-PCR analysis. As shown in Fig. 6C, the compounds had no effect on M2 RNA levels in delNS1-infected cells, indicating that their effect on M2 RNA in wild type-infected cells was indeed NS1 dependent. Furthermore, the compounds had no effect on IFN-β mRNA levels in delNS1-infected cells, a finding consistent with the data shown in Fig. 6B. The mechanism by which the compounds affect M2 RNA levels in wild type-infected cells will require further investigation; however, the data are consistent with the possibility that NS1 is allosterically regulated by the compounds so as to alter expression of M2 and other viral RNAs (see Discussion). The fact that NSC125044 had no effect on M2 RNA levels despite strong activity in terms of IFN-β mRNA accumulation (Fig. 6A) suggests that its mechanism of action differs from that of the other compounds.
A titration experiment was performed to assess the concentration dependence of the effect on IFN-β mRNA induction. Figure 6D shows that NSC109834 induced IFN-β RNA between 20 and 50 μM. NSC128164 restored induction even at the lowest concentrations tested, with nearly maximal induction between 10 and 20 μM. Also, in Fig. 6 are shown the results of RT-PCR analysis of viral RNAs from infected Vero cells challenged by each of the compounds. As expected, there was little or no effect on viral RNA expression in Vero cells due to the lack of IFN expression (Fig. 6E).
Drug activity is linked to NS1 function.
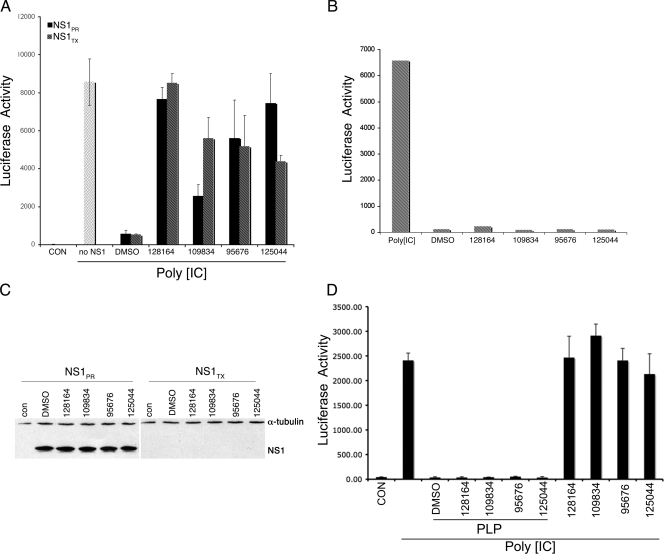
To determine whether the activities of the various compounds are specific for NS1 function, a transfection experiment was performed. Human 293 cells were cotransfected with a luciferase reporter driven by the IFN-β promoter and also expression plasmids encoding the NS1 protein from either PR or A/TX/36/91 (TX). As shown in Fig. 7A, in the absence of NS1 expression the IFN-β reporter was fully inducible by poly[IC] (second lane), whereas in the presence of either NS1PR8 or NS1TX, induction was almost completely inhibited (“DMSO” lane), as has been shown previously (28). Significantly, each compound efficiently restored induction of the IFN-β reporter, as shown in Fig. 7A. NSC128164 was the most efficient, increasing IFN-β promoter activity to maximal levels through inhibition of both NS1PR8 and NS1TX. NSC95676 also strongly restored induction. NSC109834 and NSC125044 differentially affected NS1PR8 and NS1TX activity, perhaps suggesting a structural difference between these proteins as they relate to the compounds. Importantly, none of the compounds triggered induction of the IFN-β reporter construct in the absence of cotransfected NS1 (Fig. 7B). This demonstrates that NS1 protein is absolutely required for the function of the compounds and is consistent with the idea that NS1 is their direct target. These results are also fully consistent with those presented in Fig. 6 for induction of cellular IFN-β mRNA in virus-infected cells, where induction was only observed in the presence of virus infection. As an additional control, transfected cells were analyzed for NS1 protein expression by Western blotting to determine whether drug treatment altered the cellular levels of NS1. As shown in Fig. 7C, treatment with the compounds had no effect on NS1 protein levels, indicating that restoration of reporter activity was not due to drug-induced effects on NS1 gene expression or protein turnover. In performing the Western blot experiments we successfully reproduced data from Kochs et al. (28) that showed that NS1TX protein dramatically inhibits its own expression in transfection experiments (Fig. 7C). Despite the low expression of NS1TX this protein clearly was active in inhibition of IFN-β promoter activity (Fig. 7A, DMSO lane). Therefore, we conclude that drug-dependent induction of IFN-ββ promoter activity in the case of NS1TX, as for NS1PR, was due to the inhibition of NS1-dependent function by the compounds under study. In addition, we used the compounds to challenge an independent inhibitor of IFN-β induction, the SARS-CoV PLP (9), as shown in Fig. 7D. Consistent with published results, cells cotransfected with the IFN-β reporter and a SARS-CoV PLP expression plasmid were significantly inhibited for IFN-β reporter induction by poly[IC] (9). However, none of the anti-NS1 compounds had any effect in restoring IFN-β induction that had been inhibited by SARS-CoV PLP. These data demonstrate the specificity of these compounds for influenza virus NS1 in restoration of IFN-β induction.
FIG. 7.
Specific inhibition of NS1-dependent function. (A) 293 cells were cotransfected with a firefly luciferase reporter driven by the human IFN-β promoter and expression constructs encoding the indicated NS1 proteins. At 16 h after transfection the cells were treated with 50 μg of poly[IC]/ml and incubated for an additional 24 h prior to harvesting for determination of the luciferase activity. Drug treatment with 50 μM concentrations of the indicated compounds was for the final 24 h of the experiment. “Con” indicates the luciferase activity of untransfected cells; “DMSO” indicates treatment with 1% DMSO. (B) 293 cells were transfected with the IFN-β reporter and treated with poly[IC] (first lane only), 1% DMSO, or 50 μM concentrations of the indicated anti-NS1 compounds in the absence of poly[IC]. (C) 293 cells were transfected with constructs to express the indicated NS1 proteins and harvested for Western blot analysis 40 h later. Drug treatment with 50 μM concentrations of the indicated compounds was for the final 24 h of the experiment. Whole-cell extracts were blotted for NS1 or α-tubulin, whose positions are indicated. “Con” indicates the signal from untransfected cells. “DMSO” indicates DMSO control. (D) 293T cells were transfected with the IFN-β luciferase plasmid alone or together with a SARS PLP-expressing plasmid. At 6 h posttransfection cells were treated with the indicated drugs, incubated for an additional 18 h and then treated with poly[IC] for 6 h. The cells were then lysed and measured for luciferase activity.
DISCUSSION
It is widely appreciated that global influenza pandemics represent a considerable risk to human health and the economy, and new therapeutic approaches and drug targets are needed to protect the public health, especially against RNA viruses that evolve quickly in response to chemotherapeutic intervention (25, 26, 50, 51). In the present study compounds were identified based on their ability to suppress the phenotypic effect of NS1 in S. cerevisiae. The precise mechanism of growth inhibition by NS1 in yeast is not known, although Ward et al. showed that regions within the N-terminal and C-terminal domains are required for toxicity in yeast (72). Regardless, the fact that the compounds identified in the yeast screen also inhibit virus replication and specifically reverse NS1-dependent inhibition of IFN-β mRNA induction strongly indicates that the mechanism of growth inhibition in yeast is directly related to the function of NS1 during infection. This illustrates an advantage of the yeast-based drug discovery assay used here: it is not necessary to know the precise mechanism(s) of the target in order to identify relevant inhibitory compounds, so long as expression of the target protein induces a phenotypic change in yeast that can be incorporated into an assay for drug discovery, and physiologically relevant models (i.e., virus infection and IFN-β mRNA induction) can be used as secondary assays. This situation also suggests that compounds with entirely different mechanisms of action can be derived from the same screen, assuming that multiple features of the target contribute to the phenotypic effect. Additional advantages of the yeast system include identification of compounds that are able to enter living cells and the fact that the selection is positive for growth, which eliminates compounds that are severely toxic, at least in yeasts.
An impressive list of functional domains of NS1 has been established that includes regions that mediate RNA binding (10, 34, 70), binding to RIG-I (20, 41, 52), inhibition of nuclear RNA export (53, 61), binding to CPSF (28, 45), and other activities (14). Additional experiments will determine which domain(s) of NS1 are involved in the response to the compounds under study. A method for high-throughput screening against the RNA-binding activity of NS1 was reported recently (39). It is important to emphasize that, based on the design of the yeast screen, direct targets of the compounds can be either NS1 itself or possibly cellular proteins. Whereas our results firmly establish NS1 as a required participant in drug-mediated effects (Fig. 6 and 7), they do not rule out the possibility that the direct target is a cellular protein that regulates NS1 or one that can be caused to bypass its activity. This question is the subject of ongoing investigation.
NS1 has been shown to inhibit induction of IFN-β by several independent mechanisms. Two of these are transcriptional mechanisms involving the N-terminal domain of NS1 and are triggered by binding to dsRNA or to RIG-I (10, 20, 41, 52, 71). An additional mechanism is posttranscriptional and is carried out by binding of the “effector” domain of NS1 to CPSF (28, 45). As reported by Kochs et al., both NS1PR and NS1Tx are able block IFN-β induction in virus-infected cells and can also bind to RIG-I and interfere with activation of IRF3 (28). This indicates that both proteins share the capacity to inhibit transcriptional activation of IFN-β. Interestingly, Kochs et al. also found that only NS1Tx, but not NS1PR, was able to suppress cellular gene expression posttranscriptionally through binding to CPSF (28). Since we found that both NS1PR and NS1Tx were sensitive to the effects of the compounds reported here, this suggests that the compounds are acting, at least in part, to counter the transcriptional mechanism(s) carried out by the N-terminal domain of NS1 and not the mechanism involving CPSF. We also observed that the compounds were active only in cells that express NS1 and that also have an intact IFN system (Fig. 5 to 7). In the absence of NS1, the compounds failed to induce IFN-β mRNA or promoter activity on their own (Fig. 6 and 7). Also, when cells were treated with poly[IC] in the absence of NS1 to activate IFN-β transcription or were infected with delNS1, which allowed activation of IFN-β, the compounds did not cause a further increase in activation. These data indicate that the compounds are not simply targeting cellular components of the IFN-β activation pathway that are poised to activate IFN-β in the absence of NS1. Furthermore, our experiments showed that the compounds did not restore IFN-β induction that had been inhibited by the SARS-CoV PLP protein (Fig. 7D). This also suggests that they are not acting at the level of a common signaling molecule in the activation of IFN-β transcription. Rather, our data suggest a direct, NS1-dependent function for the drugs. As mentioned above, this could involve direct binding to NS1 or to a cellular function that regulates it.
As shown in Fig. 6A, three of the compounds (NSC128164, NSC109834, and NSC95676) triggered a significant reduction in viral M2 and NS1-specific RNAs, as judged by RT-PCR assay. However, NSC125044 did not affect viral RNA levels despite triggering a significant increase in IFN-β mRNA (Fig. 6A) and a significant reduction in overall viral production, as the other three compounds also did (Fig. 3). These data suggest that at least two antiviral mechanisms are at play here. Interestingly, in contrast to the decrease in viral RNAs caused by NSC128164, NSC109834, and NSC95676, complete deletion of NS1 sequences in virus delNS1 did not cause the same decrease in viral RNA levels (Fig. 6A and 6C). This indicates that in a wild-type infected cell that is treated with NSC128164, NSC109834, or NSC95676, NS1 has a negative effect on viral RNA expression that is not observed in cells infected with the delNS1 virus. Since these compounds are only active in the presence of NS1 protein and are not simply mimicking the absence of NS1, this suggests that there may be interactions between these compounds and NS1 that allosterically regulate the ability of NS1 to control transcription, RNA stability, or other processes. As reported previously, several temperature-sensitive mutants of NS1 affect viral RNA levels. For instance, an Arg-to-Lys mutation at position 25 was shown to lead to a significant decrease in M1 and HA mRNAs (36). Other studies have implicated NS1 in the function or regulation of the viral polymerase complex (38, 43), a process that could be affected directly by some of the compounds reported here. On the other hand, another temperature-sensitive mutant of NS1, also with a change in the N-terminal domain at amino acid 11, showed a drastic decrease in formation of virus particles without large changes in transcription of viral RNAs. Thus, allosteric effects of the compounds on NS1 function may trigger a variety of effects that result in reduction of overall virus replication. Therefore, it is anticipated that the compounds identified here may be of use in elucidating these or novel interactions between NS1 and viral or host functions, in addition to their potential clinical utility.
Supplementary Material
Acknowledgments
We thank Dan Burke, Tom Braciale, Gail Wertz, Luis Martinez-Sobrido, Adolfo Garcia-Sastre, and Peter Palese for many valuable reagents, including cells, viruses, antibodies, and DNA clones.
This study was supported by Public Health Service grant R01AI071341 to D.A.E.
Footnotes
Published ahead of print on 3 December 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abdel-Ghafar, A. N., T. Chotpitayasunondh, Z. Gao, F. G. Hayden, D. H. Nguyen, M. D. de Jong, A. Naghdaliyev, J. S. Peiris, N. Shindo, S. Soeroso, and T. M. Uyeki. 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358261-273. [DOI] [PubMed] [Google Scholar]
- 2.Amberg, D., D. J. Burke, and J. N. Strathern. 2005. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Basler, C. F., and P. V. Aguilar. 2008. Progress in identifying virulence determinants of the 1918 H1N1 and the Southeast Asian H5N1 influenza A viruses. Antivir. Res. 79166-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 982746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucher, E., H. Hemmes, P. de Haan, R. Goldbach, and M. Prins. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85983-991. [DOI] [PubMed] [Google Scholar]
- 6.Burger, H., H. Steuler, and C. Scholtissek. 1985. A mutant of fowl plague virus (influenza A) with an enhanced electrophoretic mobility of RNA segment 8. J. Gen. Virol. 661679-1686. [DOI] [PubMed] [Google Scholar]
- 7.Carrat, F., and A. Flahault. 2007. Influenza vaccine: the challenge of antigenic drift. Vaccine 256852-6862. [DOI] [PubMed] [Google Scholar]
- 8.Desmyter, J., J. L. Melnick, and W. E. Rawls. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devaraj, S. G., N. Wang, Z. Chen, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 28232208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 7713257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 726437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43247-252. [DOI] [PubMed] [Google Scholar]
- 13.Falcon, A. M., A. Fernandez-Sesma, Y. Nakaya, T. M. Moran, J. Ortin, and A. Garcia-Sastre. 2005. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 862817-2821. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Sesma, A. 2007. The influenza virus NS1 protein: inhibitor of innate and adaptive immunity. Infect. Disord. Drug Targets 7336-343. [DOI] [PubMed] [Google Scholar]
- 15.Fortes, P., A. Beloso, and J. Ortin. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gambotto, A., S. M. Barratt-Boyes, M. D. de Jong, G. Neumann, and Y. Kawaoka. 2008. Human infection with highly pathogenic H5N1 influenza virus. Lancet 3711464-1475. [DOI] [PubMed] [Google Scholar]
- 17.Garaigorta, U., A. M. Falcon, and J. Ortin. 2005. Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles. J. Virol. 7915246-15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252324-330. [DOI] [PubMed] [Google Scholar]
- 19.Gog, J. R., S. Afonso Edos, R. M. Dalton, I. Leclercq, L. Tiley, D. Elton, J. C. von Kirchbach, N. Naffakh, N. Escriou, and P. Digard. 2007. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 351897-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, Z., L. M. Chen, H. Zeng, J. A. Gomez, J. Plowden, T. Fujita, J. M. Katz, R. O. Donis, and S. Sambhara. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36263-269. [DOI] [PubMed] [Google Scholar]
- 21.Hatada, E., and R. Fukuda. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 733325-3329. [DOI] [PubMed] [Google Scholar]
- 22.Hatada, E., M. Hasegawa, K. Shimizu, M. Hatanaka, and R. Fukuda. 1990. Analysis of influenza A virus temperature-sensitive mutants with mutations in RNA segment 8. J. Gen. Virol. 711283-1292. [DOI] [PubMed] [Google Scholar]
- 23.Hatada, E., S. Saito, N. Okishio, and R. Fukuda. 1997. Binding of the influenza virus NS1 protein to model genome RNAs. J. Gen. Virol. 781059-1063. [DOI] [PubMed] [Google Scholar]
- 24.Hatada, E., T. Takizawa, and R. Fukuda. 1992. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA in vitro. J. Gen. Virol. 7317-25. [DOI] [PubMed] [Google Scholar]
- 25.Hayden, F. G. 2006. Antivirals for influenza: historical perspectives and lessons learned. Antivir. Res. 71372-378. [DOI] [PubMed] [Google Scholar]
- 26.Hayden, F. G., and A. T. Pavia. 2006. Antiviral management of seasonal and pandemic influenza. J. Infect. Dis. 194(Suppl. 2)S119-S126. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, D., M. J. Hossain, D. Hickman, D. R. Perez, and R. A. Lamb. 2008. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. USA 1054381-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kochs, G., A. Garcia-Sastre, and L. Martinez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 817011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kochs, G., I. Koerner, L. Thiel, S. Kothlow, B. Kaspers, N. Ruggli, A. Summerfield, J. Pavlovic, J. Stech, and P. Staeheli. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 881403-1409. [DOI] [PubMed] [Google Scholar]
- 30.Koennecke, I., C. B. Boschek, and C. Scholtissek. 1981. Isolation and properties of a temperature-sensitive mutant (ts 412) of an influenza A virus recombinant with a ts lesion in the gene coding for the nonstructural protein. Virology 11016-25. [DOI] [PubMed] [Google Scholar]
- 31.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309181-189. [DOI] [PubMed] [Google Scholar]
- 32.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 34913-21. [DOI] [PubMed] [Google Scholar]
- 33.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 1011350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4896-899. [DOI] [PubMed] [Google Scholar]
- 35.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214222-228. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig, S., U. Vogel, and C. Scholtissek. 1995. Amino acid replacements leading to temperature-sensitive defects of the NS1 protein of influenza A virus. Arch. Virol. 140945-950. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. Garcia-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 7611166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marion, R. M., T. Zurcher, S. de la Luna, and J. Ortin. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 782447-2451. [DOI] [PubMed] [Google Scholar]
- 39.Maroto, M., Y. Fernandez, J. Ortin, F. Pelaez, and M. A. Cabello. 2008. Development of an HTS assay for the search of anti-influenza agents targeting the interaction of viral RNA with the NS1 protein. J. Biomol. Screen. 13581-590. [DOI] [PubMed] [Google Scholar]
- 40.McKimm-Breschkin, J. L., and I. H. Holmes. 1982. Conditions required for induction of interferon by rotaviruses and for their sensitivity to its action. Infect. Immun. 36857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligonucleotide (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 1037100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min, J. Y., S. Li, G. C. Sen, and R. M. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363236-243. [DOI] [PubMed] [Google Scholar]
- 44.Monto, A. S., and R. J. Whitley. 2008. Seasonal and pandemic influenza: a 2007 update on challenges and solutions. Clin. Infect. Dis. 461024-1031. [DOI] [PubMed] [Google Scholar]
- 45.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30-kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1991-1000. [DOI] [PubMed] [Google Scholar]
- 46.Nichol, K. L., and J. J. Treanor. 2006. Vaccines for seasonal and pandemic influenza. J. Infect. Dis. 194(Suppl. 2)S111-S118. [DOI] [PubMed] [Google Scholar]
- 47.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307386-395. [DOI] [PubMed] [Google Scholar]
- 48.Olsen, B., V. J. Munster, A. Wallensten, J. Waldenstrom, A. D. Osterhaus, and R. A. Fouchier. 2006. Global patterns of influenza a virus in wild birds. Science 312384-388. [DOI] [PubMed] [Google Scholar]
- 49.Oomens, A. G., A. G. Megaw, and G. W. Wertz. 2003. Infectivity of a human respiratory syncytial virus lacking the SH, G, and F proteins is efficiently mediated by the vesicular stomatitis virus G protein. J. Virol. 773785-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palese, P. 2004. Influenza: old and new threats. Nat. Med. 10S82-S87. [DOI] [PubMed] [Google Scholar]
- 51.Pekosz, A., and G. E. Glass. 2008. Emerging viral diseases. Md Med. 911-16. [PMC free article] [PubMed] [Google Scholar]
- 52.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 53.Qian, X. Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 682433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian, X. Y., C. Y. Chien, Y. Lu, G. T. Montelione, and R. M. Krug. 1995. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA. 1948-956. [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 682425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1304-316. [PMC free article] [PubMed] [Google Scholar]
- 57.Reed, L., and H. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27943-947. [Google Scholar]
- 58.Reid, A. H., J. K. Taubenberger, and T. G. Fanning. 2001. The 1918 Spanish influenza: integrating history and biology. Microbes Infect. 381-87. [DOI] [PubMed] [Google Scholar]
- 59.Rogers, B., A. Decottignies, M. Kolaczkowski, E. Carvajal, E. Balzi, and A. Goffeau. 2001. The pleiotropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 3207-214. [PubMed] [Google Scholar]
- 60.Rothberg, M. B., S. D. Haessler, and R. B. Brown. 2008. Complications of viral influenza. Am. J. Med. 121258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Satterly, N., P. L. Tsai, J. van Deursen, D. R. Nussenzveig, Y. Wang, P. A. Faria, A. Levay, D. E. Levy, and B. M. Fontoura. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA 1041853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaw, M. W., E. W. Lamon, and R. W. Compans. 1982. Immunologic studies on the influenza A virus nonstructural protein NS1. J. Exp. Med. 156243-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shimizu, K., M. G. Mullinix, R. M. Chanock, and B. R. Murphy. 1983. Temperature-sensitive mutants of influenza A/Udorn/72 (H3N2) virus. III. Genetic analysis of temperature-dependent host range mutants. Virology 12435-44. [DOI] [PubMed] [Google Scholar]
- 64.Stephenson, I., K. G. Nicholson, J. M. Wood, M. C. Zambon, and J. M. Katz. 2004. Confronting the avian influenza threat: vaccine development for a potential pandemic. Lancet Infect. Dis. 4499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 747989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 974309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taubenberger, J. K., A. H. Reid, T. A. Janczewski, and T. G. Fanning. 2001. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos. Trans R. Soc. Lond. B Biol. Sci. 3561829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson, W. W., L. Comanor, and D. K. Shay. 2006. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J. Infect. Dis. 194(Suppl. 2)S82-S91. [DOI] [PubMed] [Google Scholar]
- 69.Wang, W., and R. M. Krug. 1998. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA 455-64. [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 7411566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, A. C., A. A. Azad, and I. G. Macreadie. 1994. Expression and characterisation of the influenza A virus nonstructural protein NS1 in yeast. Arch. Virol. 138299-314. [DOI] [PubMed] [Google Scholar]
- 73.Whitley, R. J., J. Bartlett, F. G. Hayden, A. T. Pavia, M. Tapper, and A. S. Monto. 2006. Seasonal and pandemic influenza: recommendations for preparedness in the United States. J. Infect. Dis. 194(Suppl. 2)S155-S161. [DOI] [PubMed] [Google Scholar]
- 74.Wolstenholme, A. J., T. Barrett, S. T. Nichol, and B. W. Mahy. 1980. Influenza virus-specific RNA and protein syntheses in cells infected with temperature-sensitive mutants defective in the genome segment encoding nonstructural proteins. J. Virol. 351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.World Health Organization. Fact sheet 211, revised 2003. World Health Organization, Geneva, Switzerland.
- 76.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 171087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.