Abstract
Ligand-bound nuclear receptors (NR) activate transcription of the target genes. This activation is coupled with histone modifications and chromatin remodeling through the function of various coregulators. However, the nature of the dependence of a NR coregulator action on the presence of the chromatin environment at the target genes is unclear. To address this issue, we have developed a modified position effect variegation experimental model system that includes an androgen-dependent reporter transgene inserted into either a pericentric heterochromatin region or a euchromatic region of Drosophila chromosome. Human androgen receptor (AR) and its constitutively active truncation mutant (AR AF-1) were transcriptionally functional in both chromosomal regions. Predictably, the level of AR-induced transactivation was lower in the pericentric heterochromatin. In genetic screening for AR AF-1 coregulators, Drosophila CREB binding protein (dCBP) was found to corepress AR transactivation at the pericentric region whereas it led to coactivation in the euchromatic area. Mutations of Sir2 acetylation sites or deletion of the CBP acetyltransferase domain abrogated dCBP corepressive action for AR at heterochromatic areas in vivo. Such a CBP corepressor function for AR was observed in the transcriptionally silent promoter of an AR target gene in cultured mammalian cells. Thus, our findings suggest that the action of NR coregulators may depend on the state of chromatin at the target loci.
Sex steroid hormones exert a wide variety of biological actions through the transcriptional control of a particular set of target genes. This transcriptional control is mediated by nuclear steroid hormone receptors that act as hormone-dependent transcription factors. These hormone receptors are members of the nuclear receptor (NR) gene superfamily (36, 48). The NR is functionally and structurally divided into domains A through E. The C-terminal E domain encompasses the ligand binding domain (LBD) and the ligand-dependent transactivation function mutant AF-2. The N-terminal A/B domain harbors a ligand-independent activation function mutant (AF-1). Both AF-1 and AF-2 serve as docking sites for transcriptional coregulators (34, 41). For hormone-induced transcriptional regulation by NRs, a number of coregulators/coregulator complexes are required in addition to the basic transcriptional machinery. The two major functions of NR coregulators/coregulator complexes are chromatin remodeling (3, 30, 35) and histone modifications (15). Each of the nuclear events involving NR-mediated gene regulation appears to be facilitated by several classes of coregulator complexes (19, 36, 48). Particularly, histone-modifying enzyme coregulator complexes are diverse in terms of covalent modifications of histone proteins. The histone acetyltransferases (HATs), such as CREB-binding protein (CBP) and p160 member proteins, in their cognate complexes were the first major NR coactivators identified (41). Consequently, these HAT coactivators were shown to be global coactivators that activated chromatin through hyperacetylation of histones (36, 48). In addition, it has been reported that Drosophila CBP (dCBP) may regulate the formation of the chromatin state through interactions with some chromatin-associated factors (4, 5) and through functions in DNA metabolic events (54). On the other hand, the complexes containing histone deacetylase (HDAC) are known to corepress non-ligand-bound NRs through hypoacetylation of chromatin areas around NR binding sites (45, 67). Histone methylases/demethylases also appear for the other classes of major coregulators as nuclear complexes for NRs (22, 37). Together with histone acetylation, histone methylation and demethylation at specific sites in the histone molecules constitute a significant part of the “histone code.” Histone modifications define the state of chromatin (32). Methylation of histone H3-K4 triggers activation of the chromatin state into the euchromatin state, while histone H3-K9 methylation evokes a transition of the chromatin state from euchromatin into inactive chromatin (7, 24). During chromatin silencing induced by H3-K9 methylation, HP1 is recruited as a component to establish heterochromatin (14, 25). Nucleosome arrays are rearranged through ATP-dependent chromatin remodeling in response to histone modifications. The roles of each of the histone-modifying enzymes in chromatin remodeling and how the various chromatin states affect histone modifications are not completely understood.
To study the function of histone-modifying coregulators in modulation of sex hormone receptor transactivation during the chromatin state transition, we have developed a modified position effect variegation (PEV) experimental system associated with an androgen-dependent reporter transgene (ARE-GFP-white) inserted into the pericentric heterochromatin or euchromatic loci in Drosophila melanogaster flies by use of a genetic approach. In this PEV system, the inserted reporter transgene encodes the green fluorescence protein (GFP) controlled by a basal promoter linked with eight upstream copies of consensus sequences of androgen receptor (AR) response elements (ARE) and the white protein driven by its endogenous promoter. We demonstrated that dCBP corepressed AR- or AR AF-1-mediated transactivation at the pericentric region. Using truncation mutants and dCBP strategies, we determined that the C terminus of dCBP, including the HAT domain, was required for its repressive function. In vitro and in vivo acetylation assays showed that Drosophila Sir2 (dSir2) was acetylated by CBP. In transgenic flies, mutations of Sir2 acetylation sites or deletion of the dCBP HAT domain abrogated dCBP corepression action in the AR transactivation at heterochromatic area in vivo. Furthermore, a corepressive function of CBP for AR was also observed together with SIRT1 recruitment in the transcriptionally silent AR target gene promoter in MCF-7 cells. Taken together, these results demonstrate that CBP represses transcription of genes inserted in heterochromatin regions through its interaction with and acetylation of Sir2. Our findings suggest that the roles of NR coregulators are distinct and dependent on chromatin state.
MATERIALS AND METHODS
Drosophila stocks, P-element mobilization, and genetics.
All Drosophila stocks were raised on cornmeal sucrose-based media. The nej3/FM7C and UAS-dCBP/TM3 strains were provided by S. Ishii (Institute of Physical and Chemical Research, Tsukuba Life Science Center, Japan). UAS-dCBP Δ HQ/TM3 and UAS-dCBP Δ BHQ/CyO fly strains were kindly provided by J. P. Kumar (Department of Biology, Indiana University, Bloomington, IN). Other fly stocks were obtained from the Bloomington Stock Center. All crosses were performed at 25°C. Flies of similar ages were used for all comparisons.
The AR expression and ARE-GFP-white reporter constructs made for transgenic flies were previously reported (57). Flies with a P-element construct carrying the ARE-GFP-white reporter gene inserted into the euchromatin of the third chromosome were used as the starting line for the P-element mobilization scheme (63). Homozygous females from the starting line were crossed to w/Y, SbΔ2-3/TM6 males, with Δ2-3 serving as a genomic source of transposase (31). The male progeny containing the SbΔ2-3 chromosome were crossed to females of the white mutant host stock yw. Male progeny showing PEV of white expression and lacking the SbΔ2-3 chromosome potentially had insertions into heterochromatin. Most of the flies showing a uniform red eye phenotype had insertions into euchromatin. Fly lines carrying the reporter gene at euchromatic or heterochromatic loci were crossed to make homozygous lines or stable lines. The sites of insertion were determined by inverse PCR experiments.
Antibodies.
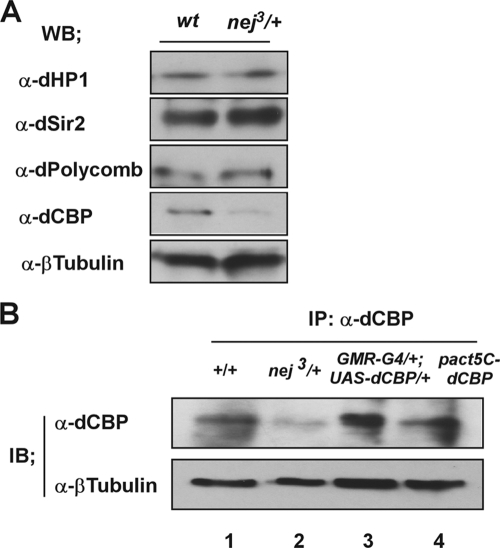
To produce an antibody against dCBP, the dCBP CREB binding domain (amino acids [aa] 825 to 1043) amplified by PCR was subcloned into the pET-29a protein expression plasmid. The His6-tagged recombinant dCBP protein (aa 825 to 1046) was expressed in Escherichia coli. The protein purified by using nickel-nitrilotriacetic spin columns (Qiagen, Chatsworth, CA) and confirmed by CBB staining was used as an immunogen in rats. Antibodies from the rat serum samples were affinity purified by using the purified dCBP protein (aa 825 to 1043). The specificity of antibodies against dCBP was confirmed by detecting the corresponding ectopically expressed proteins in fly or S2 cells (see Fig. 2B).
FIG. 2.
The expression levels of proteins involved in heterochromatin silencing are not affected in dCBP mutant lines. (A) Whole-cell extracts were made from wt (+/+) and dCBP mutant (nej3/+) third instar larva eye discs. A fraction (2%) of whole-cell extracts was analyzed by immunoblotting using the indicated antibodies. WB, Western blot. (B) Antibodies generated against dCBP peptides specifically recognized by detecting the endogenous and exogenous proteins in flies or S2 cells. Whole-cell extracts from a wt fly (+/+) (lane 1), a dCBP mutant (nej3/+) (lane 2), gain-of-function dCBP mutant flies expressing dCBP in the developing eye by the use of the UAS-GAL4 misexpression system (lane 3), and cultured S2 cells transfected with a pAct5C-dCBP expression construct (lane 4) were analyzed by Western blotting. Specific signals corresponding to full-length dCBP proteins were obtained using a polyclonal antibody against dCBP. β-Tubulin was used as the control. IB, immunoblot.
The other antibodies used in this study were anti-FLAG (M2 and rabbit; Sigma) and anti-acetyl-lysine (Ac-K103 [catalog no. 9681]; Cell Signaling Technology), anti-trimethyl H3-K9, anti-AcH3, anti-trimethyl H3-K27, anti-dimethyl H4-K20, anti-AcH4-K12, anti-AcH4-K16, and anti-AcH4 (Upstate Biotechnology), anti-HP1 C1A9 (Developmental Studies Hybridoma Bank at the University of Iowa), anti-Polycomb (d-220), anti-β-tubulin (dN-17), anti-dSir2 (dF-16), and anti-dSir2 (dC-16) (Santa Cruz Biotechnology), anti-AR (N-20 and 441) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-CBP (CBP 5I001 and AC238; Abcam), and anti-SIRT1 (E54; Abcam).
Immunostaining of polytene chromosomes and Eye disc histology analysis.
Wandering third instar larvae of D. melanogaster were collected, and salivary glands were dissected and fixed in 3.7% formaldehyde-50% acetic acid for 2 min and squashed in 45% acetic acid on subbed slides (42). The slides were frozen in liquid nitrogen and blocked in blocking buffer (1× phosphate-buffered saline [PBS], 5% nonfat dry milk, 0.1% Tween 20) and were then incubated in blocking buffer containing primary antibodies at 4°C overnight. The primary antibodies used were rat polyclonal anti-dCBP (1:80), purified murine A6 (Covance) (1:80), rabbit polyclonal anti-FLAG (Sigma) (1:80), mouse polyclonal anti-HP1 C1A9 (University of Iowa) (1:70), and rabbit polyclonal anti-AR (N-20) (Santa Cruz Biotechnology, Santa Cruz, CA) (1:100). Incubation was followed by washing in wash buffer (1× PBS, 0.1% Tween 20) three times. The slides were then incubated with secondary fluorescein isothiocyanate-, Cy3-, or Cy5-conjugated antibodies (Jackson Immunoresearch Laboratories Inc.) (1:1,000) together with DAPI (4′,6-diamidino-2-phenylindole) (Roche) (1:5,000 in PBS) for 2 h at room temperature. Slides were again washed in wash buffer three times and mounted in Vectashield antifade mounting medium (Vector Laboratories). Confocal microscopy was carried out on a Zeiss 510 confocal laser scanning system, and results were assessed with Adobe Photoshop 5.0 software (Adobe).
Eye disc histology analysis was performed as previously described (27, 57).
Plasmids.
A dSir2 cDNA clone was produced by use of an Open biosystem. Murine Sir2α (mSir2) cDNA in the pUSEamp vector was provided by A. Fukamizu (Center for Tsukuba Advanced Research Alliance, University of Tsukuba, Japan). For the production of Sir2 expression constructs, cDNA sequences encoding mSir2 or dSir2 were amplified by PCR. The PCR products were cloned into a pcDNA3.1 (Invitrogen) or a pcDNA3.1 derivate containing a sequence encoding a FLAG epitope upstream of the multiple cloning sites to generate pcDNA3-mSir2 and pcDNA3-FLAG-dSir2. To make a UAS-FLAG-dSir2 construct, a PCR product of dSir2 cDNA was cloned into a pCaSpeR3 derivate containing a sequence encoding a FLAG epitope upstream of the cloning sites. The dSir2 acetylation site mutations and a catalytically inactive mutation of dSir2 (H331Y) were generated by site-directed mutagenesis. The constructs of glutathione S-transferase (GST)-p300-HAT, GST-Pcaf-HAT, and pAct5C-dCBP were described previously (28, 60).
IP and Western blot analysis.
For immunoprecipitation (IP) experiments, the expression plasmid encoding AR was transiently transfected into LNCaP cells and MCF-7 cells by use of Lipofectamine (Invitrogen) and whole-cell extracts were prepared 48 h after transfection as described previously (30) and were immunoprecipitated using mouse monoclonal anti-AR (441; Santa Cruz) and protein G-Sepharose (GE Healthcare). Antibody-protein G-Sepharose-bound protein complexes were washed three times with IP buffer (25 mM Tris-HCl [pH 8.0], 10% glycerol, 0.1% NP-40, 0.5 mM dithiothreitol [DTT], 5 mM MgCl2 and protease inhibitor) containing 0.5 M KCl and twice with IP buffer containing 100 mM KCl. Immune complexes were analyzed by Western blotting using the indicated primary antibodies, and chemiluminescence detection was performed according to the instructions of the manufacturer (GE Healthcare). Eye discs dissected from the third instar larvae were homogenized in TNE buffer (10 mM Tris-HCl [pH 7.5], 0.1% NP-40, 150 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10% glycerol, 10 mM β-glycerophosphate and protein inhibitors). The protein concentrations of the whole-cell extracts were determined using standard Bradford assays. Equal protein amounts were immunoprecipitated using polyclonal anti-dCBP antibody (29, 58). The crude extracts and immunoprecipitated proteins were loaded onto two separate gels, and Western blot analysis was performed by probing with anti-AR (N-20), anti-dSir2 (dF-16), and anti-dCBP antibodies.
Cell culture, transfections, and luciferase assay.
LNCaP cells were grown in phenol-red-free RPMI 1640 medium (Gibco-BRL) supplemented with 5% charcoal-treated fetal bovine serum. MCF-7 cells and HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum-2 mM glutamine-streptomycin and penicillin (10 g/liter). 293T cells were cotransfected with AR or dSir2 wild type (wt) or with a series of mutation expression plasmids and with the reporter gene carrying a mouse mammary tumor virus promoter by using SuperFect (Qiagen). pRL-CMV vector was used as the internal control. At 3 h posttransfection, the cells were rinsed and incubated in Dulbecco's modified Eagle's medium supplemented with 5% coal-treated fetal bovine serum in the absence or presence of AR ligand (dehydrotestosterone [DHT] at 10−8 M). After an additional 21 h, the cells were harvested and assayed for luciferase activities by use of a dual-luciferase reporter assay system (Promega) as described previously (70).
ChIP.
Chromatin IP (ChIP) was performed as previously described (65). LNCaP cells and MCF-7 cells were treated for 4 h in the presence or absence of DHT (10−8 M). Cells were cross-linked with 1% formaldehyde at room temperature for 10 min, resuspended in lysis buffer (1% sodium dodecyl sulfate [SDS], 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1× protease inhibitor cocktail [Roche Molecular Biochemicals]), and sonicated (model UR-20P Handy Sonic) three times for 10 s each time at the maximum setting followed by centrifugation for 10 min. Supernatants were collected and diluted in buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1]) followed by immunoclearing with 2 μg of sheared salmon sperm DNA-20 μl of preimmune serum-protein A-Sepharose (45 μl of a 50% slurry in 10 mM Tris-HCl [pH 8.1]-1 mM EDTA) for 2 h at 4°C. IPs were performed overnight at 4°C with specific antibodies. Protein A-Sepharose beads were added and then washed sequentially with low-salt buffer, high-salt buffer, LiCl buffer, and Tris-EDTA buffer. The protein-DNA complexes were eluted, and the cross-linking was reversed at 65°C for 6 h. DNA fragments were purified with a DNA purification kit (DIAquick; Qiagen) and analyzed by regular PCR. Primer sequences for the KLK2 promoter region (−343 to −90) were previously described (66). For PCR, 2 μl from a 50-μl DNA extraction mixture and 21 to 25 cycles of amplification were used.
An in-fly ChIP assay was performed as described above with a few modifications. Specific flies as indicated were crossed to a UAS-dCBP/TM3 mutant or a dCBP mutant (nej3/FM7C). A white mutant host line (yw) was used as the control. Eye discs dissected from the third instar larvae were cross-linked in 1% formaldehyde for 20 min at room temperature, washed twice with cold PBS, and resuspended in IP buffer (50 mM Tris-HCl [PH 8.0], 0.3% Triton X-100, 150 mM NaCl, 10% glycerol, and protease inhibitors) and sonicated (model UR-20P Handy Sonic) until an average DNA fragment size of 200 to 500 bp was achieved (40). IPs were performed using specific antibodies and protein A-Sepharose. After reversing cross-links, DNA fragments were purified with a DNA purification kit and analyzed by regular PCR. Primer sequences for the promoter region of the ARE-GFP-white reporter gene were as follows: 5′-TTCTAGTGGATCTCTCTAGA-3′ and 5′-TCACCATCTAATTCAACAAG-3′. Primer sequences for the insertion regions (66E7 and 79E5) in the Drosophila chromosome were as follows: 5′-AAGGTTAGATCGTCTAGTGG-3′ and 5′-TTATTTCTGTTGACGGCTCC-3′ (66E7 region); 5′-TGGTCGGCTGTCATCGCACC-3′ and 5′-CTCACTCGGGTGACCTGCAC-3′ (79E5); and 5′-CGGATCGATATGCTAAGCTG-3′ and 5′-GAACGCAGGCGACCGTTGGGG-3′ (rp49). For PCR, 2 μl from 50 μl of DNA extraction and 30 to 35 cycles of amplification was used.
ChIP re-IP.
ChIP re-IP experiments were performed essentially as described previously (51). Complexes were eluted from the primary IP by incubation with 10 mM DTT at 37°C for 30 min and diluted 1:50 in buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1]) followed by re-IP with the second antibodies. For regular PCR detection, primer sequences for the promoter region of ARE-GFP-white reporter gene were the same as those used in the ChIP assay.
siRNA transfection, RNA isolation, RT, quantitative PCR.
On-Target Plus Smart Pool small interfering RNA (siRNA) against CBP, SIRT1 (Dharmacon), and a control siRNA (Ambion) were transfected in MCF-7 cells and LNCaP cells by use of Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Total RNA was isolated using Trizol reagent (Invitrogen). Reverse transcription (RT) was performed using SuperScript II (Invitrogen) and random hexamers according to the manufacturer's instructions. cDNAs were quantified by real-time PCR using SYBR green PCR master mix (Qiagen) and a LightCycler 480 instrument (Roche). The primers used to quantify mRNA from cells were as follows: for AR, 5′-CTCTCACTGTGGAAGCTGCAAG-3′ and 5′-TTTCCGAAGACGACAAGATGGAC-3′; for KLK2, 5′-GCGGGTTCTGACTCTTATGCT-3′ and 5′-AGTGTGGGCATGAGGACTATT-3′; and for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-GCACCGTCAAGGCTGAGAAC-3′ and 5′-TGGTGAAGACGCCAGTGGA-3′. The primers used to quantify mRNA from flies carrying the flanking genes at the 66E7 and 79E5 regions were as follows: for UGP, 5′-CACTTGACCGTGAGCGGAGA-3′ and 5′-AGATGCGCATATTGCCAGATACA-3′; for PGRP-LF, 5′-GCAGCTGGCTATTTCATGTGG-3′ and 5′-AAGCTTAAGGTGCGGGTACTTC-3′; for PGRP-LC, 5′-AATGGTGGTCCCAGCACGA-3′ and 5′-GTCCGACAGTGTTGGGCAGA-3′; for SPoCK, 5′-CGGCTGATATGATCCTGGTCAA-3′ and 5′-AAGCGCACGAAGTTTCGAATG-3′; and for rp49, 5′-CCCAAGGGTATCGACAACAG-3′ and 5′-CAATCTCCTTGCGCTTCTTG-3′.
The primers used to quantify mRNA from flies carrying the white gene were as follows: 5′-TGCACATCGTCGAACACCAC-3′ and 5′-GAGAATGGCCAGACCCACGTA-3′.
In vitro and in vivo acetylation assays.
In vitro acetylation assays were performed as described previously (8) with some modifications. The 30-μl reaction mixtures containing HAT buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 10 mM sodium butyrate), 1 μl of [14C]acetyl coenzyme A ([14C]acetyl-CoA) (Amersham) (55 mCi/mmol), 2 μl of substrate proteins, and the recombinant proteins (100 ng of p300 and 2.5 μg of GST-p300 HAT or GST-PCAF-HAT) were incubated at 30°C for 1 h. The substrate proteins, mSir2, human HP1α, and SUV39H1, were synthesized by use of an in vitro transcription/translation kit following the manufacturer's instructions. GST fusion proteins were expressed in E. coli and extracted with BC500 buffer (20 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 500 mM KCl, 20% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride) containing 1% NP-40 and purified on glutathione-Sepharose (Pharmacia). Full-length p300 protein was purified from Sf9 insect cells as previously described (30). For electrophoretic assays, the reaction mixture was subjected to the use of SDS-polyacrylamide gel electrophoresis and autoradiography (20, 39). Gels containing [14C]acetyl-labeled proteins were fixed with 10% glacial acetic acid and 40% methanol for 1 h and were enhanced by impregnation with a commercial fluorography-enhancing solution (Amplify; Amersham) for 30 min. Gels were then dried, and autoradiography was performed at −70°C for 3 days. [35S]-labeled synthesized substrate proteins were used as the inputs in the assays.
In vivo acetylation assays were performed using transfected HEK293T cells with an expression vector encoding dSir2 wt or a series of dSir2 mutation proteins together with CBP expression vector. The cell lysates were harvested and immunoprecipitated with anti-acetylated lysine antibody (Ac-K103 [catalog no. 9681[; Cell Signaling Technology).
The immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis and blotted with rabbit anti-FLAG antibody (Sigma).
SIRT1/Sir2 deacetylase fluorometric assay (HDAC assay).
SIRT/Sir2 NAD+-dependent HDAC assays were carried out as indicated in the manufacturer's protocol (Cyclex Co., Ltd.) and as previously described (13). The assay was performed with 100 μl of NAD+ HDAC buffer containing 50 mM Tris-HCl (pH 8.8), 4 mM MgCl2, and 0.5 mM DTT combined with 20 μM of a fluorescent-labeled acetylated lysine (substrate). NAD+ (Sigma) (250 μM) was added to each reaction as indicated. For inhibition experiments, 5 mM nicotinamide (Sigma) was added to the reactions. HDAC activity was obtained by measuring the fluorescence intensity emitted from a fluorescence-labeled acetylated lysine substrate with a fluorescence reader for microtiter plates. Assays were performed in triplicate.
RESULTS
dCBP corepresses AR or AR AF-1 at the pericentric heterochromatin.
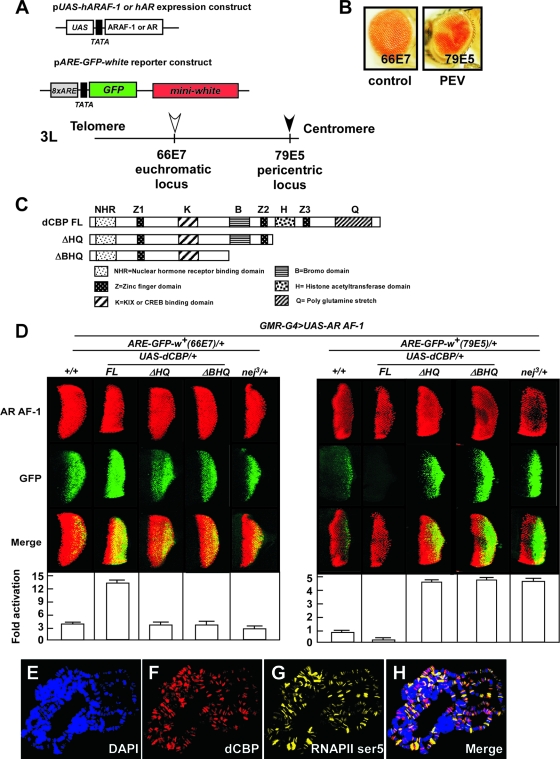
To evaluate AR function in the euchromatic and heterochromatic areas, we have developed two transgenic fly models expressing human AR or its constitutively active activation function (AF-1) domain (AR AF-1) proteins in the Drosophila eye together with an AR-dependent reporter gene (ARE-GFP-white) at euchromatic or pericentric loci (Fig. 1A). The expression constructs included AR or ARAF-1 driven by a UAS promoter. The reporter construct harbors a GFP reporter gene controlled by eight copies of AREs and a white reporter gene driven by its endogenous promoter (57). The reporter gene was randomly integrated in the various chromatin regions by a P-element mobilization scheme using the fly line carrying the reporter gene inserted in the euchromatin region as the starting line. When the reporter transgene was inserted at the pericentric region (79E5), the fly displayed a “mosaic red eye” phenotype, defined as a PEV (10, 63), whereas the control fly carrying the reporter transgene inserted in the euchromatic region (66E7) showed a uniform red eye phenotype (Fig. 1B). Inverse PCR was performed to verify the localization of the chromosomal locus of the reporter gene (see Fig. S1A in the supplemental material). Expression of a human AR or AR AF-1 was controlled by a GMR-GAL4 driver in a UAS-GAL4 misexpression system. AR or AR AF-1 was expressed in the PEV fly (pericentric model) and in the control fly (euchromatic model) (Fig. 1D). AR-induced GFP expression in the eye disc of the third instar larvae is ligand dependent (see Fig. S2 in the supplemental material). Since AR AF-1 constitutively activated the reporter gene in a ligand-independent manner, it was consequently used to analyze the modulation of AR-dependent transcriptional activity in different chromatin contexts (69).
FIG. 1.
dCBP corepresses AR AF-1-induced transactivation at the pericentric region. (A) Schematic representation of the expression and reporter constructs and cytogenetic location of the integrated reporter gene in the chromosome in newly generated PEV fly lines. The expression constructs include the human AR or ARAF-1 driven by a UAS promoter. The reporter construct harbors the GFP reporter gene controlled by eight ARE copies and the white reporter gene driven by its endogenous promoter. (B) The eye of a novel PEV fly carrying the reporter transgene (ARE-GFP-white) inserted at a pericentric region by P-element mobilization and indicated as a filled arrow in panel A (79E5) shows a mosaic phenotype (right panel). In contrast, the control fly carrying a euchromatic transgene, indicated as an open arrow in panel A (66E7), has a uniformly red eye (left panel). The insertion sites were confirmed by inverse PCR. (C) Diagram of constructs for the full-length dCBP (dCBP FL) and dCBP truncation mutants. dCBP ΔHQ was used to produce the truncated HAT domain and mutations lacking a glutamine-rich stretch; dCBP ΔBHQ was used to produce the truncated bromodomain, HAT domain, and mutations lacking a glutamine-rich stretch. Each dCBP mutant was expressed within the developing eye by the use of a UAS-GAL4 misexpression system. (D) dCBP corepresses AR AF-1-induced transactivation at the pericentric area. Fly lines carrying a gain-of-function mutation of dCBP (GMR-G4/+; UAS-dCBP FL/+), truncation mutations (GMR-G4/+; UAS-dCBP ΔHQ/+ and GMR-G4/UAS-dCBP ΔBHQ), or a loss-of-function mutation (nej3/+) were crossed to two fly models expressing AR AF-1 proteins and harboring a ARE-GFP-white reporter gene in the euchromatic or pericentric region. Expression of the AR AF-1 controlled by a GMR-GAL4 (GMR-G4) driver in the UAS-GAL4 system in the third instar larvae eye imaginala discs was assessed by immunostaining using an anti-AR antibody (upper panels). The effects of dCBP and dCBP mutations on AR AF-1-mediated transactivation were assessed by examination of GFP expression (middle panels). Merged images are shown in the lower panels. Quantification of GFP expression revealed by color intensity gradations with Adobe Photoshop (histogram) is shown at the bottom. Error bars indicate standard deviations. (E to H) Polytene chromosomes from the third instar larvae of wt flies were dissected and stained with rat polyclonal anti-dCBP antibodies (red [F]) and mouse immunoglobulin M anti-RNAPII ser5 antibodies (yellow [G]) and with DAPI to visualize DNA (blue [E]). H, merged images. dCBP is ubiquitously present on the polytene chromosome, though it is weakly stained at the centromere. RNAPII ser5 and dCBP only partially merge, revealing the presence of dCBP in nontranscribed regions.
The expression of GFP induced by AR AF-1 was clearly lower at the pericentric region (79E5) than at the euchromatic region (66E7) (Fig. 1D), suggesting that the AR AF-1 transactivation was influenced by heterochromatin-mediated silencing at the pericentric region (63). To investigate the local chromatin structure at the intact areas (66E7 and 79E5) prior to the insertion of the reporter gene, ChIP assays were performed to show an active chromatin state at the 66E7 locus and a condensed chromatin state at the 79E5 area. Our data showed that dCBP was not recruited to either area whereas dSir2 was present at the 79E5 region (see Fig. S1B in the supplemental material). We investigated whether the reporter gene insertion affected the neighboring endogenous genes flanking the transgene P elements. We analyzed expression of the genes adjacent to the 66E7 (PGRP-LC, PGRP-LF, and UGP) and 79E5 (SPoCK-RA) areas (see Fig. S1A in the supplemental material) by real-time RT-PCR experiments. The results showed that the reporter gene insertions at the 66E7 or 79E5 locus did not affect expression of the tested endogenous genes (see Fig. S1C in the supplemental material).
The transactivation function of NRs is mediated by a number of coregulators and coregulator complexes (19, 68). Some fly homologues of human NR coregulators are functionally conserved across species and share properties such as histone-modifying enzyme activity (53). Consequently, we genetically screened the coregulators to identify those modulating AR function in Drosophila flies. As expected, ectopic expression of dCBP (GMR-G4/+; UAS-dCBP/+) coactivated transcriptional activity of the AR AF-1 at the euchromatic locus. However, unexpectedly, in the pericentric area, dCBP acted as a corepressor of AR AF-1 (Fig. 1D, right panel). The similar effects of dCBP on AR-induced GFP expression were observed to occur in a ligand-dependent manner (see Fig. S2 in the supplemental material).
The transrepressive function of dCBP for AR AF-1 at the pericentric area is mediated by its C terminus, including the HAT domain.
CBP contains several functional domains, including a region that binds hormone receptors and a domain (KIX) that binds the CREB transcription factor in the N terminus. The C-terminal region harbors a bromodomain, a HAT domain, and a glutamine-rich stretch (Fig. 1C). The presence of multiple functional domains attests to the numerous mechanisms of CBP-mediated gene regulation (2).
We searched for domains that enable dCBP to transrepress AR or AR AF-1 in pericentric heterochromatin. dCBP full-length mutations (FL) and dCBP truncation mutations (dCBP ΔHQ or Δ\'42HQ) were individually introduced into the pericentric fly model. Compared with the dCBP FL, the mutations (dCBP ΔHQ or Δ\'42HQ) lacking the C terminus containing the HAT domain significantly impaired the transrepressive function of dCBP on AR AF-1 (Fig. 1D, right panel), whereas these domains appeared to mediate the dCBP coactivator function at the euchromatic locus (Fig. 1D, left panel). The C-terminal region of dCBP is therefore indispensable for transcriptional repression of AR AF-1 at the pericentric heterochromatin site in vivo.
To confirm the unexpected corepressive effect of dCBP on AR- or AR AF-1-induced transactivation, we then examined the endogenous dCBP localization on the polytene chromosome by use of immunostaining (Fig. 1F). In good agreement with a previous report (38), we observed that dCBP presented throughout the polytene chromosome, though it was less stained in the centromere. Phosphorylation of the C-terminal domain of the largest subunit of RNA polymerase II at Ser5 was proposed to occur early in the transcription cycle. Our data showed that dCBP also localized in some regions where RNAPII Ser5 was absent (Fig. 1H), suggesting that dCBP is also involved in additional processes regulating the transcription other than the initiation of transcription in active chromatin. The specificity of this antibody against dCBP could be confirmed by detecting the endogenous and exogenous dCBP (Fig. 2B).
The dCBP repression effect on heterochromatin was then analyzed by monitoring the expression levels of several heterochromatin proteins, including HP1, dSir2, and Polycomb, by Western blotting. However, the expression levels of HP1, dSir2, and Polycomb for both the wt and nej3/+ fly lines looked similar (Fig. 2A). These results suggest that the AR repression effect of dCBP for heterochromatin is not through affecting transcription levels of proteins such as HP1, dSir2, and Polycomb.
The presence of dCBP in pericentric heterochromatin associates with histone H3K9 methylation and histone H4K16 hypoacetylation.
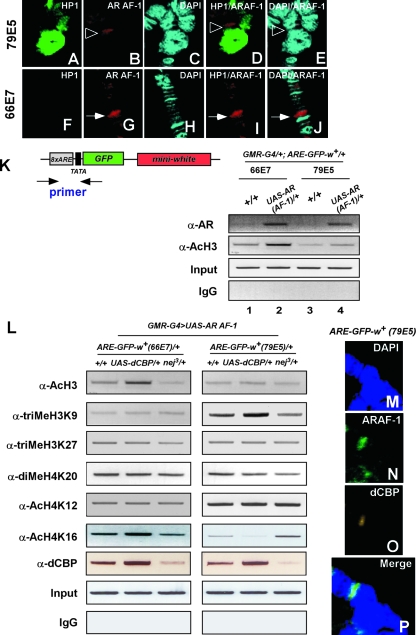
To test whether AR AF-1 indeed binds to the ARE-GFP-white reporter gene at both areas, immunostaining of polytene chromosome was performed. The AR AF-1 proteins localized at the pericentric region (79E5) (Fig. 3B) or the euchromatic (66E7) region (Fig. 3G). We then analyzed whether AR AF-1 was recruited to the promoter region of the reporter gene. A ChIP assay using anti-AR (N-20) showed that AR AF-1 was recruited to the promoters of the AR-dependent reporter gene (Fig. 3K). These results suggest that dCBP corepresses AR or AR AF-1 at the pericentric area.
FIG. 3.
dCBP in pericentric heterochromatin stimulates histone H3K9 methylation and deacetylation of histone H4K16. (A to J) Polytene chromosomes from the third instar larvae of pericentric (A to E) or euchromatic (F to J) models using flies carrying a UAS-AR AF-1 expression plasmid, a Blk-GAL4 driver gene, and the reporter transgene were dissected and stained with mouse polyclonal anti-HP1 antibodies (green [A and F]) and rabbit polyclonal anti-AR (N-20) antibodies (red [B and G]) and with DAPI to visualize DNA (blue [C and H]). In the pericentric fly model, the AR AF-1 protein was detected at the 79E5 locus, which was adjacent to the pericentric region stained by HP1 (B and D), whereas in the euchromatic fly model, the localization of the AR AF-1 protein is at the 66E7 locus, exactly at the insertion location of ARE-GFP-white reporter gene (G and J). The localization of AR AF-1 at the 79E5 area is indicated with open arrowheads, and, for the 66E7 area, with filled arrows. (K and L) A ChIP assay was performed using eye discs from the third instar larvae of the progeny with the indicated antibodies. The precipitated chromatin was amplified by PCR using primers flanking the promoter region of the reporter gene as indicated (K, upper left panel). (K) Flies carrying the ARE-GFP-white reporter gene inserted in different chromatin regions (66E7 or 79E5) were crossed to a wt fly (+/+) or to flies carrying a UAS-AR AF-1 expression construct. A ChIP assay was performed using the progeny. IgG, immunoglobulin G. (L) The flies used for the euchromatic and pericentric models represented in Fig. 1D were crossed to wt (+/+), UAS-dCBP/+, or nej3/+ flies. A ChIP assay was performed using progeny with the antibodies as indicated. (M to P) Polytene chromosomes from the third instar larvae of the pericentric fly line were dissected and stained with DAPI to visualize DNA (blue [M]) or with rabbit polyclonal anti-AR (N-20) antibodies (green [N]) or newly generated rat polyclonal anti-dCBP antibodies (yellow [O]). In the pericentric fly line, dCBP colocalizes with AR AF-1 at the 79E5 locus.
We then asked how dCBP influences histone modifications at pericentric heterochromatin. To this end, we first examined the acetylation level of histone H3 on the reporter (ARE-GFP-white) transgene inserted in different chromatin contexts by use of a ChIP assay. As shown in Fig. 3K, acetylation of histone H3 was detected on the reporter transgene located at the euchromatic locus (66E7). The acetylation was enhanced with binding of AR AF-1 (Fig. 3K, lanes 1 and 2). In contrast, the histone H3 was hypoacetylated at the pericentric area (79E5). Binding of AR AF-1 nevertheless slightly increased the level of acetylation even in the heterochromatic locus (Fig. 3K, lanes 3 and 4). These results are in good agreement with the finding that AR AF-1 transactivation was lower at the pericentric area than that seen at the euchromatic locus.
Next, we analyzed the effect of dCBP on histone modifications by use of GMR-G4/+; UAS-dCBP/+ and dCBP (nej3/+) mutants (1, 64) in our euchromatic and pericentric models (Fig. 3L). Although H3K27 trimethylation (46), H4K20 dimethylation (50), and H4K12 acetylation (56) in GMR-G4/+; UAS-dCBP/+ or nej3/+ did not differ from the results seen with the wt flies (+/+) at both chromatin sites, H3K9 trimethylation was prominently increased in GMR-G4/+; UAS-dCBP/+ mutants and decreased in nej3/+ mutants at the pericentric area (79E5) but not at the euchromatic region (66E7).
Though acetylation of histone H4K16 in GMR-G4/+; UAS-dCBP/+ mutants was enhanced and in nej3/+ mutants was decreased at the euchromatic area as expected (33), interestingly, H4K16 acetylation (47) was significantly decreased in GMR-G4/+; UAS-dCBP/+ mutants and increased in nej3/+ mutants at the 79E5 locus. From these results, it seems likely that dCBP induces gene silencing through stimulation of histone modifications characteristic of repressed chromatin.
A ChIP assay was then performed to confirm that dCBP was recruited to the AR-dependent reporter gene at both the euchromatic and pericentric loci (Fig. 3L). Immunostaining in polytene chromosome experiments further showed that dCBP located together with AR AF-1 at the pericentric region (79E5) (Fig. 3M to P).
dCBP genetically interacts with dSir2 in Drosophila flies; the transrepressive function of dCBP at the pericentric locus requires dSir2 activity.
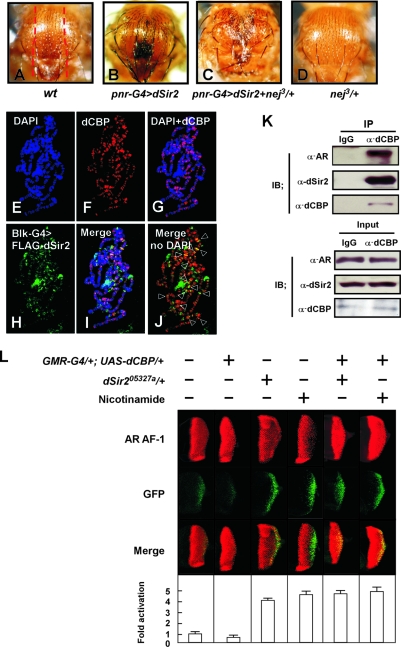
The fact that dCBP has been implicated as a cofactor for repression of transcription in pericentric heterochromatin suggests that dCBP may play a corepressor role for NR by associating with other proteins important for transcription repression. Our data further showed that the inhibitory effects of dCBP on AR AF-1-induced transactivation at the pericentric heterochromatin were coupled with increased trimethylation of histone H3K9 (52) and deacetylation of histone H4K16 (9) (Fig. 3L), and these histone modifications have been reported to be modulated by SIRT1 in human cells (62). The members of Sir2 family are conserved from Saccharomyces species to humans (17). dSir2 is a NAD+-dependent HDAC and plays a role in heterochromatic silencing as a suppressor of PEV (6, 38, 43, 47). By two-hybrid screening, it has been previously shown that dSir2 interacts with dCBP (38). Thus, we tested whether there was a genetic interaction between dCBP and dSir2 in Drosophila flies. The notum of the adult fly contains a regular pattern of small (microchaetae) and large (macrochaetae) dorsal bristles (Fig. 4A). Overexpression of the dSir2 transgene (UAS-dSir2) in the developing notum by use of a GAL4 insertion in the pannier (pnr) gene (44) resulted in a dominant alteration of bristle formation. dSir2 altered the distribution of both micro- and macrochaetae and caused defects in the midline of the notum, scutellum, and abdomen (Fig. 4B). Expression of UAS-dSir2 with the same GAL4 driver in nej3/+ mutants induced a severe phenotype (Fig. 4C), whereas pnr-GAL4 alone in nej3/+ mutants had no effect on the notum (Fig. 4D). This supports a potentially genetic interaction of dCBP with dSir2 in Drosophila flies.
FIG. 4.
Transrepressive action of dCBP at the pericentric locus requires dSir2. (A to D) Photomicrographs of the adult dorsal thorax from wt (A), UAS-dSir2/+; pnr-GAL4/+ (B), nej3/+; UAS-dSir2/+; pnr-GAL4/+ (C), and nej3/+; pnr-GAL4/+ (D) flies. (B) Ectopic expression of UAS-dSir2 with the pnr-GAL4 driver induced an alteration in the distribution of microchaetae (small bristles) and macrochaetae (large bristles), particularly the dorsocentrals and scutellars. (C) Expression of UAS-dSir2 with the pnr-GAL4 in nej3/+ mutants induced a stronger phenotype than that of either alone. (D) nej3/+ mutants with pnr-GAL4 did not show an alteration in the distribution of micro- and macrochaetae. The red dashed lines in panel A show the approximate area where the phenotype is located in panels A to D. (E to J) Polytene chromosomes from the third instar larvae of flies carrying a UAS-FLAG-dSir2 transgene with a BLK-GAL4 driver were dissected and stained with DAPI to visualize DNA (blue [E]) or with rat polyclonal anti-dCBP antibodies (red [F]) or rabbit polyclonal anti-FLAG antibodies (green [H]) to visualize dSir2 tagged with FLAG. (J) The regions in chromosomes showing the colocalization of dCBP and FLAG-dSir2 are indicated with open arrowheads. (K) dCBP physically interacts with both dSir2 and AR AF-1 in flies carrying the reporter gene at the pericentric region. Whole-cell protein extracts from flies expressing AR AF-1 in the eye by the use of a UAS-GAL4 system and carrying an ARE-GFP-white reporter in the pericentric heterochromatin (pericentric fly model) were immunoprecipitated (IP) with antibody against dCBP or control preimmune immunoglobulin G (IgG). Fractions (5%) of the input material (lower panels) and the precipitated proteins (upper panels) were analyzed by Western blotting using anti-dSir2 (dF-16), anti-AR (N-20), and anti-dCBP antibodies as indicated. IB, immunoblot. (L) Flies from the pericentric fly model line expressing ARAF-1 proteins and carrying an ARE-GFP-white reporter gene in the pericentric region were crossed to flies carrying UAS-dCBP alone or together with a dSir2 loss-of-function mutation (dSir205327a/+) as indicated. Overexpression of AR AF-1 in the eye disc of the third instar larvae was assessed by immunostaining using an anti-AR antibody (upper panels). The effect of dCBP or loss of function of dSir2 on AR AF-1-mediated transactivation was assessed by examination of GFP expression (middle panels). Merged images are shown in the lower panels. The effect of dSir205327a was further confirmed by treatment with nicotinamide (Sir2 inhibitor) (20 mM).
Immunostaining of polytene chromosomes from the third instar larvae carrying the UAS-FLAG-dSir2 transgene showed that endogenous dCBP ubiquitously diffused in active or condensed chromatin regions (Fig. 4E to G). dCBP and overexpressed dSir2 colocalized in parts of polytene chromosomes, including the pericentric regions (Fig. 4I and J). To examine the physical association of dCBP with dSir2 and AR AF-1, we performed IP experiments using the pericentric fly model. As shown in Fig. 4K, both the AR AF-1 and dSir2 proteins were detected in the dCBP precipitate, suggesting that dCBP physically associates with dSir2 and AR AF-1.
We then set out to investigate whether dSir2 activity is necessary for the dCBP function that is repressive for AR AF-1. To this end, we examined how a loss-of-function mutation of dSir2 affects dCBP-mediated AR AF-1 transrepression in vivo by use of the pericentric model. The dSir2 locus is located on the second chromosome (2L) at cytological position 34A and encodes a single transcript with one intron (47). dSir205327a is the result of a PZ element insertion within the dSir2 mRNA at position +14 (47). In good agreement with our ChIP assay results (Fig. 3L) and genetic and biochemical interaction studies (Fig. 4C and K), dSir2 mutation (dSir205327a/+) or nicotinamide (dSir2 inhibitor) treatment blocked the corepressive activity of dCBP (Fig. 4L; also see Fig. S3A in the supplemental material). From these results, it appears that dCBP overexpression in the setting of a dSir2 mutation failed to transrepress AR AF-1- or AR-mediated transactivation, indicating that dSir2 is required for the transrepressive function of dCBP at pericentric loci.
The pattern of recruitment of AR, dCBP, and dSir2 on the promoter of AR-dependent reporter gene at euchromatic and heterochromatic loci.
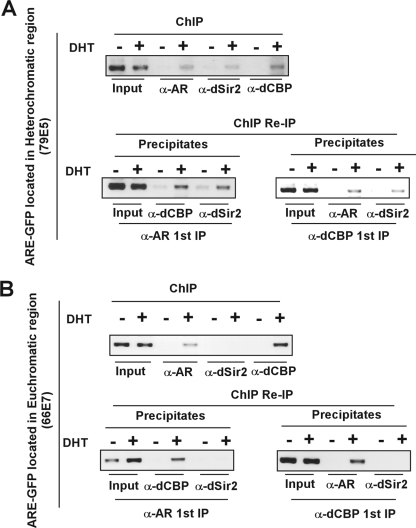
The results detailed above showed that dCBP genetically and physically interacted with dSir2 in vivo. We next asked whether dCBP, dSir2, and AR form a complex in different chromatin contexts. Double consecutive ChIP assays were performed in eye discs of third instar larvae from euchromatic or pericentric fly models. To investigate whether dCBP, dSir2, and AR are recruited to the promoter of a AR-dependent reporter gene inserted into euchromatic or heterochromatic loci, ChIP assays were first performed with antibodies against one of the proteins in the soluble chromatin derived from DHT-treated or untreated flies. Then, the precipitates from ChIP with anti-AR or -dCBP antibodies were subjected to re-IP (via ChIP) with antibodies against a second protein.
As shown in Fig. 5, the presence of all three proteins (dCBP, dSir2, and AR) at the promoter of the ARE-dependent reporter gene was detected at the heterochromatic locus (Fig. 5A upper panel), whereas dSir2 was absent at the promoter at the euchromatic area (Fig. 5B). Consistently, dSir2 was found in the secondary ChIP immunocomplexes in experiments using the heterochromatin model flies (Fig. 5A, lower panel). These results suggest that dCBP, dSir2, and AR might form a functional unit in the condensed chromatin context in the presence of DHT. However, because our system carries an AR-responsive reporter gene artificially inserted in the heterochromatic region, these findings could not exclude the possibility that this may also take place at euchromatic loci.
FIG. 5.
AR, dCBP, and dSir2 are predominantly recruited to the ARE at heterochromatic loci in the presence of androgen. Larvae from flies expressing AR protein and harboring an ARE-GFP-white reporter transgene at heterochromatic loci (A) or euchromatic loci (B) were grown on the media with vehicle or DHT (10−5 M) for 5 days. ChIP assays and ChIP/re-IP experiments were performed using specific antibodies against AR, dCBP, and dSir2 as indicated.
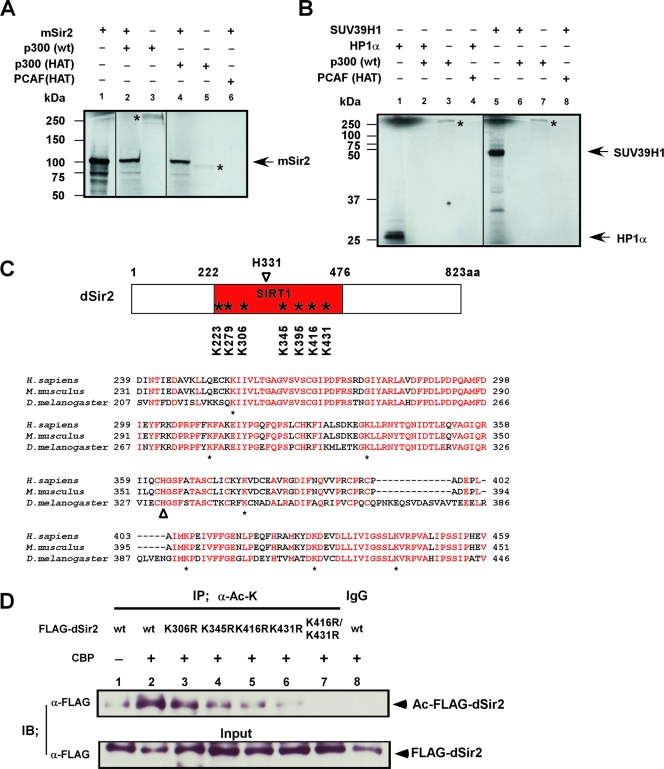
CBP acetylates Sir2 in vitro and in vivo at the conserved HDAC catalytic domain of Sir2.
Since the C-terminal dCBP region harboring the HAT domain was necessary to suppress AR AF-1 transactivation at the pericentric site (Fig. 1D), we reasoned that dCBP facilitated repression of transcription of genes inserted in heterochromatin regions through acetylation of the proteins. We performed an in vitro histone acetylation assay using recombinant dCBP human homologue (p300), p300/CBP (HAT), or PCAF (HAT) proteins and synthesized mSir2, HP1α, or Suv39H1 substrate proteins (Fig. 6A and B). These substrate proteins are thought to play key roles in the maintenance of heterochromatin structure. Nicotinamide was added to the acetylation reaction to prevent possible autodeacetylation. As is consistent with the results showing that dCBP interacts with dSir2 (Fig. 4C and K), p300 or p300/CBP (HAT) clearly acetylated Sir2 protein (Fig. 6A) but not HP1α and Suv39H1 protein (Fig. 6B). Thus, Sir2 appeared to be an acetylation substrate for p300/CBP in vitro.
FIG. 6.
dSir2 is acetylated by dCBP in vivo and in vitro at its conserved HDAC catalytic core region. (A and B) dCBP human homologue (p300) or p300/CBP HAT acetylates Sir2 in vitro. The p300, GST-P300/CBP HAT, and GST-PCAF HAT proteins were subjected to in vitro acetylation assays with in vitro-synthesized substrate proteins, including mSir2, human HP1α, and SUV39H1. Reaction products were analyzed by autoradiography (14C). The autoacetylated p300 or p300/CBP HAT is indicated with a star. Fractions of 35S-labeled in vitro-synthesized substrate proteins (2.5%) were used as the input (panel A, lane 1; panel B, lanes 1 and 5). (C) (Upper panel) Schematic representation of Sir2 orthologues in Drosophila flies (dSir2); the conserved lysine sites at its catalytic core regions (SIRT1 domain) are indicated with stars. H331 is a well-defined catalytic site of dSir2 (open arrowhead). (Lower panel) Sequence alignment of the conserved core region with the SIRT domains from several species as indicated. Conserved residues with 100% identity are indicated with red characters. Below the alignment, conserved lysine (K) residues are labeled (stars); histidine (H) residues (marked with an open arrowhead) represent a well-defined catalytic site of Sir2. (D) 293T cells were transfected with FLAG-dSir2 wt or a series of FLAG-dSir2 mutation expression plasmids together with CBP expression vector as indicated. The cell lysates were immunoprecipitated (IP) with anti-acetylated lysine antibody (α-Ac-K [mouse monoclonal]) or with control preimmune mouse immunoglobulin G (IgG). Precipitated proteins and fractions (5%) of the input cell lysates were analyzed by immunoblotting (IB) with anti-FLAG antibody.
To map the acetylation sites on dSir2 protein, we generated a series of dSir2 mutants. dSir2 encompasses seven lysine residues within its highly conserved catalytic HDAC core region. We then point mutated these lysine residues to arginine (Fig. 6C). Luciferase assays were performed to test the corepressive effect of FLAG-dSir2 mutants on AR-induced transactivation. Since the mutants (K223R, K279R, and K395R) showed similar corepressive effects to that seen with wt dSir2 (data not shown), these mutants were not used for the additional experiments.
In vivo acetylation assays were performed using 293T cells to identify the acetylation sites in dSir2. Cells were transfected with FLAG-dSir2 wt or a series of FLAG-dSir2 mutant expression plasmids together with CBP expression vector. In anti-acetylated lysine IP experiments, we showed that dCBP was able to acetylate dSir2 in vivo (Fig. 6D, lane 2) and that the acetylation of dSir2 by CBP was slightly reduced with the dSir2 K345R mutation (Fig. 6D, lane 2 and 4) and severely reduced with either the dSir2 K416R or the K431R mutation (Fig. 6D, lanes 2, 5, and 6) but did not change with the dSir2 K306R mutation. Moreover, a double mutation, dSir2 K416R/K431R, resulted in the loss of acetylation of dSir2 by CBP (Fig. 6D, lane 7). These results show that dSir2 is acetylated by dCBP in vivo at its HDAC catalytic core region, suggesting that acetylation of Sir2 by CBP may be important for Sir2-mediated HDAC activity.
Acetylation of dSir2 by dCBP influences dSir2 HDAC function, and dCBP corepressive action requires acetylation of dSir2.
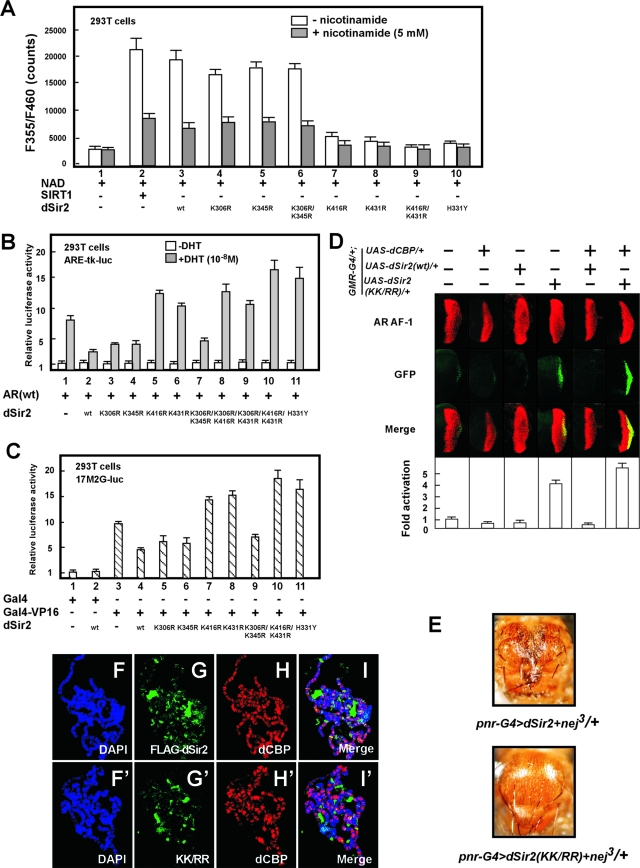
To test this activity, we constructed an expression vector encoding a dSir2 mutation in which a conserved histidine (H331) residue in the catalytic domain was mutated into tyrosine (H331Y) (Fig. 6C). Such a mutation in the catalytic domain of mSir2 has been shown to abolish the HDAC activity (12). This inactive dSir2 mutation did not show NAD+-dependent dSir2 HDAC activity (Fig. 7A, lane 10). We further measured the HDAC activity of a series of dSir2 mutants (K306R, K345R, K306R/K345R, K416R, K431R, and K416R/K431R). In similarity to the effect seen with the recombinant SIRT1 (dSir2 human homologue), nuclear extracts from the cells expressing wt dSir2 showed a prominent enhancement of a NAD+-dependent HDAC activity, and such activity was inhibited by nicotinamide (SIR2 inhibitor) (Fig. 7A, lanes 1 to 3). Significantly, dSir2 HDAC activity was clearly impaired by the acetylation site mutations of dSir2 at K416 or K431 or at both lysines (K416/K431) (Fig. 7A, lanes 3 and 7 to 9) but not at K306, K345, or K306/K345 (Fig. 7A, lanes 3 to 6). Furthermore, the attenuation of the HDAC activity in dSir2 acetylation site mutants (K416R, K431R, or K416R/K431R) was similar to that seen with the inactive dSir2 catalytic mutant (H331Y). Taken together, these results demonstrate that acetylation of dSir2 at K416 and K431 by CBP is required for the HDAC domain catalytic activity of dSir2.
FIG. 7.
dCBP corepressor function requires acetylation of dSir2. (A) 293T cells were transfected with equivalent amounts of wt dSir2 and dSir2 mutant expression constructs as indicated. The nuclear extracts from the transfected cells were tested for Sir2 NAD+-dependent HDAC activity in the absence (open columns) or presence (filled columns) of 5 mM nicotinamide. (B and C) dSir2 suppresses Gal4-VP16- and AR-mediated transactivation, and defective acetylation of dSir2 impairs its repression activity. pcDNA3-dSir2 H331Y represents an inactive catalytic mutation. The relative luciferase units shown represent the mean values of the results of triplicate experiments. (B) 293T cells were cotransfected with plasmids expressing the AR together with dSir2 wt or dSir2 mutations as indicated. (C) 293T cells were cotransfected with a series of plasmids, including 0.2 μg of 17M2Gluc (lanes 1 to 11), 0.02 μg of pPM containing Gal4-DBD (Gal4) (lanes 1 and 2), 0.02 μg of pPM-VP16 containing Gal4-DBD-VP16 (Gal4-VP16) (lanes 3 to 11), 0.1 μg of pcDNA3 (lanes 1 and 3), and 0.1 μg of pcDNA3-dSir2 (wt) or pcDNA3-dSir2 mutants as indicated. (D) The dCBP corepressive function requires the acetylation of dSir2. Flies of the pericentric model used as described for Fig. 3L were individually crossed to GMR-G4/+; UAS-dCBP/+ or GMR-G4/+; UAS-dSir2 (wt)/+ or GMR-G4/+; UAS-dSir2 K416R/K431R/+ fly lines as indicated. Analysis of AR AF-1 transactivation was performed on the eye disc of the third instar larvae from the progeny as described for Fig. 3L. (E) Photomicrographs of the adult dorsal thorax from nej3/+; UAS-dSir2/+; pnr-GAL4/+ flies (upper panel) and nej3/+; UAS-dSir2KK/RR/+; pnr-GAL4/+ flies (lower panel). Ectopic expression of UAS-dSir2 with the pnr-GAL4 driver in nej3/+ flies induced a severe alteration in the distribution of microchaetae (small bristles) and macrochaetae (large bristles), particularly the dorsocentrals and scutellars (upper panel). Overexpression of UAS-dSir2KK/RR in nej3/+ flies did not alter the phenotype of bristle formation on the developing notum (lower panel). (F to I′) The acetylation sites of dSir2 are required for dSir2 localization in polytene chromosomes. Polytene chromosomes from the third instar larvae of flies carrying a UAS-FLAG-dSir2 or UAS-FLAG-dSir2KK/RR (KK/RR) transgene with the BLK-GAL4 driver were dissected and stained with DAPI (blue) (F and F′), with rabbit polyclonal anti-FLAG to visualize dSir2 tagged with FLAG (green) (G and G′), or with rat polyclonal anti-dCBP (red) (H and H′). Merged images, I and I′.
We then asked whether the acetylation of dSir2 is required for repression of transcription. To this end, we examined how mutations of dSir2 in its acetylation sites affect AR-mediated transactivation in 293T cells. As shown in Fig. 7B, dSir2 wt suppressed ligand-induced AR transactivation (lane 1 and 2). In good agreement with our HDAC assay experiments performed using 293T cells (Fig. 7A), mutations of dSir2 at K416 or K431 or at both lysines (K416/K431) significantly abrogated the repressive action of dSir2. As is consistent with these results, we also showed that acetylation site mutations of dSir2 (K416R, K431R, or K416R/K431R) abolished the repressive action of dSir2 for general transcriptional machinery-mediated transactivation (Gal4-VP16 system) (Fig. 7C, lanes 4, 7, 8, and 10). These results suggest that acetylation of dSir2 is indispensable for dSir2 HDAC activity.
In order to further determine whether the acetylation of dSir2 is necessary for the repressive function of CBP in AR transactivation in vivo, we generated transgenic flies expressing a wt dSir2 (dSir2 wt) or dSir2 K416R/K431R (dSir2 KK/RR) mutation lacking HDAC activity (Fig. 7D). In agreement with our results obtained with luciferase reporter assays, overexpression of dSir2 alone suppressed AR AF-1 transactivation in the pericentric fly model (Fig. 7B and C), and the dSir2 KK/RR mutation alone impaired the repression activity of dSir2. In addition, dSir2 KK/RR in the setting of GMR-G4/+; UAS-dCBP abolished the dCBP transrepressive function for the AR AF-1 transactivation (Fig. 7D) or AR transactivation (see Fig. S3B in the supplemental material). Taken together, these findings demonstrate that dSir2 may act as a mediator of the dCBP corepressor function for pericentric heterochromatin.
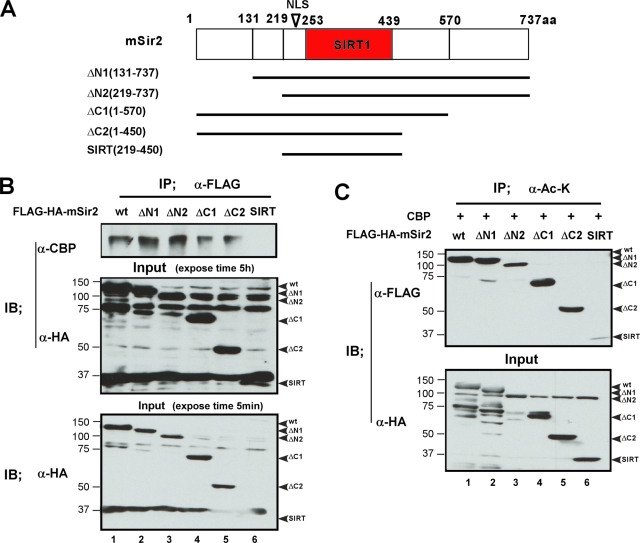
We then asked whether the acetylation of Sir2 by CBP is mediated by the interaction between CBP and Sir2. To address this issue, we made a series of mSir2 truncation mutants and performed IP experiments and an in vivo acetylation assay in parallel (Fig. 8A to C). The results showed that both the N and C termini of mSir2 appeared to be required for the association of mSir2 and CBP and that the acetylation of mSir2 by CBP was decreased when both the N-terminal mSir2 and the C-terminal mSir2 were truncated (Fig. 8C, lane 6). Thus, these data suggest that the interaction between CBP and Sir2 is necessary for the acetylation of Sir2 by CBP.
FIG. 8.
The acetylation of mSir2 by CBP is mediated by the association between mSir2 and CBP. (A) Schematic representation of mSir2 and its truncation mutations. (B) Anti-FLAG IP was performed on extracts from 293T cells expressing FLAG-HA-mSir2 or its truncation mutations. Immunoprecipates (IP) prepared using anti-FLAG and whole-cell extracts (Input) were analyzed by immunoblotting using the indicated antibodies. Binding of CBP to mSir2 fragments is lost when both the N and C termini of mSir2 are truncated (SIRT) (lane 6), suggesting that both the N and C termini of mSir2 are required for the association between mSir2 and CBP. (C) 293T cells were cotransfected with FLAG-HA-mSir2 wt or a series of truncation mutation expression plasmids together with CBP expression vector as indicated. The whole-cell lysates were immunoprecipitated with anti-acetylated lysine antibody (α-Ac-K). Precipitated proteins and fractions (5%) of the input cell lysates were analyzed by immunoblotting (IB) with anti-FLAG or anti-HA antibodies.
Having established that the functional association between dSir2 and dCBP is required for suppression of AR AF-1 transactivation in pericentric heterochromatin, we investigated whether this association also plays a role in dorsal bristle formation of Drosophila flies. As shown in Fig. 4C, overexpression of UAS-dSir2 wt with the pnr-GAL4 driver in the setting of the nej3/+ mutation induced a severe alteration of dorsal bristle formation. We further examined the effect of dSir2 KK/RR in nej3/+ on the developing notum. Supporting these observations, overexpression of UAS-dSir2 KK/RR in nej3/+ mutants did not alter dorsal bristle formation on developing notum (Fig. 7E). These in vivo data strongly suggest that the biological function of dSir2 might be mediated through its K416/K431 acetylation by CBP.
To investigate the role of dSir2 acetylation in the accumulation of dSir2 on the polytene chromosome, immunostaining experiments were performed using transgenic flies harboring UAS-FLAG-dSir2 or UAS-FLAG-dSir2 KK/RR (KK/RR) with a BLK-GAL4 driver. The staining of ectopically expressed dSir2 showed that dSir2 was predominantly localized in the heterochromatic areas, although dSir2 was also present in some euchromatic regions (Fig. 7G). The accumulation of dSir2 on the polytene chromosome was defective in the KK/RR mutant (Fig. 7G′), suggesting that acetylation of dSir2 is potentially essential for the localization of dSir2 on the chromosomes. Importantly, the localization of dCBP was not altered in flies carrying dSir2 KK/RR mutations (Fig. 7H′).
Involvement of CBP or SIRT1 in regulation of endogenous AR target gene expression.
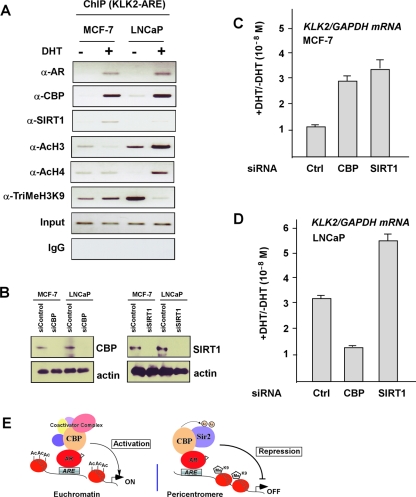
Having shown in our Drosophila model system that dCBP exerts its corepressive function on AR- or AR AF-1-induced transactivation at the pericentric locus, next we set out to confirm its transrepressive function in a mammalian experimental cell model. After searching the cell lines in which AR target gene promoters were in an inactive chromatin state, we chose the MCF-7 cell line (human breast cancer cell line) as a model with an AR-dependent gene (KLK2) in an apparently repressed state. Real-time RT-PCR experiments showed that in MCF-7 cells, AR and KLK2 genes were expressed at low levels, but their expression was not altered in response to the stimulation by DHT, whereas expression of the KLK2 gene was strongly induced by the treatment with DHT in LNCaP cells (see Fig. S4A in the supplemental material). We then asked how the chromatin structure is situated on the promoter of the endogenous AR target gene (KLK2 promoter) in both MCF-7 and LNCaP cell lines. ChIP assays were performed with antibodies recognizing chromatin state markers (Fig. 9A). The results showed an inactive chromatin state at the KLK2 promoter in MCF-7 cells, whereas in LNCaP cells, the chromatin state of the KLK2 promoter appeared to be in an active state. Furthermore, in agreement with the ChIP re-IP results presented in Fig. 5, CBP, SIRT1, and AR were recruited to the KLK2 promoter in MCF-7 cells (Fig. 9A). However, such DHT-induced SIRT1 recruitment was not clearly observed in the KLK2 promoter in LNCaP cells treated with DHT.
FIG. 9.
Expression of endogenous AR target genes is regulated by CBP and Sir2. (A) ChIP assays with the indicated antibodies and PCR at the promoter of KLK2 gene were performed using MCF-7 and LNCaP cells treated with DHT for 4 h or left untreated. IgG, immunoglobulin G. (B) Western blotting measurement of the expression levels of CBP and SIRT1 in MCF-7 and LNCaP cells after transfection with the indicated siRNA. (C and D) MCF-7 (C) and LNCaP (D) cells were transfected with the indicated siRNA and treated with DHT (10−8 M) or left untreated. Induction of KLK2 expression is expressed as the ratio of KLK2 mRNA levels normalized to GAPDH levels between cells treated with vehicle or DHT. (E) Schematic illustration of CBP coregulator functions depending on the chromatin contexts. Based on the chromatin states, various transcription factors or coregulator complexes may be recruited to perform different functions in the euchromatic and pericentric regions. In the heterochromatic context, CBP and Sir2 recruited to AR target genes suppress the AR-mediated transactivation. A possible explanation for the repressive effect of CBP at the condensed environment is that CBP contributes to epigenetic silencing by acetylating chromatin proteins, such as Sir2, rather than histone. The CBP-mediated acetylation enhanced Sir2 HDAC activity and led to consecutive histone deacetylation.
We next examined whether endogenous CBP or SIRT1 is required for the AR-dependent gene activation in vivo. For this, we analyzed the androgen-induced expression of the endogenous AR target gene (KLK2) after knockdown of CBP or SIRT1 by siRNA assays using MCF-7 or LNCaP cells (Fig. 9B). The increase in induction of the KLK2 gene by DHT treatment of MCF-7 cells transfected with control siRNA was only marginal at 1.2-fold but was significantly enhanced when CBP (2.9-fold) or SIRT1 (3.3-fold) (Fig. 9C) was knocked down. On the other hand, in LNCaP cells, the increase in induction of KLK2 expression by DHT induction was 3.2-fold (siRNA) but was significantly (1.3-fold) reduced by CBP knockdown. SIRT1 knockdown enhanced the induction of KLK2 expression by the DHT treatment (5.3-fold) (Fig. 9D). These results again supported the idea that CBP and SIRT1 are recruited together to the promoters of AR target genes in response to the presence of androgen.
We further analyzed the alteration of histone modification with the recruitment of each protein (AR, CBP, or Sir2) in the silent and active chromatin context by means of ChIP assay experiments using cultured mammalian cells (MCF-7 and LNCaP cells) and flies (see Fig. S5A and B in the supplemental material). From the results of the knockdown assay using siRNA in the transcriptionally silent AR target gene (KLK2) promoter in MCF-7 cells (see Fig. S5A in the supplemental material), Sir2 and CBP appeared to be required for chromatin inactivation, as indicated by enhanced histone H3 K9 methylation and deacetylation of histone H4 K16. On the other hand, CBP was indispensable for activating histone modification in the transcriptionally active gene promoter in LNCaP cells (see Fig. S5A in the supplemental material). Similar roles of dCBP and dSir2 in epigenetic alteration were observed in the results obtained with flies (see Fig. S5B in the supplemental material).
DISCUSSION
The AR or AR AF-1 is transcriptionally functional at a pericentric heterochromatic site.
We established a Drosophila experimental system by introducing the AR-dependent reporter gene carrying the ARE at the euchromatic or pericentric locus. The AR or AR AF-1 potently activated transcription through the ARE in the both regions. The integrated ARE is composed of a palindromic element of the two 5′-AGGTCA-3′ core motifs. It binds the AR or AR AF-1 homodimer. It is unlikely that endogenous fly NRs activate transcription through the ARE, since insect endogenous NRs bind to directly repeated motifs as heterodimers (59). AR AF-1 effectively activated transcription through the ARE at the euchromatic locus, suggesting that endogenous fly coregulators facilitate its transcriptional activation. At the pericentric heterochromatin site, AR AF-1 was also transcriptionally active but its transcriptional activity was lower. Apparently, given the observed transcriptional activity, AR AF-1 potently bound to the ARE, conceivably through chromatin reorganization. Thus, the AR DBD recognizes and stably binds to the ARE even when it is located within a heterochromatic region. The classes of ATP-dependent chromatin remodeling complexes that assist this binding remain to be identified. These findings confirm that AR is able to reach and anchor to its specific target DNA sequences even in the inactive areas of chromatin.
Sir2 HDAC activity is required for the transrepressive function of dCBP on AR-mediated transactivation at the pericentric heterochromatin site.
Sir2 is the most evolutionarily conserved HDAC, with homologs in animals, plants, fungi, and bacteria. Sir2 is a NAD+-dependent deacetylase that targets histone H4 at lysine 16 (62) and facilitates increases in levels of histone H3K9me3 (52, 61). dSir2 mutations have been shown to be required for heterochromatic silencing (47), to perturb PEV (38), and to disrupt silencing of a mini-white reporter transgene mediated by a Polycomb response element (21). We have demonstrated here that the recruitment of dCBP to the promoter region of the transgenic AR reporter gene at the pericentric locus leads to hypoacetylation of histone H4K16 and hypermethylation of histone H3K9 at the same chromatin area. Since dSir2 has been identified as a dCBP-interacting protein (38), it was tempting to speculate that a functional link between dSir2 and dCBP may exist.
In the present study, we detected dSir2 in the dCBP immunocomplexes precipitated from Drosophila tissues. A potential genetic interaction between dCBP and dSir2 in the developing notum was also detected using a pnr-GAL4 driver. Moreover, we showed that acetylation of Sir2 by CBP was necessary for Sir2 HDAC activity and the accumulation of dSir2 on the polytene chromosome. In the dSir2 acetylation site mutant (dSir2 K416R/K431R) lacking dSir2 HDAC activity, the dCBP corepressor activity at the pericentric site was lost. The coactivator activity of dCBP at the euchromatic site, however, was unaltered. Thus, the corepressor function of dCBP for AR-dependent transactivation at the pericentric locus appears to be dependent on its HAT activity and associates with dSir2 HDAC activity. However, it is unclear how dSir2 is recruited to dCBP at the pericentric heterochromatin area, and the mediators between dSir2 and dCBP at the pericentric site need to be further identified.
SIRT1, a human homologue of Sir2, has been identified as a DHT-dependent corepressor of AR, and SIRT1 repression of DHT-induced AR transactivation requires the deacetylation of AR by SIRT1 on the LBD of AR (18). In the present study, however, we used a constitutively active AR AF-1 mutant lacking the LBD, and in in vitro assays, deacetylation of the AR AF-1 by Sir2 was not seen in our study (data not shown), suggesting that the deacetylation of histones rather than of AR AF-1 is necessary for the repressive effect of dSir2 on AR AF-1 in our Drosophila experimental system.
Moreover, in our Drosophila experimental models carrying the reporter gene at euchromatic or heterochromatic loci, the results obtained in double ChIP assays suggest that AR, dCBP, and dSir2 mainly form a functional unit at the heterochromatic area, though these results do not exclude the possibility that this may also take place at euchromatic loci. To test whether such association occurs in mammalian cells, we set out to look for the cell lines with silent AR-responsive genes. We chose the MCF-7 cell line as a cell model in which AR target genes are in an inactive chromatin state even in the presence of androgen. Conversely, the LNCaP cell line was taken as a cell model in which AR target genes are in an active chromatin conformation. Using these two cell lines with functionally different states of ARE-dependent genes, we showed, following the treatment with DHT, that AR, CBP, and SIRT1 were predominantly recruited to the promoter of transcriptionally silent AR-responsive genes in MCF-7 cells, whereas SIRT1 did not occupy the ARE on the promoter of AR target genes in LNCaP cells. These observations support the recent report that the function of AR in the form of the binding of an AR antagonist mediates transrepression of SIRT1 and NCoR through an AR-responsive promoter in LNCaP cells (11). Thus, it appears that in addition to the results seen with the Drosophila experimental system, the functional link between AR, CBP repression function, and SIRT1 activity is present in mammalian cells.
Dual actions of CBP in transcriptional regulation.
CBP/p300 is a well-established coactivator for several classes of sequence-specific transcriptional factors, including NRs. However, CBP/p300 also transrepresses transcription through protein acetylation by CBP/p300 (16, 26). As is consistent with previous findings (55), a molecular dissection of p300 recovered the presence of a transrepressive domain. This domain is sumoylated to associate with HDAC6 for transrepression (23). In an elegant approach using in vitro chromatin transcription with reconstituted factors, p300 was found to have a chromatin-specific, transcriptional repression activity (49). Interestingly, in the reconstituted system, an additional acetyl-CoA converted the p300 corepressive activity into a coactivator function. Irrespective of the key role of acetyl-CoA in the p300 coregulator function, the acetyltransferase region of p300 was not required for the p300-mediated transrepressive function. Instead, the p300 bromodomain mediated the transrepression. Thus, it has been proposed that p300-mediated repression is distinct from that mediated by condensing chromatin (49). These findings clearly suggest that the coregulator activities of p300/CBP are reversible, depending on the promoter context and the cellular conditions. This is a reflection of the multiple function domains of p300/CBP. In this study, we have created a Drosophila experimental system with which we can look at dCBP activity in heterochromatin. In a suppressive environment, dCBP acts as a repressor. Thus, the results of our study suggest that the action of CBP/p300 may depend on the state of chromatin at the target loci.
Supplementary Material
Acknowledgments
We thank J. P. Kumar for kindly providing the UAS-dCBP Δ HQ/TM3 and UAS-dCBP Δ BHQ/CyO fly lines, S. Ishii for help with the UAS-dCBP/TM3 and nej3/FM7C fly stains, and A. Fukamizu for the generous gift of the mSir2 plasmid. We are grateful to S. Hirose and M. Sato for helpful discussions. The anti-HP1 polyclonal antibody (C1A9) developed by investigators was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biology Science, Iowa City, IA 52242.
This work was supported by a grant-in-aid for Exploratory Research for Advanced Technology (ERATO) and an Invitation Fellowship from the Japan Society for Promotion of Science (JSPS) (A.P.K.).
Footnotes
Published ahead of print on 15 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akimaru, H., Y. Chen, P. Dai, D. X. Hou, M. Nonaka, S. M. Smolik, S. Armstrong, R. H. Goodman, and S. Ishii. 1997. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386735-738. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J., R. Bhandari, and J. P. Kumar. 2005. A genetic screen identifies putative targets and binding partners of CREB-binding protein in the developing Drosophila eye. Genetics 1711655-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 2005. Reversing histone methylation. Nature 4361103-1106. [DOI] [PubMed] [Google Scholar]
- 4.Bantignies, F., R. H. Goodman, and S. M. Smolik. 2000. Functional interaction between the coactivator Drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers. Mol. Cell. Biol. 209317-9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bantignies, F., R. H. Goodman, and S. M. Smolik. 2002. The interaction between the coactivator dCBP and Modulo, a chromatin-associated factor, affects segmentation and melanotic tumor formation in Drosophila. Proc. Natl. Acad. Sci. USA 992895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barlow, A. L., C. M. van Drunen, C. A. Johnson, S. Tweedie, A. Bird, and B. M. Turner. 2001. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp. Cell Res. 26590-103. [DOI] [PubMed] [Google Scholar]
- 7.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90569-580. [DOI] [PubMed] [Google Scholar]
- 9.Chiani, F., F. Di Felice, and G. Camilloni. 2006. SIR2 modifies histone H4-K16 acetylation and affects superhelicity in the ARS region of plasmid chromatin in Saccharomyces cerevisiae. Nucleic Acids Res. 345426-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csink, A. K., and S. Henikoff. 1996. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature 381529-531. [DOI] [PubMed] [Google Scholar]
- 11.Dai, Y., D. Ngo, L. W. Forman, D. C. Qin, J. Jacob, and D. V. Faller. 2007. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol. Endocrinol. 211807-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daitoku, H., M. Hatta, H. Matsuzaki, S. Aratani, T. Ohshima, M. Miyagishi, T. Nakajima, and A. Fukamizu. 2004. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. USA 10110042-10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter, K. Struhl, and P. Spierer. 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384589-591. [DOI] [PubMed] [Google Scholar]
- 14.Elgin, S. C., and S. I. Grewal. 2003. Heterochromatin: silence is golden. Curr. Biol. 13R895-R898. [DOI] [PubMed] [Google Scholar]
- 15.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15172-183. [DOI] [PubMed] [Google Scholar]
- 16.Fonte, C., J. Grenier, A. Trousson, A. Chauchereau, O. Lahuna, E. E. Baulieu, M. Schumacher, and C. Massaad. 2005. Involvement of β-catenin and unusual behavior of CBP and p300 in glucocorticosteroid signaling in Schwann cells. Proc. Natl. Acad. Sci. USA 10214260-14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frye, R. A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273793-798. [DOI] [PubMed] [Google Scholar]
- 18.Fu, M., M. Liu, A. A. Sauve, X. Jiao, X. Zhang, X. Wu, M. J. Powell, T. Yang, W. Gu, M. L. Avantaggiati, N. Pattabiraman, T. G. Pestell, F. Wang, A. A. Quong, C. Wang, and R. G. Pestell. 2006. Hormonal control of androgen receptor function through SIRT1. Mol. Cell. Biol. 268122-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiki, R., M. S. Kim, Y. Sasaki, K. Yoshimura, H. Kitagawa, and S. Kato. 2005. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 243881-3894. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Fukuda, T., K. Yamagata, S. Fujiyama, T. Matsumoto, I. Koshida, K. Yoshimura, M. Mihara, M. Naitou, H. Endoh, T. Nakamura, C. Akimoto, Y. Yamamoto, T. Katagiri, C. Foulds, S. Takezawa, H. Kitagawa, K. Takeyama, B. W. O'Malley, and S. Kato. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 9:604-611. [DOI] [PubMed] [Google Scholar]
- 21.Furuyama, T., R. Banerjee, T. R. Breen, and P. J. Harte. 2004. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr. Biol. 141812-1821. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Bassets, I., Y. S. Kwon, F. Telese, G. G. Prefontaine, K. R. Hutt, C. S. Cheng, B. G. Ju, K. A. Ohgi, J. Wang, L. Escoubet-Lozach, D. W. Rose, C. K. Glass, X. D. Fu, and M. G. Rosenfeld. 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 111043-1054. [DOI] [PubMed] [Google Scholar]
- 24.Grewal, S. I., and S. C. Elgin. 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grewal, S. I., and S. Jia. 2007. Heterochromatin revisited. Nat. Rev. Genet. 835-46. [DOI] [PubMed] [Google Scholar]
- 26.Guidez, F., L. Howell, M. Isalan, M. Cebrat, R. M. Alani, S. Ivins, I. Hormaeche, M. J. McConnell, S. Pierce, P. A. Cole, J. Licht, and A. Zelent. 2005. Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 255552-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito, S., K. Takeyama, A. Yamamoto, S. Sawatsubashi, Y. Shirode, A. Kouzmenko, T. Tabata, and S. Kato. 2004. In vivo potentiation of human oestrogen receptor alpha by Cdk7-mediated phosphorylation. Genes Cells 9983-992. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 198136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley, R. L., I. Solovyeva, L. M. Lyman, R. Richman, V. Solovyev, and M. I. Kuroda. 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81867-877. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa, H., R. Fujiki, K. Yoshimura, Y. Mezaki, Y. Uematsu, D. Matsui, S. Ogawa, K. Unno, M. Okubo, A. Tokita, T. Nakagawa, T. Ito, Y. Ishimi, H. Nagasawa, T. Matsumoto, J. Yanagisawa, and S. Kato. 2003. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell 113905-917. [DOI] [PubMed] [Google Scholar]
- 31.Konev, A. Y., C. M. Yan, D. Acevedo, C. Kennedy, E. Ward, A. Lim, S. Tickoo, and G. H. Karpen. 2003. Genetics of P-element transposition into Drosophila melanogaster centric heterochromatin. Genetics 1652039-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, M. G., J. Norman, A. Shilatifard, and R. Shiekhattar. 2007. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128877-887. [DOI] [PubMed] [Google Scholar]
- 33.Ludlam, W. H., M. H. Taylor, K. G. Tanner, J. M. Denu, R. H. Goodman, and S. M. Smolik. 2002. The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 223832-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6838-849. [DOI] [PubMed] [Google Scholar]
- 36.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 37.Metzger, E., M. Wissmann, N. Yin, J. M. Muller, R. Schneider, A. H. Peters, T. Gunther, R. Buettner, and R. Schule. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437436-439. [DOI] [PubMed] [Google Scholar]
- 38.Newman, B. L., J. R. Lundblad, Y. Chen, and S. M. Smolik. 2002. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics 1621675-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtake, F., A. Baba, I. Takada, M. Okada, K. Iwasaki, H. Miki, S. Takahashi, A. Kouzmenko, K. Nohara, T. Chiba, Y. Fujii-Kuriyama, and S. Kato. 2007. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446562-566. [DOI] [PubMed] [Google Scholar]
- 40.Ohtake, F., K. Takeyama, T. Matsumoto, H. Kitagawa, Y. Yamamoto, K. Nohara, C. Tohyama, A. Krust, J. Mimura, P. Chambon, J. Yanagisawa, Y. Fujii-Kuriyama, and S. Kato. 2003. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 423545-550. [DOI] [PubMed] [Google Scholar]
- 41.Oñate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 42.Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra, J. A. Birchler, and S. C. Elgin. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303669-672. [DOI] [PubMed] [Google Scholar]
- 43.Parsons, X. H., S. N. Garcia, L. Pillus, and J. T. Kadonaga. 2003. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proc. Natl. Acad. Sci. USA 1001609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peña-Rangel, M. T., I. Rodriguez, and J. R. Riesgo-Escovar. 2002. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics 1601035-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116511-526. [DOI] [PubMed] [Google Scholar]
- 46.Ringrose, L., H. Ehret, and R. Paro. 2004. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16641-653. [DOI] [PubMed] [Google Scholar]
- 47.Rosenberg, M. I., and S. M. Parkhurst. 2002. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109447-458. [DOI] [PubMed] [Google Scholar]
- 48.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 201405-1428. [DOI] [PubMed] [Google Scholar]
- 49.Santoso, B., and J. T. Kadonaga. 2006. Reconstitution of chromatin transcription with purified components reveals a chromatin-specific repressive activity of p300. Nat. Struct. Mol. Biol. 13131-139. [DOI] [PubMed] [Google Scholar]
- 50.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 181251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103843-852. [DOI] [PubMed] [Google Scholar]
- 52.Shankaranarayana, G. D., M. R. Motamedi, D. Moazed, and S. I. Grewal. 2003. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 131240-1246. [DOI] [PubMed] [Google Scholar]
- 53.Shao, W., S. Halachmi, and M. Brown. 2002. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol. Cell. Biol. 223358-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smolik, S., and K. Jones. 2007. Drosophila dCBP is involved in establishing the DNA replication checkpoint. Mol. Cell. Biol. 27135-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snowden, A. W., L. A. Anderson, G. A. Webster, and N. D. Perkins. 2000. A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol. Cell. Biol. 202676-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swaminathan, J., E. M. Baxter, and V. G. Corces. 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 1965-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeyama, K., S. Ito, A. Yamamoto, H. Tanimoto, T. Furutani, H. Kanuka, M. Miura, T. Tabata, and S. Kato. 2002. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron 35855-864. [DOI] [PubMed] [Google Scholar]
- 58.Takezawa, S., A. Yokoyama, M. Okada, R. Fujiki, A. Iriyama, Y. Yanagi, H. Ito, I. Takada, M. Kishimoto, A. Miyajima, K. Takeyama, K. Umesono, H. Kitagawa, and S. Kato. 2007. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 26764-774. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Thomas, H. E., H. G. Stunnenberg, and A. F. Stewart. 1993. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature 362471-475. [DOI] [PubMed] [Google Scholar]
- 60.Thummel, C. S., A. M. Boulet, and H. D. Lipshitz. 1988. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene 74445-456. [DOI] [PubMed] [Google Scholar]
- 61.Vaquero, A., M. Scher, H. Erdjument-Bromage, P. Tempst, L. Serrano, and D. Reinberg. 2007. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450440-444. [DOI] [PubMed] [Google Scholar]
- 62.Vaquero, A., M. Scher, D. Lee, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 1693-105. [DOI] [PubMed] [Google Scholar]
- 63.Wallrath, L. L., and S. C. Elgin. 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 91263-1277. [DOI] [PubMed] [Google Scholar]
- 64.Waltzer, L., and M. Bienz. 1998. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature 395521-525. [DOI] [PubMed] [Google Scholar]
- 65.Wang, Q., J. S. Carroll, and M. Brown. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19631-642. [DOI] [PubMed] [Google Scholar]
- 66.Wissmann, M., N. Yin, J. M. Muller, H. Greschik, B. D. Fodor, T. Jenuwein, C. Vogler, R. Schneider, T. Gunther, R. Buettner, E. Metzger, and R. Schule. 2007. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9347-353. [DOI] [PubMed] [Google Scholar]
- 67.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9140-147. [DOI] [PubMed] [Google Scholar]
- 68.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell 9553-562. [DOI] [PubMed] [Google Scholar]
- 69.Zhao, Y., K. Goto, M. Saitoh, T. Yanase, M. Nomura, T. Okabe, R. Takayanagi, and H. Nawata. 2002. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment. Identification of the p102 U5 small nuclear ribonucleoprotein particle-binding protein as a coactivator for the receptor. J. Biol. Chem. 27730031-30039. [DOI] [PubMed] [Google Scholar]
- 70.Zhao, Y., G. Lang, S. Ito, J. Bonnet, E. Metzger, S. Sawatsubashi, E. Suzuki, X. Le Guezennec, H. G. Stunnenberg, A. Krasnov, S. G. Georgieva, R. Schule, K. Takeyama, S. Kato, L. Tora, and D. Devys. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 2992-101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.