Abstract
Aurora-A is a conserved kinase implicated in mitotic regulation and carcinogenesis. Aurora-A was previously implicated in mitotic entry and spindle assembly, although contradictory results prevented a clear understanding of the roles of Aurora-A in mammals. We developed a conditional null mutation in the mouse Aurora-A gene to investigate Aurora-A functions in primary cells ex vivo and in vivo. We show here that conditional Aurora-A ablation in cultured embryonic fibroblasts causes impaired mitotic entry and mitotic arrest with a profound defect in bipolar spindle formation. Germ line Aurora-A deficiency causes embryonic death at the blastocyst stage with pronounced cell proliferation failure, mitotic arrest, and monopolar spindle formation. Aurora-A deletion in mid-gestation embryos causes an increase in mitotic and apoptotic cells. These results indicate that murine Aurora-A facilitates, but is not absolutely required for, mitotic entry in murine embryonic fibroblasts and is essential for centrosome separation and bipolar spindle formation in vitro and in vivo. Aurora-A deletion increases apoptosis, suggesting that molecular therapies targeting Aurora-A may be effective in inducing tumor cell apoptosis. Aurora-A conditional mutant mice provide a valuable system for further defining Aurora-A functions and for predicting effects of Aurora-A therapeutic intervention.
The equal partitioning of chromosomes at mitosis is critical for avoiding aneuploidy, a condition associated with spontaneous miscarriage, developmental disorders, and cancer (50). Mitosis requires coordinated completion of multiple events including nuclear envelope breakdown, chromosome condensation and congression to the metaphase plate, centrosome separation, spindle formation, chromosome-spindle attachment and error correction, sister chromatid separation, and cytokinesis. Multiple regulators, many of which are kinases, are required to ensure that each event is completed in a timely fashion and in the proper order (reviewed in reference 46). Although a number of mitotic kinases have been identified, their targets and the intricacies of mitotic signal transduction pathways are just beginning to be understood.
The Aurora kinases are key mitotic regulators in eukaryotes (reviewed in reference 45). The Aurora family includes a single member in yeasts (Saccharomyces cerevisiae Ipl1p, Schizosaccharomyces pombe Ark1), two members each in Caenorhabditis elegans and Drosophila, and two or three members in vertebrates. Although originally given a variety of names, Aurora kinases in multicellular eukaryotes have subsequently been classified into A, B, and C groups based on patterns of mitotic subcellular localization and homology, which also appear to reflect functional distinctions (8, 46). Aurora-A kinases are observed at centrosomes and adjacent spindle fibers, and current evidence supports key roles in regulating protein localization and function at centrosomes, as well as regulation of the assembly, stability, and function of the mitotic spindle (reviewed in reference 43). Aurora-B kinases display “chromosomal passenger” localization, residing on mitotic chromosomes and subsequently moving to the spindle midzone after separation of sister chromatids. Aurora-B family members have been implicated in the regulation of kinetochore-spindle attachment, the spindle checkpoint, and cytokinesis (reviewed in references 1 and 8). Aurora-C kinases, which have only been identified in mammals, have a limited expression pattern and appear to have functions that overlap those of Aurora-B (7, 53).
The human Aurora-A kinase (hAurA) was first identified because of its overexpression in cancer cell lines (5, 58). The hAurA gene (stk15) resides on chromosome 20q13, a region frequently amplified in human cancers (5, 58). hAurA has been dubbed an oncogene because of the fact that its overexpression transforms immortalized rodent fibroblasts (5, 70). Polymorphisms in hAurA are associated with an increased risk of colon cancer, while murine AurA (mAurA) polymorphisms confer increased susceptibility to experimentally induced skin tumors (14). The mAurA gene is frequently amplified in radiation-induced lymphomas from p53 heterozygous mice, while loss of one mAurA allele has been observed in lymphomas from p53-null mice (41). Thus, aberrant AurA expression is associated with tumorigenesis, suggesting that insight into AurA functions will lead to a better understanding of tumorigenesis mechanisms.
A number of experimental observations suggest that AurA kinases are required for normal centrosome maturation and bipolar spindle assembly. The AurA ortholog in Drosophila melanogaster (Aurora) was identified in a screen for mutations that impact the centrosome cycle (21). Syncytial embryos from hypomorphic Aurora mutant females display a variety of mitotic abnormalities resulting from a failure to separate centrosomes. Aurora-null flies die at the larval stage with characteristic monopolar spindles and circular chromosome arrays in larval neuroblasts. Such monopolar spindles arise from failed centrosome separation (21). Subsequent studies of Drosophila Aurora mutant alleles revealed additional defects in centrosome maturation (including a failure to localize transforming acidic coiled-coil protein, centrosomin, and γ-tubulin at centrosomes) and in asymmetric localization of Numb protein in sensory organ precursor cells (3, 17). Similar to the case in Drosophila, disruption of the C. elegans AurA ortholog AIR-1 by RNA interference (RNAi) or mutation causes defects in centrosome maturation and monopolar spindle formation. Centrosomes undergo normal separation but collapse, leading to monopolar spindle formation (16, 24, 56). Studies of the Xenopus AurA homolog pEg2 revealed similar phenotypes after overexpression of kinase-dead mutants, antibody-mediated inhibition, or immunodepletion (18, 19, 38, 52). Furthermore, Xenopus AurA has been shown to interact with and phosphorylate Eg5, a mitotic kinesin required for bipolar spindle formation, suggesting a possible mechanism by which AurA could influence bipolar spindle formation and/or stabilization (19). Thus, existing reports from these systems are quite consistent in implicating AurA in centrosome separation and function.
In contrast to the systems described above, published reports of RNAi-mediated reduction of AurA expression in mammalian cell lines have contained conflicting results about the role of AurA in mitotic entry, bipolar spindle formation, and mitotic progression. AurA RNAi in HeLa cells was reported to block or delay mitotic entry, prompting the conclusion that AurA is essential for mitotic commitment in mammalian cells (27, 36). In contrast, other AurA RNAi studies showed accumulation of mitotic cells with monopolar spindles (12, 20, 67). These discrepancies call into question the functional conservation of AurA in mammals and highlight a need for additional studies to definitively address the roles of AurA. This is particularly critical for understanding the roles of AurA in cancer and for projecting possible effects of AurA inhibitors currently in development as anticancer agents. We used gene targeting in mouse embryonic stem (ES) cells to produce a conditional null allele at the AurA locus. Here we describe cellular phenotypes of AurA deletion in primary cells in vitro and developmental phenotypes of AurA mutant mice. We show that AurA deletion in primary embryonic fibroblasts causes delayed mitotic entry with accumulation of cells in early prophase, consistent with a role for AurA in mitotic entry. Nevertheless, AurA-deficient cells that enter prometaphase arrest with monopolar spindles and eventually exit mitosis without segregating their chromosomes. Prolonged culture of AurA-deficient cells leads to polyploidy with abnormal nuclear structure. Germ line AurA deficiency causes embryonic death at the blastocyst stage with mitotic arrest and monopolar spindle formation, while AurA deletion in mid-gestation embryos causes an increased mitotic index and increased apoptosis. Together, our findings indicate that AurA is required for timely mitotic entry and bipolar spindle formation in vitro and in vivo.
MATERIALS AND METHODS
AurA gene targeting and mice.
AurA genomic bacterial artificial chromosome clones were isolated from a 129SvEv library by hybridization screening and subsequent PCR verification. Bacterial strain EL350 (37) was used for “recombineering” to subclone a 13-kb fragment encompassing AurA exons 1 to 6 into a pUC19-based plasmid including a negative-selection thymidine kinase gene (a gift of R. Thresher, University of North Carolina animal models core facility). A loxP site was inserted into intron 2 by recombineering a loxP-flanked zeocin cassette, followed by Cre-mediated cassette removal in EL350. A loxP- and FRT-flanked neomycin resistance cassette was subsequently inserted into a unique SalI restriction site in intron 1. The targeting vector was linearized and used for gene targeting in line E14 ES cells (129P2/OlaHsd strain). ES cell targeting was verified by PCR and Southern blotting, and correctly targeted clones were injected into C57BL/6J blastocysts for chimera generation. Chimeric males were bred with C57BL/6J females, and germ line transmission of the AurAneo allele was verified by PCR. The AurAd2 and AurAf alleles were generated by crossing the AurAneo allele with an X-linked CMV-Cre strain (61). Recombined alleles were backcrossed for at least 2 generations to C57BL/6J mice before use in this study. R26CreER (2) and R26Rep (60) mice were obtained from Jackson Laboratories. For the PCR primers used and genotyping information for the AurA mutant alleles, see Table S1 in the supplemental material.
MEFs.
Murine embryonic fibroblasts (MEFs) were generated from embryonic day 14.5 (E14.5) embryos by standard protocols. MEFs were grown in Dulbecco modified Eagle medium (DMEM) with 4,500 mg/liter d-glucose and supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 55 μM 2-mercaptoethanol in a humidified incubator with 5% CO2 and passaged every 2 to 3 days. All experiments were performed with cells at passages 3 to 5. β-Galactosidase detection was performed by a standard 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining protocol (59). For immunofluorescence (IF) experiments, cells were grown on acid-washed glass coverslips coated with poly-l-lysine. Coverslips were fixed in 2% formaldehyde in phosphate-buffered saline (PBS) for 20 to 30 min and stored in PBS with 0.1% sodium azide at 4°C until used. Primary antibodies were anti-Aurora-A (IAK1; BD Biosciences no. 610938, 1:500 dilution), anti-α-tubulin (rat monoclonal, 1:200 dilution), anti-γ-tubulin (Sigma T5192, 1:1,000 dilution), anti-β-tubulin (Sigma T4026, 1:200 dilution), anti-XMad2 (affinity-purified rabbit polyclonal, 1:1,000 dilution) (66), and anti-phosphorylated histone H3 (anti-PH3; serine 10; Upstate Biotech, 06-570, 1:500 dilution). 4-Hydroxytamoxifen (OHT; Sigma H7904) was dissolved in 100% ethanol and used at 50 nM for all experiments.
Flow cytometry.
Adherent cells were removed from plates by trypsinization and pooled with cell culture supernatant containing nonadherent cells. Cells were washed once with PBS, fixed in cold 70% ethanol, and stored at −20°C until analyzed. For staining, 1 × 106 cells were washed in PBS and stained in PBS with 50 μg/ml propidium iodide, 200 μg/ml boiled RNase A, and 0.1% Triton X-100. Analyses were performed on a BD FACScan flow cytometer and analyzed with Summit software (Dako).
Time-lapse imaging.
AurA+/+; R26CreER/+ and AurAf/f; R26CreER/+ cells were incubated in DMEM-0.1% FBS for 24 h, after which 50 nM OHT was added for an additional 24 h of incubation. Cell cycle entry was stimulated by replacing the medium with fresh DMEM with 10% FBS for 14 h of incubation, followed by the addition of fresh DMEM-10% FBS-10 mM HEPES, pH 7.4, for an additional 3 h of incubation prior to live-cell imaging. Time-lapse images were captured on an Olympus IX70 microscope with a custom-built environmental chamber. Images were taken every 10 min for 50 h. Images were analyzed with OpenLab software (Improvision) and QuickTime (Apple). Mitotic entry was judged as the point of cell rounding, and mitotic exit was judged as the point at which cells flattened out. Each experiment was repeated with MEFs from two different embryos. Differences in mitotic timing were compared by Kaplan-Meier analysis. For representative movies and phenotype images, see the supplemental material.
Immunoblotting.
Cells were harvested at the indicated time points, washed in PBS, counted, and lysed by boiling in sodium dodecyl sulfate (SDS) sample buffer (60 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 100 mM dithiothreitol). Volumes representing equal cell numbers were separated by 4 to 20% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were blocked in 5% nonfat dry milk in PBS with 0.1% Tween 20 (block) and incubated with antibodies diluted in block at room temperature (RT) for 1 to 2 h or overnight at 4°C. Membranes were washed extensively in PBS with 0.1% Tween 20 and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at RT in block. Following extensive washes, targets were detected with a SuperSignal West Pico Chemiluminescent Substrate kit (Pierce).
Blastocyst culture and staining.
Timed matings were set, and the morning of vaginal plug detection was considered E0.5. Blastocysts were flushed from the uteri of pregnant females at E3.5. Blastocysts were cultured in drops of DMEM with 4,500 mg/liter d-glucose, 15% FBS, 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 55 μM 2-mercaptoethanol overlaid with mineral oil in a humidified incubator with 5% CO2. Outgrowths were photographed after 5 to 7 days and harvested for genotyping. DNA lysates were prepared by the HotShot method (63). For immunostaining, freshly isolated blastocysts were rinsed in PBS and fixed in fresh 4% formaldehyde in PBS for 30 to 60 min at RT. Blastocysts were subsequently rinsed in PBS; washed three times for 10 min each in PBS with 1% bovine serum albumin and 0.5% Triton X-100 (PBT); blocked for 1 h in PBT with 5% goat serum (Block); incubated overnight with primary antibodies diluted in block; washed three times for 10 min each in PBT; incubated for 1 h with secondary antibodies in PBT; washed three times for 10 min each in PBT, one time in PBS, once in a 1:2 mixture of glycerol-PBS, and once in 1:1 glycerol-PBS; and mounted in mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI). All incubations were performed at 25°C. The primary antibodies used were mouse monoclonal anti-Aurora-A (IAK1; BD Biosciences no. 610938, 1:500 dilution) and rat monoclonal anti-α-tubulin (1:200 dilution). Images were analyzed on a Leica SP2 laser scanning confocal microscope with a 40× objective (1.25 numerical aperture).
RESULTS
Conditional inactivation of murine Aurora-A.
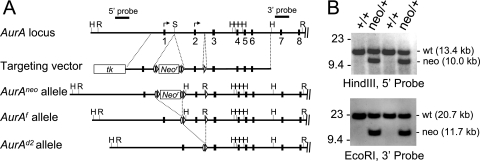
To better understand the functions of AurA in development and cancer, we generated a conditional AurA allele by gene targeting in mouse ES cells (Fig. 1). The AurA locus includes nine exons, with potential translational start codons in exons 1 and 2. The targeting strategy was designed to flank AurA exon 2 with loxP sites, allowing Cre-mediated exon 2 deletion. Exon 2 deletion was predicted to produce a null allele because of removal of the main ATG codon and by causing a frameshift of any protein produced from the upstream ATG codon after the 20th amino acid (the resulting protein would also include 13 out-of-frame amino acids prior to a stop codon). The targeting construct produced the AurAneo allele, which incorporates a loxP- and FRT-flanked neomycin resistance (neo) cassette in intron 1 and a single loxP site in intron 2. Correctly targeted ES cell clones were used to derive chimeric mice that transmitted the AurAneo allele through the germ line. AurAneo/+ mice were crossed to a Cre deleter strain [HprtTg(CMV-Cre)Brd] (61) to produce the AurAf (“floxed”) and AurAd2 alleles by deletion of the neo cassette alone or the neo cassette plus exon 2, respectively. Mice heterozygous for each of the mutant alleles, or homozygous for the AurAf allele, were viable and fertile and did not show any overt abnormalities.
FIG. 1.
Production of AurA conditional and mutant alleles. (A) Schematic representation of the AurA locus, targeting vector, and mutant alleles. Exons 1 to 8; Southern blot probes; and HindIII (H), EcoRI (R), and SalI (S) restriction sites are shown. Arrows indicate potential translational start codons. The targeting vector included a thymidine kinase gene (tk) and a neomycin resistance gene (Neor) flanked by FRT (ovals) and loxP (triangles) sites. Mice carrying the AurAneo allele were crossed to a Cre recombinase strain to generate the AurAf (conditional) and AurAd2 (null) alleles. (B) Southern blotting of ES cells indicating correct targeting event in the second and fourth clones from the left.
Conditional AurA deletion in MEFs.
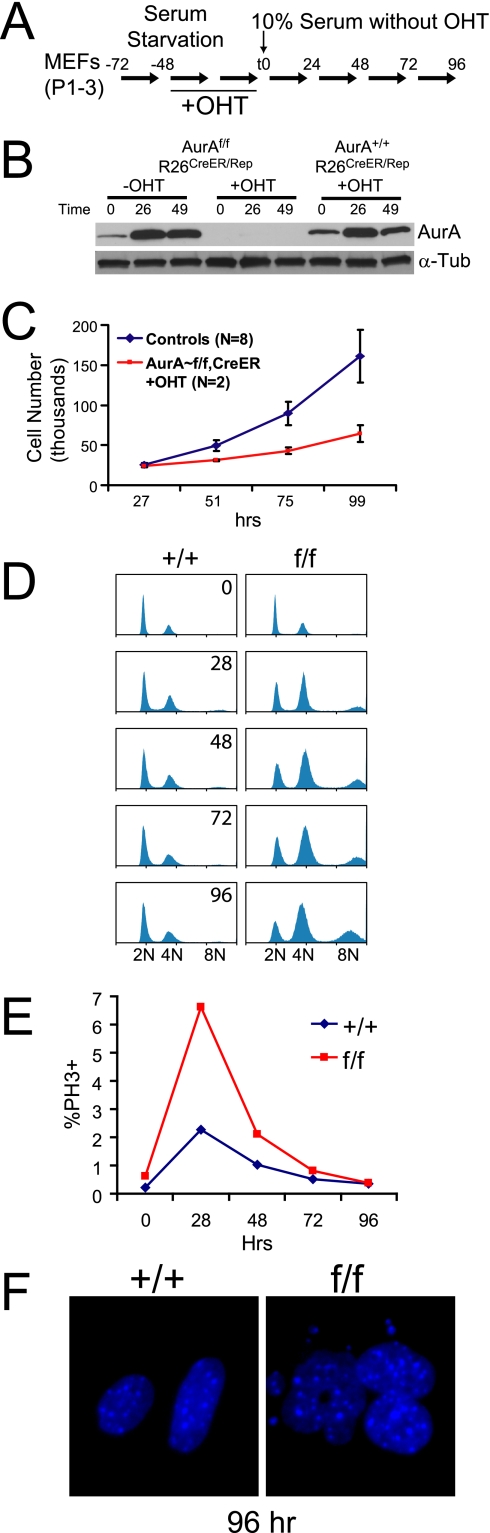
Published reports of RNAi of AurA expression in mammalian cell lines gave conflicting results about the role of AurA in mitotic entry and progression (12, 20, 27, 36, 67). RNAi usually does not produce complete inhibition of protein expression and may have off-target effects that confound interpretation (30, 35, 40). Moreover, HeLa and other established cell lines harbor multiple genetic abnormalities because of their tumor origins and selection for efficient growth in culture. Therefore, we tested the effects of genetic AurA deletion in early-passage MEFs to determine the phenotypes of AurA-null primary cells. MEFs were derived from intercrosses of AurAf mice harboring a Rosa26-Cre-ERTM transgene (R26CreER) (2). The R26CreER transgene drives ubiquitous expression of a Cre-estrogen receptor fusion protein from the endogenous Rosa26 locus. The Cre-ERTM molecule is inactive until cells are treated with tamoxifen or the analog OHT, allowing temporal control over recombinase activity. The MEF system also included a Cre-inducible Rosa26-β-galactosidase reporter allele (R26Rep), allowing β-galactosidase marking of cells that experienced Cre activity (60). To avoid potential confounding effects of Cre-mediated DNA damage during the cell cycle (39), we devised a system to delete AurA in quiescent cells and subsequently allow cell cycle progression (Fig. 2A). AurA gene expression and protein stability are regulated by the cell cycle: AurA gene expression is limited to S/G2/M cells, and AurA protein is degraded by the proteosome upon G1 entry (5, 11, 15, 22, 29, 52, 62, 64). We reasoned that if the Cre recombination occurred in G0- or G1-phase cells, the cells should lack AurA protein as they traverse subsequent cell cycle phases, providing a clean system in which to detect mitotic phenotypes. Cells were arrested by 24 h of serum starvation, followed by 2 days of additional starvation in the presence of OHT to induce Cre-ERTM activity in the quiescent cells. Cells were then fed serum-containing medium without OHT to induce cell cycle entry in the absence of Cre activity. This protocol gave efficient recombination of the R26 reporter allele (∼85%) and production of the Aurd2 allele as assessed by induction of β-galactosidase and PCR, respectively (not shown). A background of approximately 5% recombination was observed in untreated cultures (data not shown). Immunoblot analyses of control cells (AurAf/f; R26CreER/Rep without OHT and AurA+/+; R26CreER/Rep with OHT) showed the expected pattern of AurA expression (Fig. 2B). Very little AurA is detected in arrested cells; the protein is abundant at 24 h, the time when the mitotic index peaks in the population, and intermediate levels are detected at 48 h, when the population has reverted to more asynchronous growth and fewer cells are in mitosis (Fig. 2B). In contrast, AurA protein was undetectable in OHT-treated AurAf/f; R26CreER/Rep MEFs at all of the time points tested (Fig. 2B). Semiquantitative reverse transcription-PCR analysis demonstrated very similar AurA mRNA levels in OHT-treated AurAf/f; R26CreER/Rep and AurA+/+; R26CreER/Rep MEFs, although primers spanning exon 2 clearly demonstrated that the deletion had occurred in AurAf/f; R26CreER/Rep cells (Fig. S1). Thus, the deletion of exon 2 causes a loss of protein expression, as expected, despite the continued presence of the AurA mRNA with exon 2 deleted. Together, these results indicate that the R26CreER transgene provides efficient conversion of the AurAf allele to the AurAd2 allele and that the AurAd2 allele is null for protein production. The conditional AurA MEFs thus provide an excellent system in which to determine the effects of AurA deletion on cell cycle progression.
FIG. 2.
AurA mutation causes MEF growth defects and ploidy changes. (A) Experimental protocol for AurA deletion in quiescent MEFs. Early-passage MEFs were serum starved for 24 h and then serum starved for 48 h with OHT. The time of serum addition (without OHT) is defined as time zero. (B) Immunoblot analysis of AurA protein expression in OHT-treated MEFs shows that AurA protein is undetectable following Cre recombination. Cells were treated as shown in panel A, and samples were harvested at 0, 26, and 49 h after serum addition. α-Tubulin (α-Tub) detection confirmed equal loading. (C) MEF growth curve following the treatment protocol from panel A. Controls were AurAf/f; R26CreER/Rep cells with OHT treatment (n = 2), AurA+/+; R26CreER/Rep cells with or without OHT (n = 2 each), and AurAf/f; R26Rep/Rep cells with or without OHT (n = 1 each). (D) AurA deletion causes increased G2/M and endoreduplicating fractions. Cultures were treated with OHT as in panel A and analyzed by fluorescence-activated cell sorting for DNA content with propidium iodide. +/+ = AurA+/+; R26CreER/Rep; f/f = AurAf/f; R26CreER/Rep. (E) AurA deletion causes an increased PH3+ index. Cultures were treated with OHT as in panel A and analyzed by fluorescence-activated cell sorting for PH3 staining. +/+ = AurA+/+; R26CreER/Rep; f/f = AurAf/f; R26CreER/Rep. (F) Nuclear atypia following AurA deletion. DAPI-stained images from cultures 96 h after serum readdition are shown.
To determine phenotypes of AurA loss, we first tested the growth and ploidy of AurA mutant MEFs over a 5-day time course after Cre-mediated deletion. OHT-treated AurAf/f; R26CreER/Rep MEFs displayed a clear growth defect relative to controls after serum addition (Fig. 2C). Controls included AurAf/f; R26CreER/Rep cells without OHT treatment and AurA+/+; R26CreER/Rep cells with and without OHT treatment. No growth defects were observed in response to OHT treatment in controls, indicating that Cre activity in quiescent cells does not impact the subsequent cell cycle. We also examined the cell cycle and ploidy by flow cytometry (Fig. 2D). Control cells had a normal cell cycle and ploidy profile at all of the time points examined. In contrast, AurA-deficient cultures displayed a dramatic increase in cells with 4N DNA content 28 h after serum addition, and this population remained elevated throughout the 96-h observation period. AurA mutant cultures also displayed an increasing 8N population over the course of the experiment (Fig. 2D). The population of AurA mutant cells with 4N or greater (4N+) DNA content increased from ∼50% at time zero to ∼80% at 96 h, while the 4N+ population in control cells remained near 50% across all of the time points tested.
PH3 is widely used as a marker of mitotic cells. PH3 immunoreactivity is detectable in punctate foci at centromeres beginning in G2 phase and spreads across the chromatin just prior to chromosome condensation in prophase (31). To determine if the increased 4N population in AurA mutant cultures was attributable to mitotic arrest, we analyzed the PH3 index of cells with 4N and 8N DNA contents. Both wild-type (WT) and AurA-deficient cultures displayed a peak PH3 index at 28 h, with reduced levels at each subsequent time point (Fig. 2E). AurA-deficient cells showed an approximately threefold higher peak PH3 index than controls at 28 h and maintained a higher PH3 index than controls at each time point until 96 h. PH3+ cells were observed in the 4N population, as expected, but also in the 8N population, indicating that some polyploid cells enter mitosis (data not shown). AurA-deficient cells at later time points displayed large, aberrant nuclei with frequent micronuclei (Fig. 2F), indicating that AurA deficiency causes mitotic abnormalities over the course of multiple cell cycles. Together, these data suggest that AurA deficiency causes a mitotic-arrest phenotype. It is notable that the peak PH3 index in AurA mutant cultures was only ∼7%, while the 4N and 8N populations accounted for up to 80% of the total cells. This discrepancy likely reflects the fact that AurA mutant cells are delayed in mitotic entry (see below), such that the true peak mitotic index was later in mutant cultures and was not captured at the time points chosen for this experiment.
AurA-deficient MEFs display impaired mitotic entry.
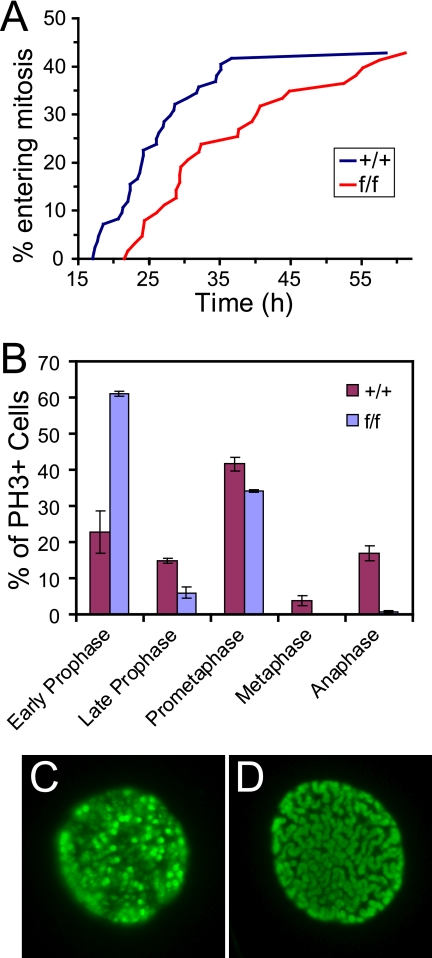
The fact that AurA-deficient cells showed peak PH3 levels around the same time as controls (Fig. 2E) suggested that AurA-deficient cells enter mitosis with approximately normal kinetics. However, AurA reduction by RNAi in HeLa cells was reported to cause a marked delay in mitotic entry (27). AurA depletion from Xenopus extracts also causes delayed mitotic entry, consistent with a role for AurA in the process (38, 55). To further investigate the mitotic entry of AurA mutant MEFs, we measured the timing of mitotic entry by live-cell imaging (Fig. 3A; see Fig. S2 in the supplemental material). About 40% of the cells imaged in both WT and AurA mutant cultures entered mitosis during the imaging interval (17 to 67 h after serum addition). However, AurA mutant cultures displayed a median 8-h delay relative to WT cells (median time to mitotic entry = 24 h for +/+ cells versus 32 h for f/f cells). At 28 h, the time at which we measured the PH3+ index in the experiment shown in Fig. 2E, only ∼12% of the AurA mutant cells had entered mitosis, compared to ∼30% of the WT cells. In this light, the mitotic index of ∼7% observed for AurA mutant cultures at 28 h suggests that AurA mutant cells are significantly delayed in mitosis, as well as prior to mitotic entry.
FIG. 3.
AurA-deficient cells have a mitotic entry defect. (A). Line graphs of time-lapse imaging data showing that AurA mutant cultures are delayed in mitotic entry after serum starvation. Graphs indicate the cumulative percentage of cells that entered mitosis from 17 to 61 h after serum addition. The experiment was repeated twice, with total numbers of 84 AurA+/+; R26CreER/+ (+/+) and 63 AurAf/f; R26CreER/+ (f/f) samples. (B). Bar graph showing the percentage of PH3+ cells in each mitotic phase 48 h after serum addition. Cells on coverslips were synchronized, treated with OHT, and released as described in the legend to Fig. 2A. Coverslips were fixed 48 h after serum addition, stained with anti-PH3 antibody, and manually scored for mitotic phase. Genotypes are as described in the legend to Fig. 2E. The experiment was performed with MEFs from two different embryos per genotype, and 200 random PH3+ cells were scored for each MEF line. t tests were used to compare percentages in each phase between genotypes: early prophase (P = 0.0108), late prophase (P = 0.0295), prometaphase (P = 0.0422), metaphase (P = 0.0572), and anaphase (P = 00092). (C). Example of PH3 antibody staining in an early prophase cell. (D). Example of PH3 antibody staining in a late prophase cell.
To better understand the delay in mitotic entry and mitotic passage of AurA-deficient MEFs, we used IF with fixed cells to quantitate the percentage of PH3+ cells in each mitotic phase at 26, 30, and 48 h after serum addition (Fig. 3B and data not shown). AurA mutant cultures had two striking differences from controls. First, we noted a marked increase in prophase cells in mutants (67% versus 38% in WT cells, P = 0.018). Moreover, the majority of AurA mutant prophase cells displayed an early prophase phenotype with moderate PH3 staining and only partially condensed chromosomes (Fig. 3C, for example). Only 6% of prophase cells in AurA mutant cultures showed a late prophase phenotype with fully condensed chromosomes (Fig. 3A and D). In contrast, control cultures had approximately equal percentages of early- and late prophase cells. Although PH3 staining is widely used as a marker of mitosis, phosphorylation of histone H3 begins in late G2 and is thus not a definitive mitotic marker (26). Cytological features such as chromosome condensation that have historically been used to define mitotic entry are also not necessarily true indicators that a cell has committed to mitosis (47). However, nuclear envelope breakdown at the prophase-prometaphase transition clearly indicates that the cell has passed the point of no return for mitosis (47). The accumulation of early prophase cells in AurA-deficient cultures is thus consistent with a role for AurA in G2 progression events that prepare cells for mitosis. Nevertheless, the fact that a significant percentage of cells enter prometaphase without AurA indicates that AurA is not absolutely required for mitotic entry in vitro.
AurA is required for bipolar spindle formation.
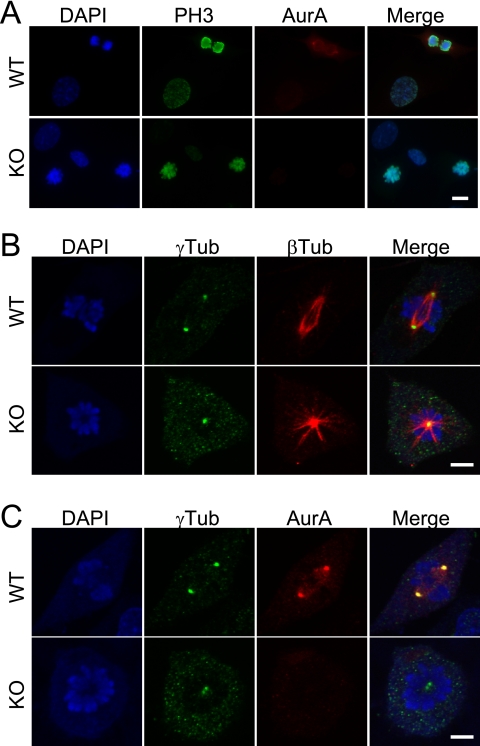
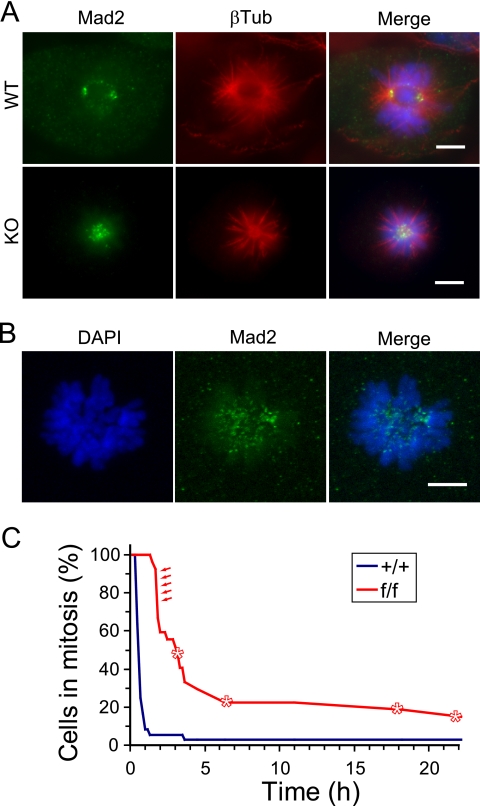
Despite the striking accumulation of AurA mutant cells in early prophase, the requirement for AurA in mitotic entry is not absolute, as prometaphase cells were also observed in AurA mutant cultures (Fig. 3A). However, in stark contrast to controls, AurA mutant cultures were nearly devoid of cells in metaphase, anaphase, and telophase. This observation suggested that AurA is required for key mitotic events, in addition to its role in facilitating mitotic entry. Examination of AurA-deficient prometaphase cells revealed a characteristic monopolar spindle pattern with closely adjacent centrosomes (γ-tubulin) in the center of a circular chromosome rosette (Fig. 4B, KO). This pattern contrasts to the well-separated centrosomes and bipolar spindles observed in control cells (Fig. 4B, WT). γ-Tubulin staining was also slightly weaker at the centrosome(s) of some AurA-deficient cells, with a concomitant increase in punctate staining throughout the cells (Fig. 4B and C). Although not dramatic, this phenotype is consistent with the reported impairment of centrosome maturation in C. elegans AurA RNAi experiments (24). Rare cells in AurAf/f; R26CreER/Rep MEFs treated with OHT displayed bipolar spindles, suggesting the possibility that AurA might not be absolutely required for bipolar spindle formation. However, AurA immunostaining demonstrated that cells with bipolar spindles always had AurA staining at centrosomes, indicating that these cells had not undergone Cre-mediated recombination to delete the AurA gene (AurA staining is shown in the WT parts of Fig. 4A and C). In contrast, cells with monopolar spindles lacked AurA staining (Fig. 4C, KO). Thus, in contrast to the dispensable role of AurA in mitotic entry, our data indicate that AurA is required for bipolar spindle formation and chromosome segregation in MEFs.
FIG. 4.
AurA-deficient cells that enter mitosis form monopolar spindles. (A) AurA-deficient cells enter mitosis and phosphorylate histone H3. DAPI, PH3, and AurA staining in WT (AurA+/+; R26CreER/Rep +OHT) and AurA-deficient (AurAf/f; R26CreER/Rep +OHT) MEFs 24 h after serum addition. Note the PH3-positive mitotic cells lacking AurA staining in the knockout (KO) part of the panel. Scale bar = 10 μm. (B) AurA-deficient cells form monopolar spindles. Confocal images of WT and KO cells stained for DAPI, γ-tubulin (γTub; centrosomes), and β-tubulin (βTub; spindles) are shown. The WT cell shows a normal bipolar prometaphase configuration. Note the characteristic monopolar spindle in the KO cell, including the circular chromosome array surrounding the single γ-tubulin focus with an astral microtubule array. Scale bar = 5 μm. (C) Centrosomes duplicate but fail to separate in AurA-deficient cells. Confocal images of WT and KO cells stained for DAPI, γ-tubulin (centrosomes), and AurA are shown. AurA is localized on centrosomes and adjacent spindle structures of the WT cell. Note the two closely associated γ-tubulin foci in the KO cell. Scale bar = 5 μm.
AurA-deficient cells are delayed in mitosis and activate the spindle checkpoint.
Monopolar spindle formation caused by the drug monastrol causes spindle checkpoint arrest because of a lack of bipolar chromosome attachment and/or lack of tension (33). To determine if monopolar spindles induced by AurA deletion in MEFs cause a similar checkpoint arrest, we stained cells for the checkpoint protein Mad2, whose localization at unattached kinetochores correlates with checkpoint arrest (reviewed in reference 44). In control cells, Mad2 staining was identified on kinetochores of chromosomes during prometaphase, when kinetochores are not attached to the mitotic spindle (Fig. 5A, WT). Control cells that achieved chromosome alignment and attachment during metaphase lost Mad2 staining at kinetochores (not shown). In contrast, Mad2 staining was prominent at kinetochores of AurA mutant cells with monopolar spindles, consistent with activation of the spindle checkpoint in these cells (Fig. 5A, KO, and B).
FIG. 5.
Monopolar spindles in AurA-deficient cells activate the spindle checkpoint. (A) Mad2 stains kinetochores of AurA-deficient cells. Cells were stained for Mad2, β-tubulin (βTub), and DAPI (shown as a merged image). WT cell shows Mad2 foci at poles and at unattached kinetochores. Multiple Mad2 foci are visible in the central region of the monopolar spindle in the AurA mutant cell, consistent with kinetochore localization (bottom). Scale bars = 5 μm. (B) Confocal maximum projection image of DAPI and Mad2 staining in an AurA mutant cell. Note the Mad2 foci in the central region near the ends of chromosomes where kinetochores are located. Scale bar = 5 μm. (C) AurA mutant cells experience mitotic delays and fail to divide. Line graph of time-lapse imaging data depicting times spent in mitosis by AurA+/+; R26CreER/+ (+/+) and AurAf/f; R26CreER/+ (f/f) cells. Asterisks indicate cells that died. Red arrows indicate f/f cells that divided. All other f/f cells died or exited mitosis without dividing, while 35/36 +/+ cells divided (see Table S2 and Fig. S2 in the supplemental material).
To further define the spindle checkpoint arrest in AurA-deficient cells, we used time-lapse microscopy to determine the mitotic transit times of +/+ and f/f cells after OHT treatment. AurA mutant cells spent significantly longer in mitosis than their WT counterparts, consistent with a spindle checkpoint arrest phenotype (Fig. 5C, P < 0.001). While 90% of the WT cells divided and exited mitosis in 1 h or less, all of the AurA mutant cells remained in mitosis for at least 1.5 h, with a median time of approximately 3 h (see Table S2 and Fig. S2 in the supplemental material). Most of the AurA mutant cells (56%) eventually exited mitosis without dividing. However, 11% failed to exit mitosis during the imaging period, remaining in mitosis for at least 20 h. An additional 15% of the AurA mutant cells died during mitosis (Fig. 5C; see Table S2 in the supplemental material). About 18% of the AurA mutant cells divided successfully, although they remained in mitosis for nearly 2 h prior to dividing (Fig. 5C, arrows; see Table S2 in the supplemental material). AurA immunostaining of a parallel culture from the same experiment indicated that approximately 20% of the f/f cells had detectable AurA staining, suggesting that Cre-mediated deletion of both AurA alleles only occurred in 80% of the cells (data not shown). This suggests that the five cells in the f/f culture that divided successfully may represent cells that retain AurA function. Together, these findings suggest that monopolar spindles formed in the absence of AurA trigger the spindle checkpoint, eliciting mitotic arrest and increasing the probability of cell death. The monopolar spindles formed in the absence of AurA are thus phenotypically similar to those caused by monastrol. These data also indicate that AurA is not required for spindle checkpoint activity, in contrast to AurB (10, 25, 32). Together, these MEF studies demonstrate that mAurA plays a conserved and essential role in centrosome separation and bipolar spindle assembly, similar to its orthologs in Drosophila and C. elegans.
AurA is essential for embryonic development.
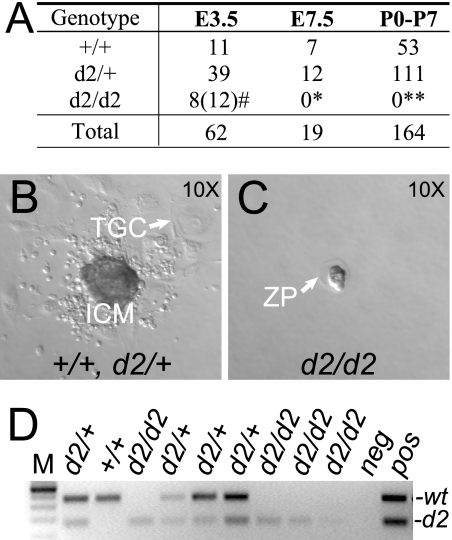
If AurA is critical for transition through mitosis, as indicated by the above MEF studies, AurA-deficient embryos should arrest very early in development. Indeed, AurAd2/d2 animals were not obtained among 164 live-born animals from AurAd2/+ intercrosses (Fig. 6A, postnatal days 0 to 7 [P0-P7]), nor among 19 embryos examined at E7.5 (Fig. 6A, E7.5). To determine if we could recover AurA-deficient blastocysts and to test their developmental capability, we harvested blastocysts at E3.5 and cultured them for 5 to 7 days to generate blastocyst outgrowths. Of 63 blastocysts harvested from AurAd2/+ intercrosses, 49 hatched from the zona pellucida, attached to the culture dish, and produced characteristic blastocyst outgrowths with a cluster of cells derived from the inner cell mass surrounded by trophoblast-derived cells (Fig. 6B). Forty-eight of the 49 hatched embryo outgrowths were successfully genotyped by PCR and shown to be of either the AurAd2/+ or the AurA+/+ genotype. In contrast, 11/63 blastocysts failed to hatch and degenerated within the zona pellucida (Fig. 6C). In addition, two blastocysts that were hatched at harvest failed to produce any outgrowth. A retrospective analysis of the blastocysts that failed to hatch or produce outgrowths showed that most appeared developmentally delayed and/or had debris in the blastocele cavity at harvest, suggesting that defects were already present in blastocyst stage embryos. Genotyping confirmed that 8 of the 13 defective blastocysts were genotyped as AurAd2/d2, 1 was genotyped as AurAd2/+, and genotypes could not be obtained for the other 4. Taken together, these results indicate that AurAd2/d2 embryos are produced at approximately Mendelian ratios but are unable to hatch and grow in vitro, while the majority of AurAd2/+ and AurA+/+ blastocysts show normal in vitro growth. In other experiments, we found additional blastocysts that hatched in utero but showed characteristic cellular defects and a lack of AurA staining by IF, suggesting that they too were AurAd2/d2 embryos (Fig. 7). This indicates that AurA is not required for hatching per se but is essential for growth and survival in vivo, such that AurAd2/d2 embryos die shortly after the blastocyst stage.
FIG. 6.
AurAd2/d2 embryos die at the blastocyst stage. (A) Genotype analysis of progeny from AurAd2/+ intercrosses at E3.5, E7.5, and postnatal stages. E3.5 embryos were cultured to generate blastocyst outgrowths and then genotyped. #, 12 embryos displayed similar in vitro phenotypes, but genotyping was only successful for 8; P > 0.05 by χ2 test for Mendelian segregation (including 12 presumed d2/d2 embryos). *, E7.5, P < 0.05. **, postnatal days 0 to 7 (P0-P7), P < 0.0001. (B) In vitro blastocyst outgrowth characteristic of the AurA+/+ and AurAd2/+ genotypes. ICM, cell clump derived from inner cell mass; TGC, trophoblast-derived giant cell. (C) AurAd2/d2 blastocyst after 7 days in culture. AurA mutant blastocysts remained unhatched within the zona pellucida (ZP). (D) Representative PCR genotyping results for blastocysts after in vitro culture. Deduced genotypes are shown at the top. Positions of WT and d2 products are indicated on the right. M, DNA ladder; neg, reactions run without template DNA; pos, positive control reactions.
FIG. 7.
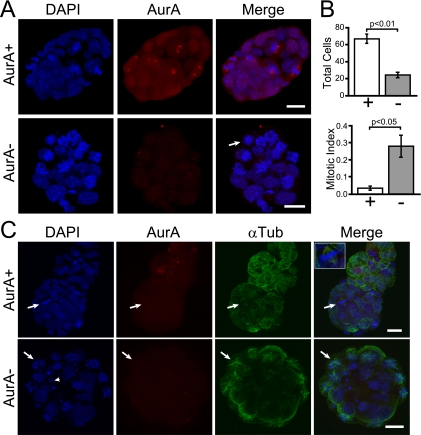
Mitotic arrest and monopolar spindles in AurAd2/d2 blastocysts. (A) Confocal maximum z projection of E3.5 blastocysts after whole-mount IF staining. Top: AurA+/+ or AurAd2/+ embryo with a clear AurA signal at the centrosomes of mitotic cells. DAPI was used to stain DNA. Mitotic cells are identifiable by condensed chromosomes. Bottom: AurAd2/d2 embryo. Note the lack of AurA staining and the presence of multiple mitotic cells with condensed chromatin. The arrow indicates a circular chromosome array indicative of a monopolar spindle. Scale bars = 20 μm. (B) AurAd2/d2 blastocysts have significantly reduced cell numbers (top) and an increased mitotic index (bottom) relative to those of WT littermates. Embryos stained as described for panel A were scored as WT (AurA+/+ or AurAd2/+) or AurAd2/d2 mutants based on the presence or absence of AurA staining. Cells were counted in serial confocal z sections and scored as mitotic or nonmitotic on the basis of chromatin condensation. Histograms show mean values for three embryos per class. Error bars indicate standard errors. P values were derived from two-tailed t tests. (C) Apoptosis and monopolar spindles in AurAd2/d2 embryos. Top: AurA+/+ or AurAd2/+ embryo with normal α-tubulin (αTub) staining. The arrow indicates a metaphase cell with a bipolar spindle. Bottom: AurAd2/d2 embryo. Note the multiple apoptotic bodies (arrowhead) and cells with circular chromosome arrays and astral microtubule patterns indicative of a monopolar spindle phenotype (arrow). Images are confocal maximum z projections of a subset of z slices for each embryo. Scale bars = 20 μm.
AurA mutant embryos display mitotic arrest and monopolar spindles.
The MEF results suggested that the hatching and outgrowth defects of AurAd2/d2 embryos were likely due to defective mitotic entry and/or defects in mitotic progression. However, as the cell culture environment is artificial, it was possible that in vivo phenotypes would be distinct. We reasoned that if the lethality of AurA-deficient embryos is due to defective mitotic entry, embryos should have few mitotic cells, as determined by chromosome condensation. Alternately, if cells enter mitosis but are unable to assemble a bipolar spindle, the embryos should have increased numbers of cells with condensed chromosomes and monopolar spindles. To test these predictions, we performed whole-mount IF staining of E3.5 blastocysts (without culture) from AurAd2/+ intercrosses. For these experiments, AurA IF staining was used to score embryos as either AurA+ (AurA+/+ or AurAd2/+) or AurA− (AurAd2/d2). Eight of the 11 embryos imaged were scored as AurA+ on the basis of the presence of one or more mitotic cells with AurA staining at spindle poles (Fig. 7A, AurA+). In contrast, three embryos lacked AurA staining, indicating that they were AurAd2/d2 (Fig. 7A, AurA−). These embryos displayed multiple cells with condensed chromatin in circular arrays characteristic of monopolar spindles (arrow in Fig. 7A). Cell counts revealed that mutant embryos contained nearly threefold fewer total cells than did AurA+ embryos (Fig. 7B, top; AurA+ mean = 67, AurA− mean = 24.3), while the mitotic index (as determined by chromosome condensation) was eightfold higher in mutants (Fig. 7B, bottom; AurA+ mean = 3.3%, AurA− mean = 28%). Some mutant embryos also had cells with condensed chromatin patterns indicative of apoptosis (Fig. 7C, arrowhead in the AurA− part). To confirm the monopolar spindle phenotype, embryos were stained with antibodies to α-tubulin. While AurA+ embryos had mitotic cells with normal bipolar spindles (Fig. 7C, AurA+), AurA− embryos displayed characteristic astral microtubule arrays emanating from chromosome rosettes, similar to those observed in MEFs (Fig. 7C, AurA−). These results clearly indicate that AurA mutant blastocysts have a monopolar spindle phenotype very similar to that observed in MEFs. The observation of large numbers of cells with condensed chromatin and a monopolar spindle phenotype suggests that the mitotic entry impairment observed in MEFs may not be a significant phenotype in vivo, at least in early embryos. The early death of AurA mutant animals is thus apparently due to an inability to complete mitosis during the critical wave of early cell divisions at the blastocyst stage.
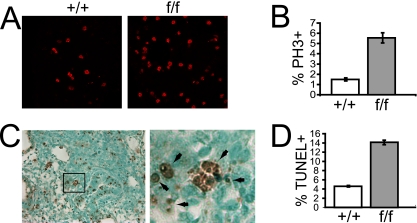
To determine if AurA is also essential and plays similar roles in later-stage embryos, we intercrossed AurAf/+; R26CreER/Rep mice and injected pregnant dams with tamoxifen at day 10.5 of pregnancy. Embryos were harvested 2 days later and analyzed histologically for PH3 and apoptosis (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling [TUNEL]) in the lung and the thymus. AurA deletion in AurAf/f; R26CreER embryos caused an approximately fourfold increase in the percentage of PH3+ cells in both the thymus and the lung compared to that in AurA+/+; R26CreER embryos, indicating that AurA deletion causes a similar mitotic arrest phenotype in later-stage embryos, as observed in blastocysts and MEFs (Fig. 8A to C; data not shown). The majority of the PH3+ cells in mutant embryos displayed condensed chromosomes and prometaphase morphology, supporting the conclusion that AurA is not essential for mitotic entry in vivo (Fig. 8B). AurA mutant embryos also showed a threefold increase in the percentage of apoptotic cells in the thymus and the lung, indicating that AurA deficiency induces apoptosis (Fig. 8D). Together, these data indicate that AurA deficiency causes mitotic arrest and apoptosis in proliferating cells in vivo.
FIG. 8.
AurA deletion in mid-gestation embryos causes an increased mitotic index and apoptosis. (A). Representative PH3 staining in lung sections from E14.5 embryos after tamoxifen treatment at E12.5. (B). Quantitation of PH3+ cells in lung tissue from E14.5 embryos after tamoxifen treatment at E12.5. (C). Left: representative TUNEL of a lung section from an E14.6 AurAf/f; R26CreER/Rep embryo after tamoxifen treatment at E12.5. Right: magnified image from the box in the left part of the panel showing examples of TUNEL+ cells (brown). (D). Quantitation of TUNEL+ cells in lung tissue from an E14.5 embryo after tamoxifen treatment at E12.5.
DISCUSSION
Here we describe the generation and characterization of a conditional null mutation in the mouse AurA gene. We show that deletion of AurA exon 2 produces a null allele suitable for definitively determining the roles of AurA in vivo and in primary cells ex vivo.
Roles of AurA in mitosis.
Mutation or depletion of AurA homologs in Drosophila, C. elegans, and Xenopus cultured cells causes accumulation of mitotic cells with monopolar spindle phenotypes, suggesting a crucial role for AurA in centrosome separation (21, 24, 52). More recent studies have also implicated AurA in timely mitotic entry in both the C. elegans and Xenopus systems (23, 38, 48, 55, 57). However, the role of mammalian AurA has previously been unclear because of conflicting results of cell culture studies. RNAi studies with HeLa cells suggested that AurA is required for mitotic entry via recruitment and activation of cdk1-cyclin B at centrosomes (27, 42). AurA was also reported to contribute indirectly to cdk1-cyclin B activation and mitotic entry via activation of the cdc25 phosphatase (13) and Polo-like kinase 1 (57). However, other studies with HeLa or other transformed cell types reported that AurA-depleted or -inhibited cells accumulated in mitosis with monopolar spindle phenotypes (12, 20, 67) and in some cases were able to complete mitosis despite a high percentage of abnormal spindles and chromosome segregation abnormalities (28). These discrepancies could be due to differences in the reagents, cell types, and conditions employed, as well as distinct levels of AurA inhibition inherent in RNAi and inhibitor studies. Our genetic model allowed us to definitively address this issue by inducing an AurA null mutation in genetically defined primary MEFs. By inducing the mutation in quiescent cells and examining phenotypes in a subsequent mitosis, we were able to ensure that most cells entering mitosis lack detectable AurA protein. We found that AurA-deficient primary MEFs experience delayed mitotic entry, as evidenced by time-lapse imaging. This delay is associated with increased numbers of early prophase cells, supporting an important role for AurA in committing cells to mitosis. However, AurA is not essential for mitotic entry, as many cells clearly enter prometaphase. On the other hand, AurA is essential for bipolar spindle formation. AurA-deficient cells that entered prometaphase displayed a monopolar spindle phenotype and were arrested by the spindle checkpoint. AurA-deficient cells were not observed in metaphase, anaphase, or telophase, and live-cell imaging indicated that most mutant cells eventually exit mitosis without cell division. Intriguingly, a subset of AurA-deficient cells remained arrested in mitosis for extended periods of time. Vertebrate cells treated for extended periods with a mitotic inhibitor such as nocodazole, taxol, or monastrol normally exit mitosis after several hours because of cyclin B degradation (6). The reason for the extended mitotic arrest of AurA-deficient cells is unclear. A subset of AurA-deficient cells also died during mitosis, suggesting that the mitotic arrest experienced by these cells sometimes triggers cell death stimuli. We also observed increased apoptosis in AurA mutant embryos, indicating that this phenotype is not unique to cultured cells. Together, our results establish a conserved role for mammalian AurA in bipolar spindle formation and confirm that AurA also plays a role in mitotic entry in vitro.
AurA is essential for development.
We also found that AurA is required for mouse embryonic development. AurA-null embryos survive to the blastocyst stage but are not recovered at E7.5 or later, indicating that AurA deficiency causes lethality at some time between E3.5 and E7.5. AurA-null blastocysts harvested at E3.5 fail to hatch and grow in vitro and display multiple cells arrested in mitosis with monopolar spindles. Similar results were very recently reported by others using an independently generated mouse AurA mutant (54). The observation of mitotic arrest and monopolar spindle formation suggests that mitotic entry defects are not the predominant phenotype of AurA deficiency in vivo, although we cannot rule out a minor role. AurA is present in mouse oocytes and early embryos, where it localizes at spindle poles (69). The survival of AurA-deficient cells to the blastocyst stage suggests that maternal AurA mRNA and/or protein stores are sufficient to sustain AurA levels through the several cell divisions of early embryogenesis. However, AurA becomes limiting at the blastocyst stage, leading to defects in mutant embryos.
AurA mutation causes mitotic arrest and apoptosis in mid-gestation.
AurA deletion in mid-gestation embryos caused a marked increase in mitotic (PH3+) cells in two of the tissue types examined (lung and thymus). As was observed in blastocysts, the bulk of PH3+ cells in mutant embryos displayed condensed chromatin patterns, indicating that the cells had entered mitosis. Together with the blastocyst data, this observation establishes that AurA is not essential for mitotic entry in vivo. However, we cannot rule out the presence of a mitotic delay, as was observed in MEFs.
The increased mitotic index in AurA mutant mid-gestation embryos was accompanied by a parallel increase in apoptotic cells. Apoptotic chromatin patterns were also observed in AurA-deficient blastocysts, indicating that this phenotype is not restricted to later gestation embryos. As discussed above, a subset of AurA-deficient MEFs died during mitosis. An important area for future work will be the determination of the signaling pathways that govern apoptosis in response to AurA mutation.
AurA as a therapeutic target in cancer.
The important roles of Aurora kinases in mitotic regulation, as well as the finding that AurA is overexpressed in many human cancers, have raised considerable interest in AurA kinase inhibitors as potential cancer therapeutics. Indeed, Aurora kinase inhibitors are currently in clinical trials (reviewed in references 9 and 34). Several currently described Aurora inhibitors display activity against both AurA and AurB in vitro (20, 67), while compounds with reported selectivity for AurA or AurB inhibition are beginning to emerge (20, 28, 68). The desirability of targeting AurA, AurB, or both is a matter of debate (9, 65, 67). Nevertheless, our results clarify the issue by clearly defining the AurA mutant phenotype. Thus, we predict that potent AurA-specific inhibitors should cause monopolar spindles, leading to mitotic arrest in cells with a competent spindle checkpoint. In contrast, inhibition of AurB alone and dual inhibition of AurA and AurB have been reported to produce similar phenotypes, including rapid mitotic exit without cell division because of loss of the spindle checkpoint and cytokinesis impairment (20, 67). Activation of the spindle checkpoint in specific AurA-inhibited cells is thus a significant functional difference that may contribute to distinct therapeutic outcomes. The long-term consequences of spindle checkpoint arrest are not fully defined but may include apoptosis, mitotic catastrophe, mitotic exit, and subsequent G1 arrest or mitotic exit followed by endoreduplication (reviewed in reference 51). Our results suggest that at least a subset of cells treated with AurA inhibitors should undergo apoptosis, although it is possible that tumor cells may respond differently from normal primary cells. Our genetic system will allow future tests of these hypotheses in vivo and in vitro.
The challenge of obtaining AurA-specific inhibitors raises the question of whether similar therapeutic effects could be achieved by targeting other molecules. Our results suggest that drugs such as monastrol that cause monopolar spindles by inhibiting the mitotic kinesin Eg5 may be good alternatives to AurA inhibitors. Several such drugs are under development (4). While these compounds may not show phenotypes identical to that produced by AurA inhibition (because of the fact that AurA has substrates involved in multiple aspects of mitosis and also regulates processes outside of mitosis [49]), they may cause a phenotypically similar arrest state with similar biological outcomes. In fact, Eg5 is a substrate of AurA in Xenopus, suggesting that Eg5 activation could be downstream of AurA. The future identification of the AurA targets mediating the monopolar spindle phenotype will also be helpful in exploring other ways to target this pathway for therapeutic benefit.
In summary, these studies provide clear evidence that mammalian AurA is essential for viability because of its key role in regulating the transition through M phase, specifically in the establishment of bipolar spindle assembly. The conditional genetic system established in this report, with paired primary cell and in vivo studies, will be instrumental in deciphering the mechanisms and pathways involved. Furthermore, the role(s) of AurA in cancer and other diseases associated with aneuploidy can now be fully explored.
Supplementary Material
Acknowledgments
We thank members of the Van Dyke, Magnuson, and Salmon laboratories for helpful discussions and technical assistance. We acknowledge the use of Michael Hooker Microscopy Core Facility equipment and resources funded by an anonymous private donor and Sundeep Kalantry, Michael Chua, and Wendy Salmon for expert assistance with confocal microscopy. We thank Allan Balmain for critical reading and comments on the manuscript.
D.O.C. was a fellow of the Leukemia & Lymphoma Foundation of America (grant 5408-02). This study was supported by NCI grant 2-R01-CA065773-11A1 to T.V.D., NIH grant GM24364 to E.D.S., and an NICHD grant to T.M.
Footnotes
Published ahead of print on 15 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andrews, P. D., E. Knatko, W. J. Moore, and J. R. Swedlow. 2003. Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 15672-683. [DOI] [PubMed] [Google Scholar]
- 2.Badea, T. C., Y. Wang, and J. Nathans. 2003. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 232314-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berdnik, D., and J. A. Knoblich. 2002. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 12640-647. [DOI] [PubMed] [Google Scholar]
- 4.Bergnes, G., K. Brejc, and L. Belmont. 2005. Mitotic kinesins: prospects for antimitotic drug discovery. Curr. Top. Med. Chem. 5127-145. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, J. R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, C. S. Chan, M. Novotny, D. J. Slamon, and G. D. Plowman. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 173052-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brito, D. A., and C. L. Rieder. 2006. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol. 161194-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. R., K. K. Koretke, M. L. Birkeland, P. Sanseau, and D. R. Patrick. 2004. Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs. BMC. Evol. Biol. 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmena, M., and W. C. Earnshaw. 2003. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4842-854. [DOI] [PubMed] [Google Scholar]
- 9.Carvajal, R. D., A. Tse, and G. K. Schwartz. 2006. Aurora kinases: new targets for cancer therapy. Clin. Cancer Res. 126869-6875. [DOI] [PubMed] [Google Scholar]
- 10.Cimini, D., X. Wan, C. B. Hirel, and E. D. Salmon. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 161711-1718. [DOI] [PubMed] [Google Scholar]
- 11.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, J., and G. J. Hannon. 2004. Suppression of p160ROCK bypasses cell cycle arrest after Aurora-A/STK15 depletion. Proc. Natl. Acad. Sci. USA 1018975-8980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutertre, S., M. Cazales, M. Quaranta, C. Froment, V. Trabut, C. Dozier, G. Mirey, J. P. Bouche, N. Theis-Febvre, E. Schmitt, B. Monsarrat, C. Prigent, and B. Ducommun. 2004. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell Sci. 1172523-2531. [DOI] [PubMed] [Google Scholar]
- 14.Ewart-Toland, A., P. Briassouli, J. P. de Koning, J. H. Mao, J. Yuan, F. Chan, L. MacCarthy-Morrogh, B. A. Ponder, H. Nagase, J. Burn, S. Ball, M. Almeida, S. Linardopoulos, and A. Balmain. 2003. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat. Genet. 34403-412. [DOI] [PubMed] [Google Scholar]
- 15.Fukuda, T., Y. Mishina, M. P. Walker, and R. P. DiAugustine. 2005. Conditional transgenic system for mouse aurora a kinase: degradation by the ubiquitin proteasome pathway controls the level of the transgenic protein. Mol. Cell. Biol. 255270-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuta, T., D. L. Baillie, and J. M. Schumacher. 2002. Caenorhabditis elegans Aurora A kinase AIR-1 is required for postembryonic cell divisions and germline development. Genesis 34244-250. [DOI] [PubMed] [Google Scholar]
- 17.Giet, R., D. McLean, S. Descamps, M. J. Lee, J. W. Raff, C. Prigent, and D. M. Glover. 2002. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156437-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giet, R., and C. Prigent. 2000. The Xenopus laevis aurora/Ip11p-related kinase pEg2 participates in the stability of the bipolar mitotic spindle. Exp. Cell Res. 258145-151. [DOI] [PubMed] [Google Scholar]
- 19.Giet, R., R. Uzbekov, F. Cubizolles, G. K. Le, and C. Prigent. 1999. The Xenopus laevis Aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 27415005-15013. [DOI] [PubMed] [Google Scholar]
- 20.Girdler, F., K. E. Gascoigne, P. A. Eyers, S. Hartmuth, C. Crafter, K. M. Foote, N. J. Keen, and S. S. Taylor. 2006. Validating Aurora B as an anti-cancer drug target. J. Cell Sci. 1193664-3675. [DOI] [PubMed] [Google Scholar]
- 21.Glover, D. M., M. H. Leibowitz, D. A. McLean, and H. Parry. 1995. Mutations in Aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 8195-105. [DOI] [PubMed] [Google Scholar]
- 22.Gopalan, G., C. S. Chan, and P. J. Donovan. 1997. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J. Cell Biol. 138643-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hachet, V., C. Canard, and P. Gonczy. 2007. Centrosomes promote timely mitotic entry in C. elegans embryos. Dev. Cell 12531-541. [DOI] [PubMed] [Google Scholar]
- 24.Hannak, E., M. Kirkham, A. A. Hyman, and K. Oegema. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 1551109-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauf, S., R. W. Cole, S. LaTerra, C. Zimmer, G. Schnapp, R. Walter, A. Heckel, J. van Meel, C. L. Rieder, and J. M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106348-360. [DOI] [PubMed] [Google Scholar]
- 27.Hirota, T., N. Kunitoku, T. Sasayama, T. Marumoto, D. Zhang, M. Nitta, K. Hatakeyama, and H. Saya. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114585-598. [DOI] [PubMed] [Google Scholar]
- 28.Hoar, K., A. Chakravarty, C. Rabino, D. Wysong, D. Bowman, N. Roy, and J. A. Ecsedy. 2007. MLN8054, a small-molecule inhibitor of Aurora A, causes spindle pole and chromosome congression defects leading to aneuploidy. Mol. Cell. Biol. 274513-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda, K., H. Mihara, Y. Kato, A. Yamaguchi, H. Tanaka, H. Yasuda, K. Furukawa, and T. Urano. 2000. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene 192812-2819. [DOI] [PubMed] [Google Scholar]
- 30.Jackson, A. L., and P. S. Linsley. 2004. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 20521-524. [DOI] [PubMed] [Google Scholar]
- 31.Juan, G., F. Traganos, W. M. James, J. M. Ray, M. Roberge, D. M. Sauve, H. Anderson, and Z. Darzynkiewicz. 1998. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry 3271-77. [DOI] [PubMed] [Google Scholar]
- 32.Kallio, M. J., M. L. McCleland, P. T. Stukenberg, and G. J. Gorbsky. 2002. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12900-905. [DOI] [PubMed] [Google Scholar]
- 33.Kapoor, T. M., T. U. Mayer, M. L. Coughlin, and T. J. Mitchison. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keen, N., and S. Taylor. 2004. Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer 4927-936. [DOI] [PubMed] [Google Scholar]
- 35.Kulkarni, M. M., M. Booker, S. J. Silver, A. Friedman, P. Hong, N. Perrimon, and B. Mathey-Prevot. 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods 3833-838. [DOI] [PubMed] [Google Scholar]
- 36.Kunitoku, N., T. Sasayama, T. Marumoto, D. Zhang, S. Honda, O. Kobayashi, K. Hatakeyama, Y. Ushio, H. Saya, and T. Hirota. 2003. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell 5853-864. [DOI] [PubMed] [Google Scholar]
- 37.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 38.Liu, Q., and J. V. Ruderman. 2006. Aurora A, mitotic entry, and spindle bipolarity. Proc. Natl. Acad. Sci. USA 1035811-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loonstra, A., M. Vooijs, H. B. Beverloo, B. A. Allak, E. Van Drunen, R. Kanaar, A. Berns, and J. Jonkers. 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA 989209-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma, Y., A. Creanga, L. Lum, and P. A. Beachy. 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443359-363. [DOI] [PubMed] [Google Scholar]
- 41.Mao, J. H., D. Wu, J. Perez-Losada, T. Jiang, Q. Li, R. M. Neve, J. W. Gray, W. W. Cai, and A. Balmain. 2007. Crosstalk between Aurora-A and p53: frequent deletion or downregulation of Aurora-A in tumors from p53 null mice. Cancer Cell 11161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marumoto, T., T. Hirota, T. Morisaki, N. Kunitoku, D. Zhang, Y. Ichikawa, T. Sasayama, S. Kuninaka, T. Mimori, N. Tamaki, M. Kimura, Y. Okano, and H. Saya. 2002. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells 71173-1182. [DOI] [PubMed] [Google Scholar]
- 43.Marumoto, T., D. Zhang, and H. Saya. 2005. Aurora-A—a guardian of poles. Nat. Rev. Cancer 542-50. [DOI] [PubMed] [Google Scholar]
- 44.May, K. M., and K. G. Hardwick. 2006. The spindle checkpoint. J. Cell Sci. 1194139-4142. [DOI] [PubMed] [Google Scholar]
- 45.Meraldi, P., R. Honda, and E. A. Nigg. 2004. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr. Opin. Genet. Dev. 1429-36. [DOI] [PubMed] [Google Scholar]
- 46.Nigg, E. A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 221-32. [DOI] [PubMed] [Google Scholar]
- 47.Pines, J., and C. L. Rieder. 2001. Re-staging mitosis: a contemporary view of mitotic progression. Nat. Cell Biol. 3E3-E6. [DOI] [PubMed] [Google Scholar]
- 48.Portier, N., A. Audhya, P. S. Maddox, R. A. Green, A. Dammermann, A. Desai, and K. Oegema. 2007. A microtubule-independent role for centrosomes and Aurora A in nuclear envelope breakdown. Dev. Cell 12515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pugacheva, E. N., S. A. Jablonski, T. R. Hartman, E. P. Henske, and E. A. Golemis. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 1291351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajagopalan, H., and C. Lengauer. 2004. Aneuploidy and cancer. Nature 432338-341. [DOI] [PubMed] [Google Scholar]
- 51.Rieder, C. L., and H. Maiato. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7637-651. [DOI] [PubMed] [Google Scholar]
- 52.Roghi, C., R. Giet, R. Uzbekov, N. Morin, I. Chartrain, R. Le Guellec, A. Couturier, M. Doree, M. Philippe, and C. Prigent. 1998. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell Sci. 111(Pt. 5)557-572. [DOI] [PubMed] [Google Scholar]
- 53.Sasai, K., H. Katayama, D. L. Stenoien, S. Fujii, R. Honda, M. Kimura, Y. Okano, M. Tatsuka, F. Suzuki, E. A. Nigg, W. C. Earnshaw, W. R. Brinkley, and S. Sen. 2004. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil. Cytoskel. 59249-263. [DOI] [PubMed] [Google Scholar]
- 54.Sasai, K., J. M. Parant, M. E. Brandt, J. Carter, H. P. Adams, S. A. Stass, A. M. Killary, H. Katayama, and S. Sen. 2008. Targeted disruption of Aurora A causes abnormal mitotic spindle assembly, chromosome misalignment and embryonic lethality. Oncogene 274122-4127. [DOI] [PubMed] [Google Scholar]
- 55.Satinover, D. L., D. L. Brautigan, and P. T. Stukenberg. 2006. Aurora-A kinase and inhibitor-2 regulate the cyclin threshold for mitotic entry in Xenopus early embryonic cell cycles. Cell Cycle 52268-2274. [DOI] [PubMed] [Google Scholar]
- 56.Schumacher, J. M., N. Ashcroft, P. J. Donovan, and A. Golden. 1998. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development 1254391-4402. [DOI] [PubMed] [Google Scholar]
- 57.Seki, A., J. A. Coppinger, C. Y. Jang, J. R. Yates, and G. Fang. 2008. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science 3201655-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sen, S., H. Zhou, and R. A. White. 1997. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene 142195-2200. [DOI] [PubMed] [Google Scholar]
- 59.Skarnes, W. C. 2000. Gene trapping methods for the identification and functional analysis of cell surface proteins in mice. Methods Enzymol. 328592-615. [DOI] [PubMed] [Google Scholar]
- 60.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 2170-71. [DOI] [PubMed] [Google Scholar]
- 61.Su, H., A. A. Mills, X. Wang, and A. Bradley. 2002. A targeted X-linked CMV-Cre line. Genesis 32187-188. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka, M., A. Ueda, H. Kanamori, H. Ideguchi, J. Yang, S. Kitajima, and Y. Ishigatsubo. 2002. Cell-cycle-dependent regulation of human aurora A transcription is mediated by periodic repression of E4TF1. J. Biol. Chem. 27710719-10726. [DOI] [PubMed] [Google Scholar]
- 63.Truett, G. E., P. Heeger, R. L. Mynatt, A. A. Truett, J. A. Walker, and M. L. Warman. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and Tris (HotSHOT). BioTechniques 2952-54. [DOI] [PubMed] [Google Scholar]
- 64.Walter, A. O., W. Seghezzi, W. Korver, J. Sheung, and E. Lees. 2000. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 194906-4916. [DOI] [PubMed] [Google Scholar]
- 65.Warner, S. L., R. M. Munoz, P. Stafford, E. Koller, L. H. Hurley, D. D. Von Hoff, and H. Han. 2006. Comparing Aurora A and Aurora B as molecular targets for growth inhibition of pancreatic cancer cells. Mol. Cancer Ther. 52450-2458. [DOI] [PubMed] [Google Scholar]
- 66.Waters, J. C., R. H. Chen, A. W. Murray, and E. D. Salmon. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1411181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, H., T. Burke, J. Dempsey, B. Diaz, E. Collins, J. Toth, R. Beckmann, and X. Ye. 2005. Mitotic requirement for Aurora A kinase is bypassed in the absence of Aurora B kinase. FEBS Lett. 5793385-3391. [DOI] [PubMed] [Google Scholar]
- 68.Yang, J., T. Ikezoe, C. Nishioka, T. Tasaka, A. Taniguchi, Y. Kuwayama, N. Komatsu, K. Bandobashi, K. Togitani, H. P. Koeffler, H. Taguchi, and A. Yokoyama. 2007. AZD1152, a novel and selective Aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood 1102034-2040. [DOI] [PubMed] [Google Scholar]
- 69.Yao, L. J., Z. S. Zhong, L. S. Zhang, D. Y. Chen, H. Schatten, and Q. Y. Sun. 2004. Aurora-A is a critical regulator of microtubule assembly and nuclear activity in mouse oocytes, fertilized eggs, and early embryos. Biol. Reprod. 701392-1399. [DOI] [PubMed] [Google Scholar]
- 70.Zhou, H., J. Kuang, L. Zhong, W. L. Kuo, J. W. Gray, A. Sahin, B. R. Brinkley, and S. Sen. 1998. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20189-193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.